Permutation Results on CD8+ T Cell CROP-seq Data
– In-house scripts
Yifan Zhou (zhouyf@uchicago.edu)
2022-09-19
We separate the cells into 2 groups:
Stimulated: 14278 cells;
Unstimulated stage: 10677 cells.
We modified GSFA so that the associations (\(\beta\)) between factors and perturbations are estimated for stimulated and unstimulated cells separately.
Here, we summarize the permutation results for GSFA, scMAGeCK, DESeq2, and MAST.
1 GSFA Permutation
Within both the stimulated and unstimulated cells, cell labels in the expression data were permuted randomly so that they are no longer correlated with the knock-down conditions. Then GSFA was performed still using all conditions as guides. Factor-guide association as well as the LFSR of each gene were evaluated as usual.
In total, 10 random permutation rounds like this were conducted.
1.1 Factor ~ KO Association
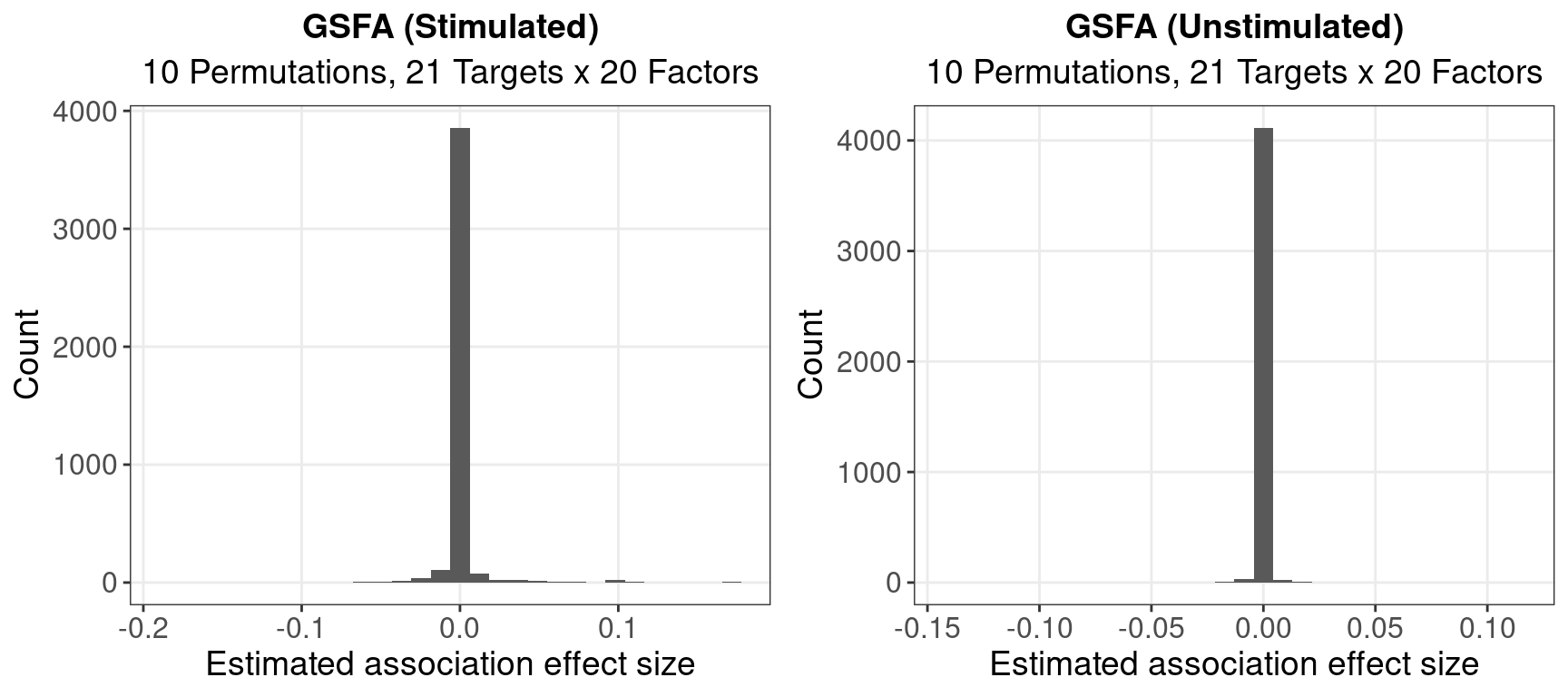
1.1.1 Posterior Mean of Beta
Unstimulated cells, # of |beta|’s > 0.05: 8
Stimulated cells, # of |beta|’s > 0.05: 60
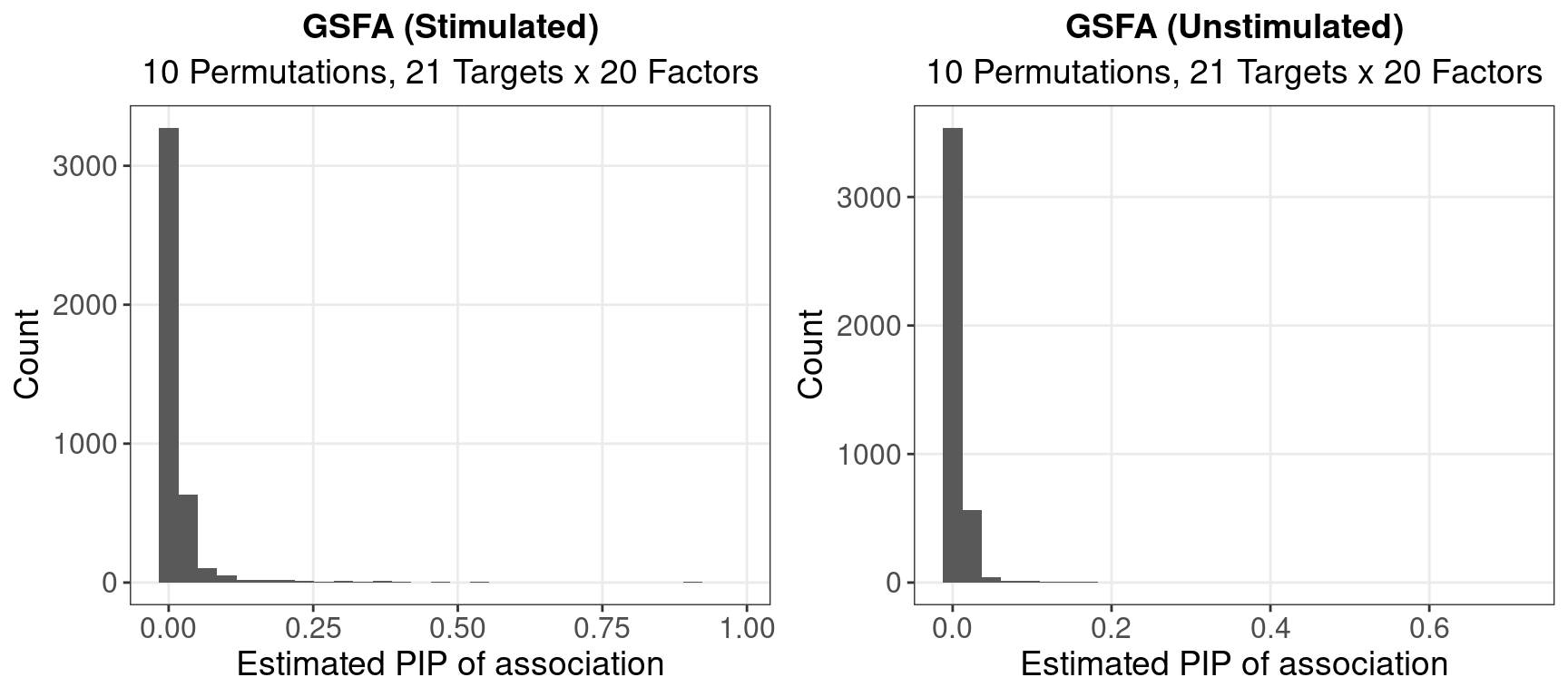
1.1.2 Beta PIP
Unstimulated cells, # of PIPs > 0.8: 0
Stimulated cells, # of PIPs > 0.8: 8
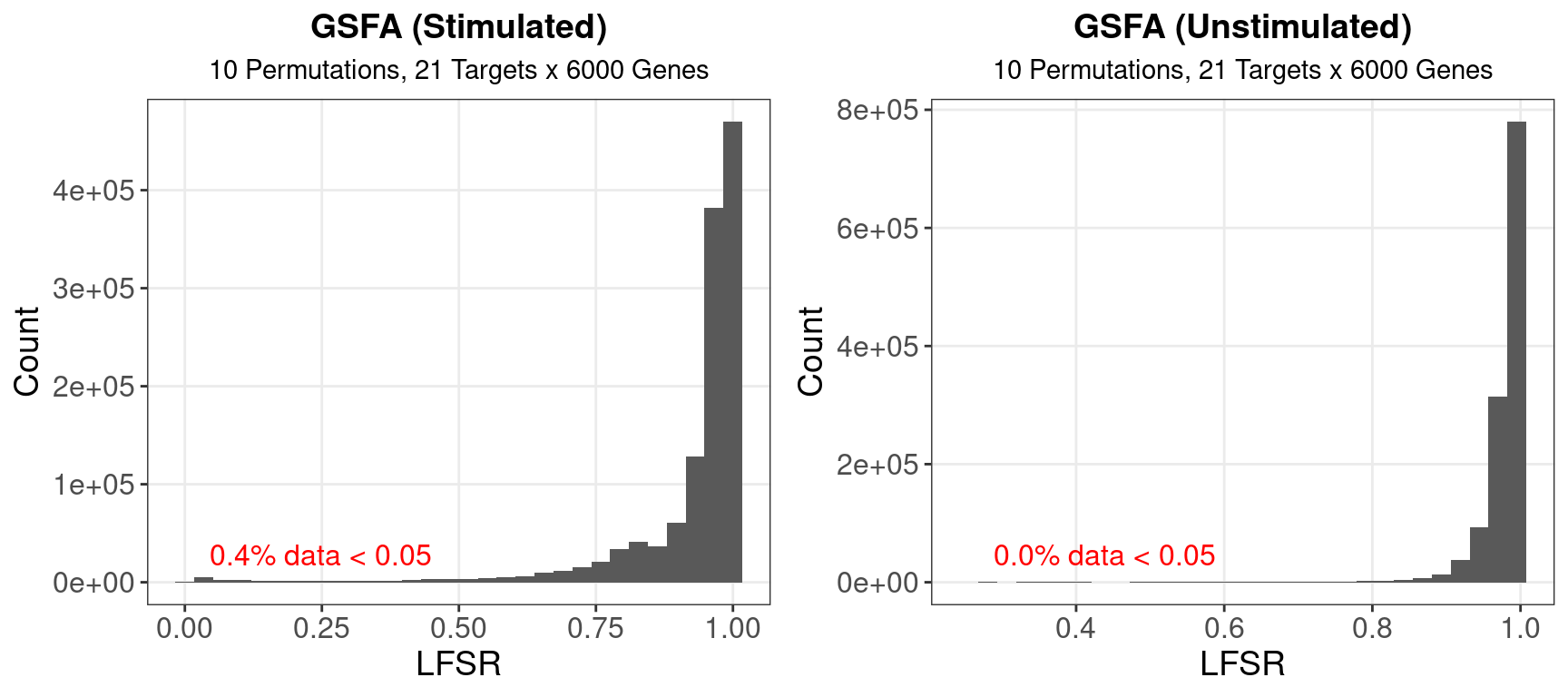
1.2 LFSR
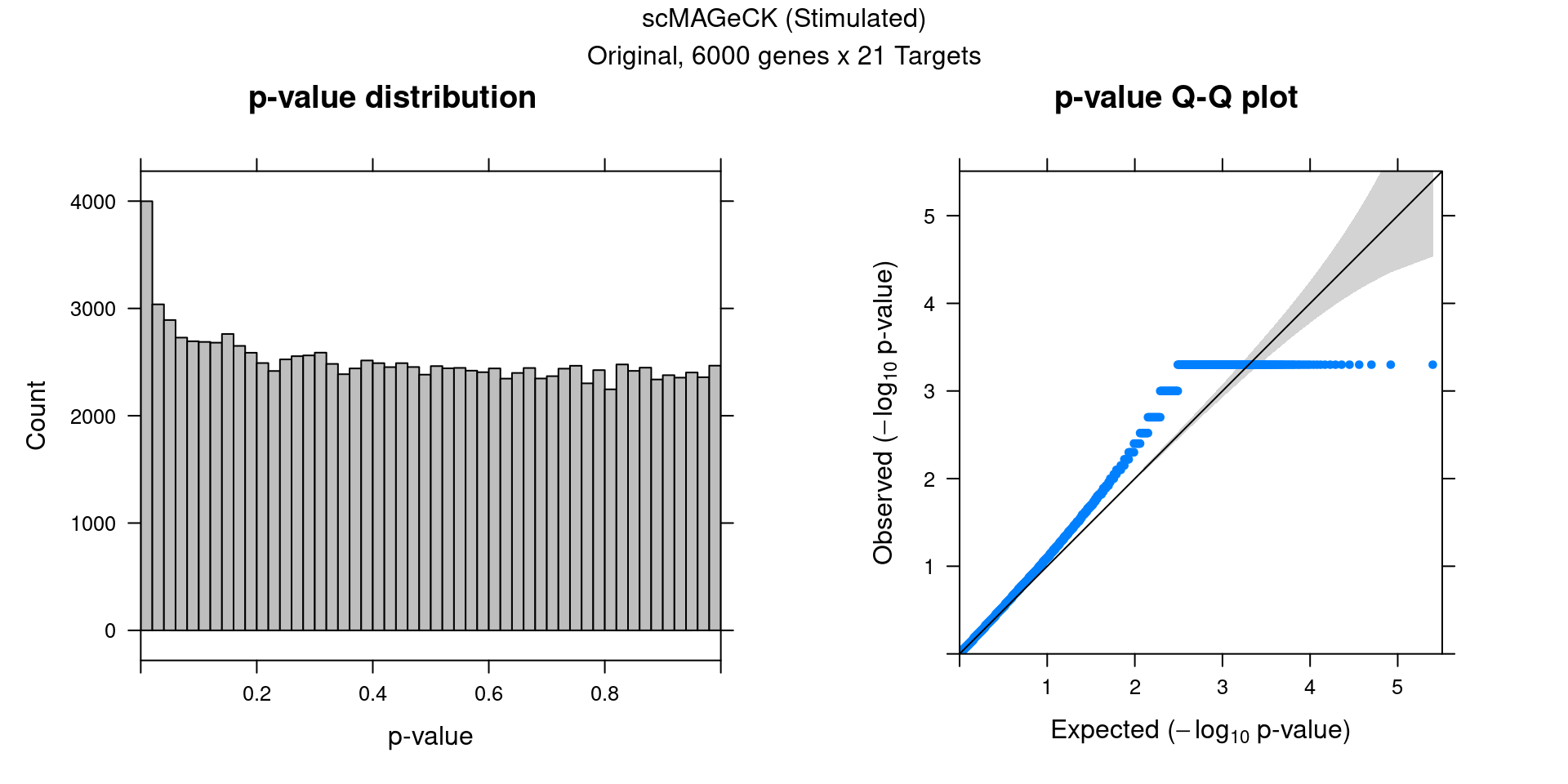
2 scMAGeCK Permutation (Stimulated)
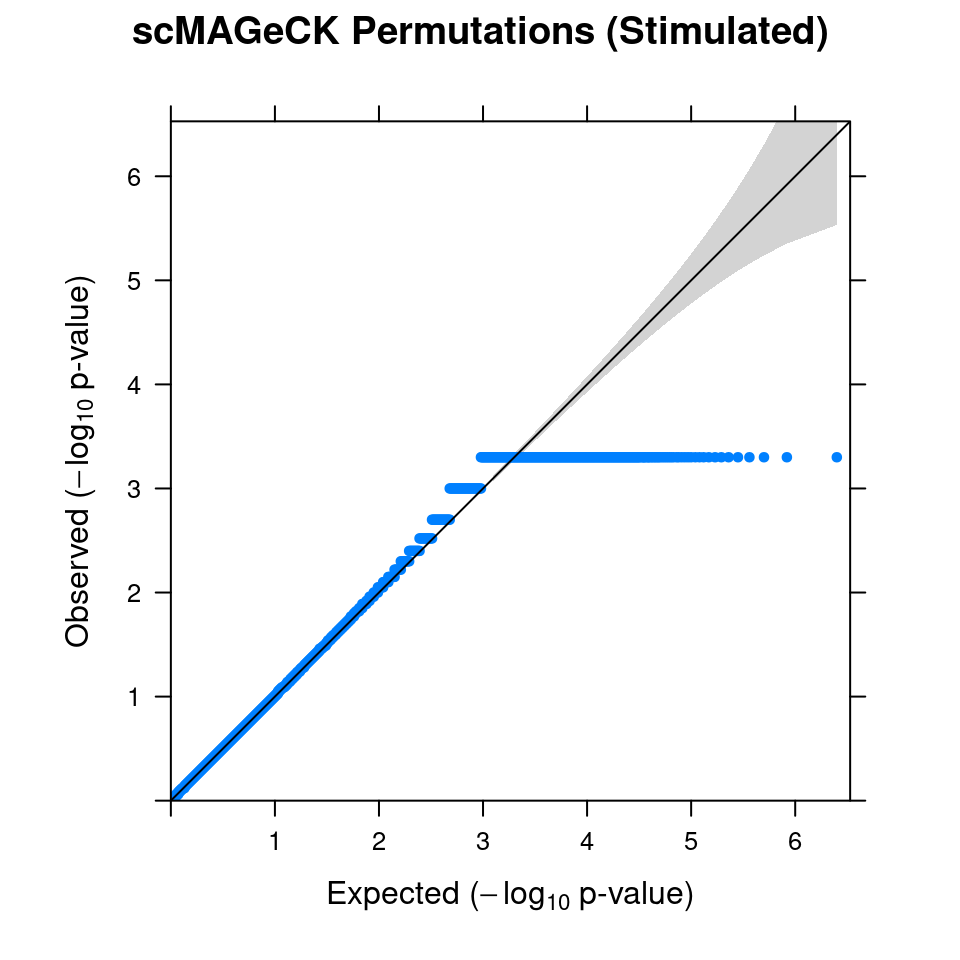
Within stimulated cells, cell labels in the expression data were permuted randomly so that they are no longer correlated with the knock-down conditions. Then scMAGeCK-LR test was performed for all guides at once.
In total, 10 random permutation rounds like this were conducted.
The outputs are empirical p values, some of them equal to 0 exactly, and had to be replaced with 0.0005 for QQ-plot.
2.1 Combined from 10 permutations
2.2 Original scMAGeCK result
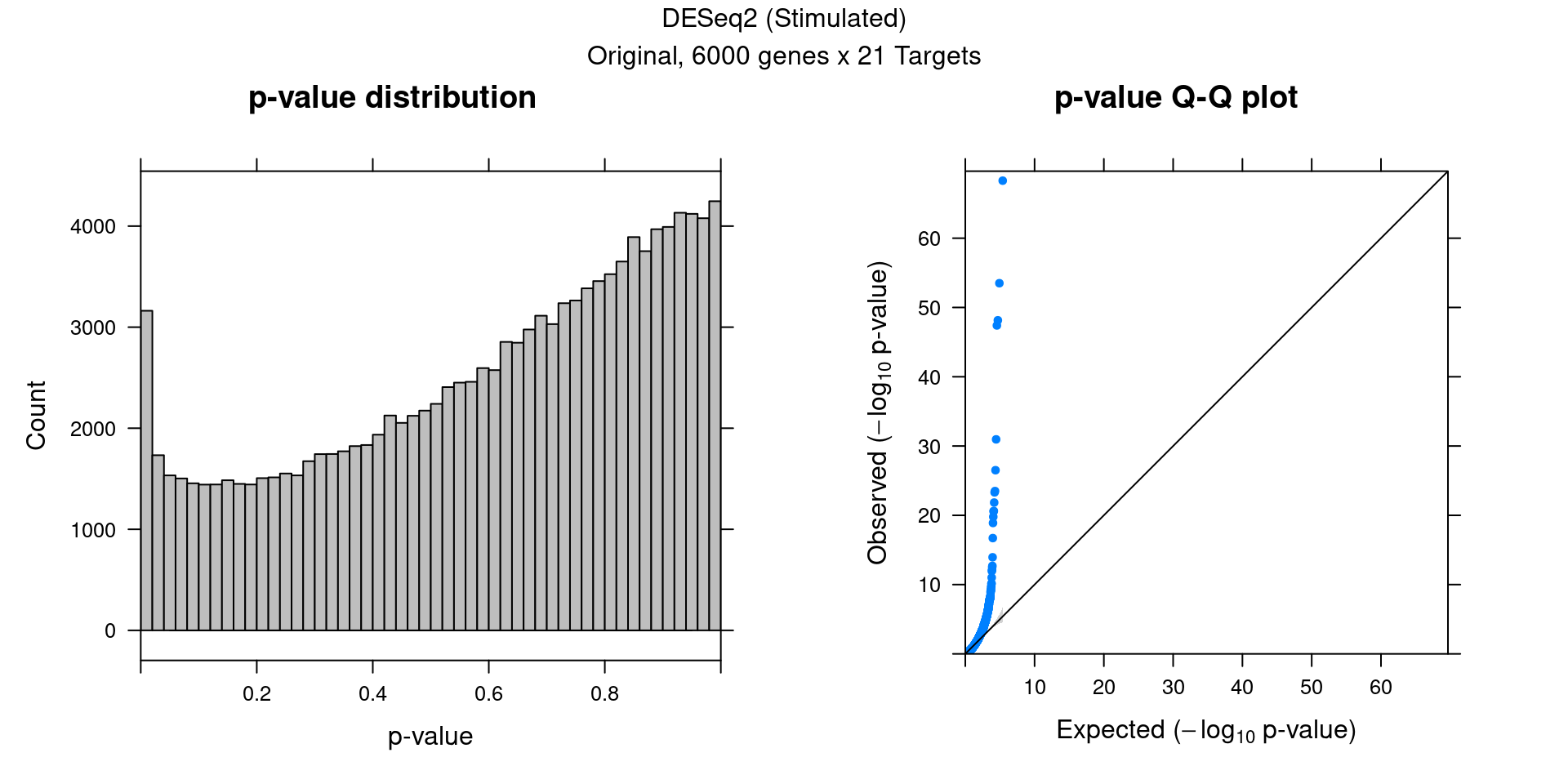
3 DESeq2 Permutation (Stimulated)
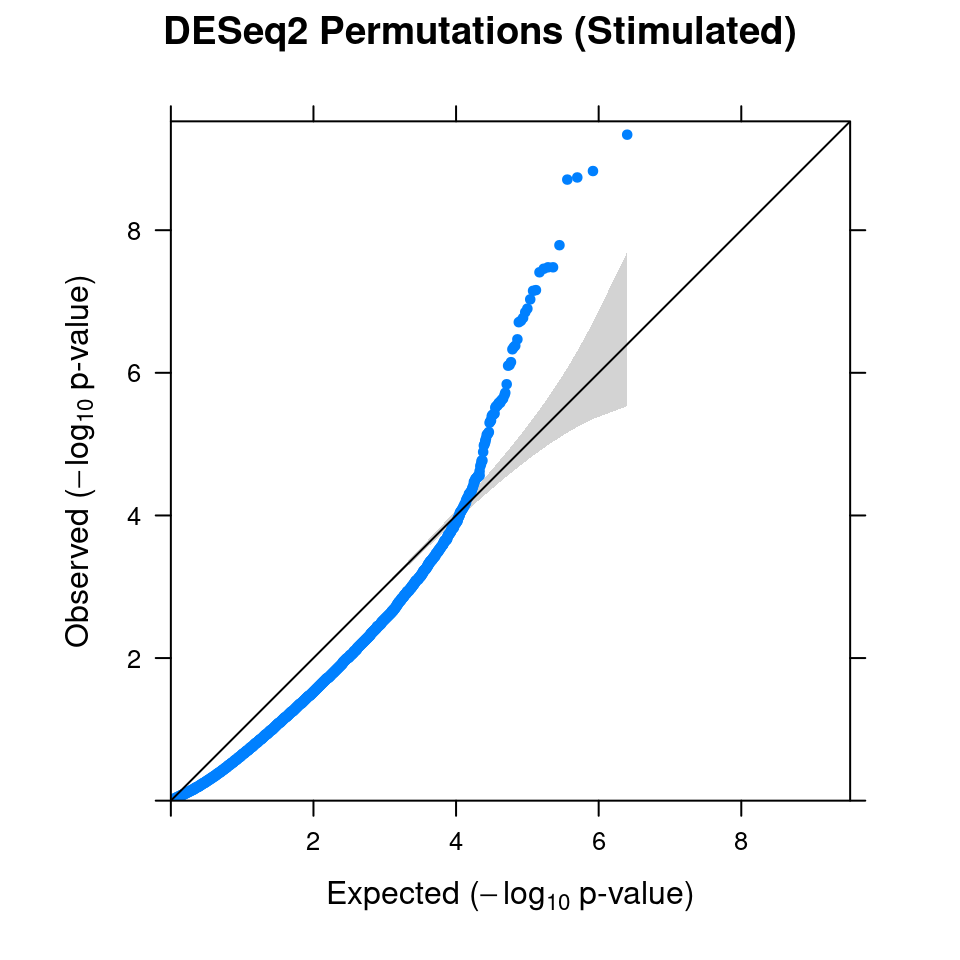
Within stimulated cells, cell labels in the expression data were permuted randomly so that they are no longer correlated with the knock-down conditions. Then DESeq2 test was performed under each guide.
In total, 10 random permutation rounds like this were conducted.
3.1 Combined from 10 permutations
3.2 Original DESeq2 DGE result
4 MAST Permutation (Stimulated)
Within stimulated cells, cell labels in the expression data were permuted randomly so that they are no longer correlated with the knock-down conditions. Then MAST likelihood ratio test was performed under each guide.
In total, 10 random permutation rounds like this were conducted.
4.1 Combined from 10 permutations
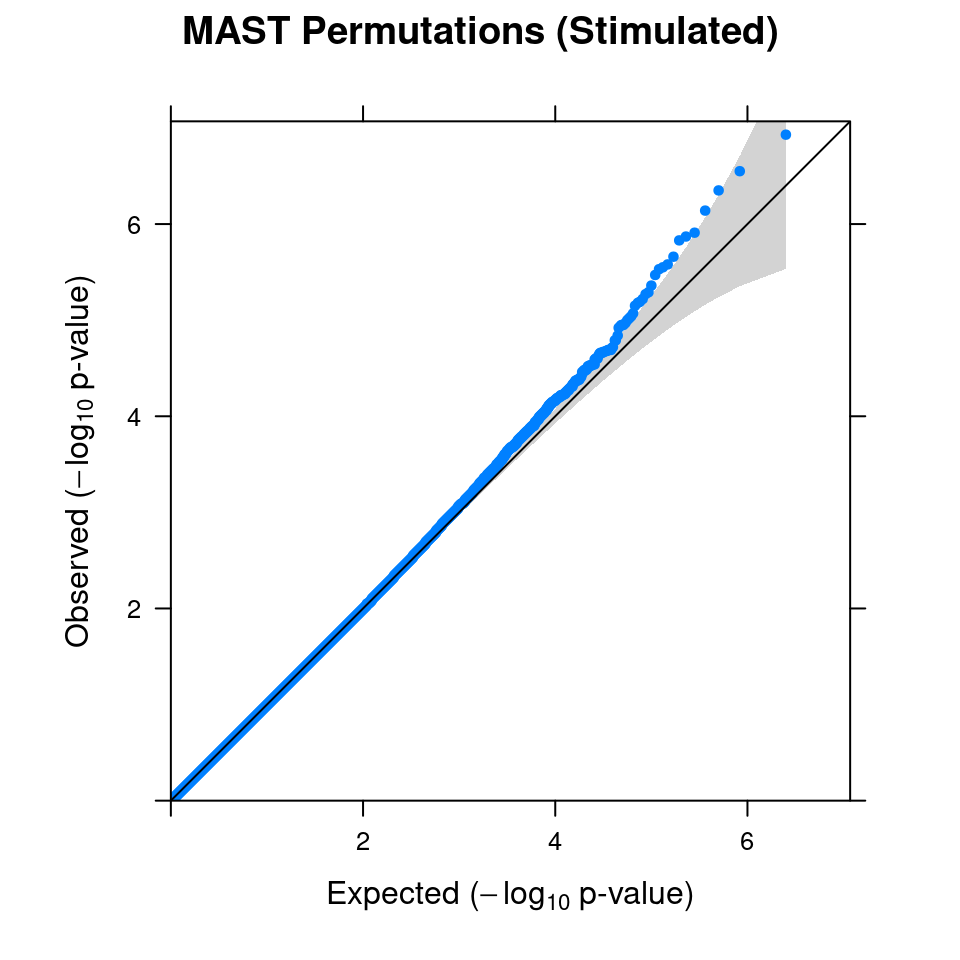
4.2 Original MAST DGE result
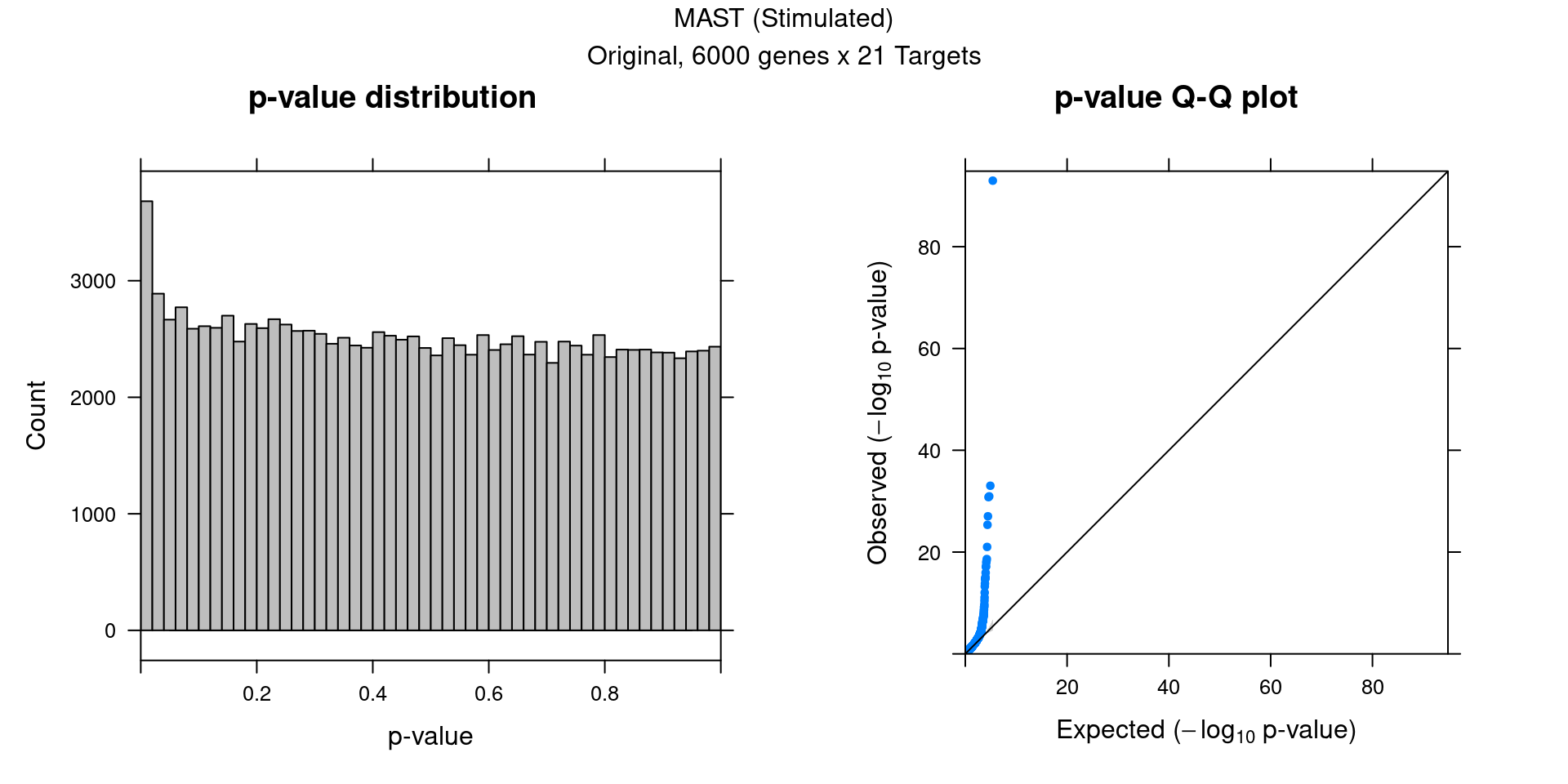
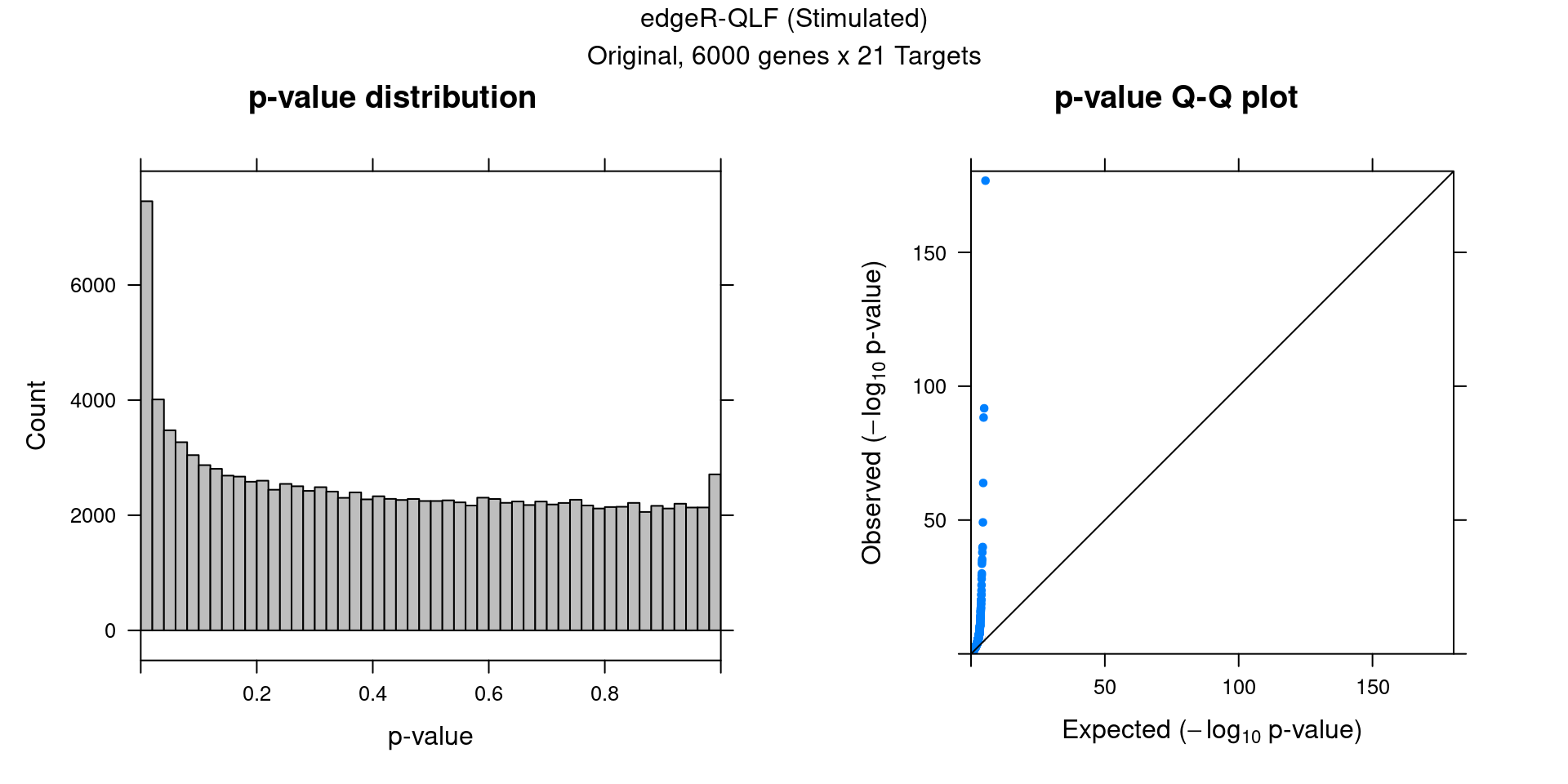
5 edgeR Permutation (Stimulated)
Within stimulated cells, cell labels in the expression data were permuted randomly so that they are no longer correlated with the knock-down conditions. Then edgeR QLF test was performed under each guide.
In total, 10 random permutation rounds like this were conducted.
5.1 Combined from 10 permutations
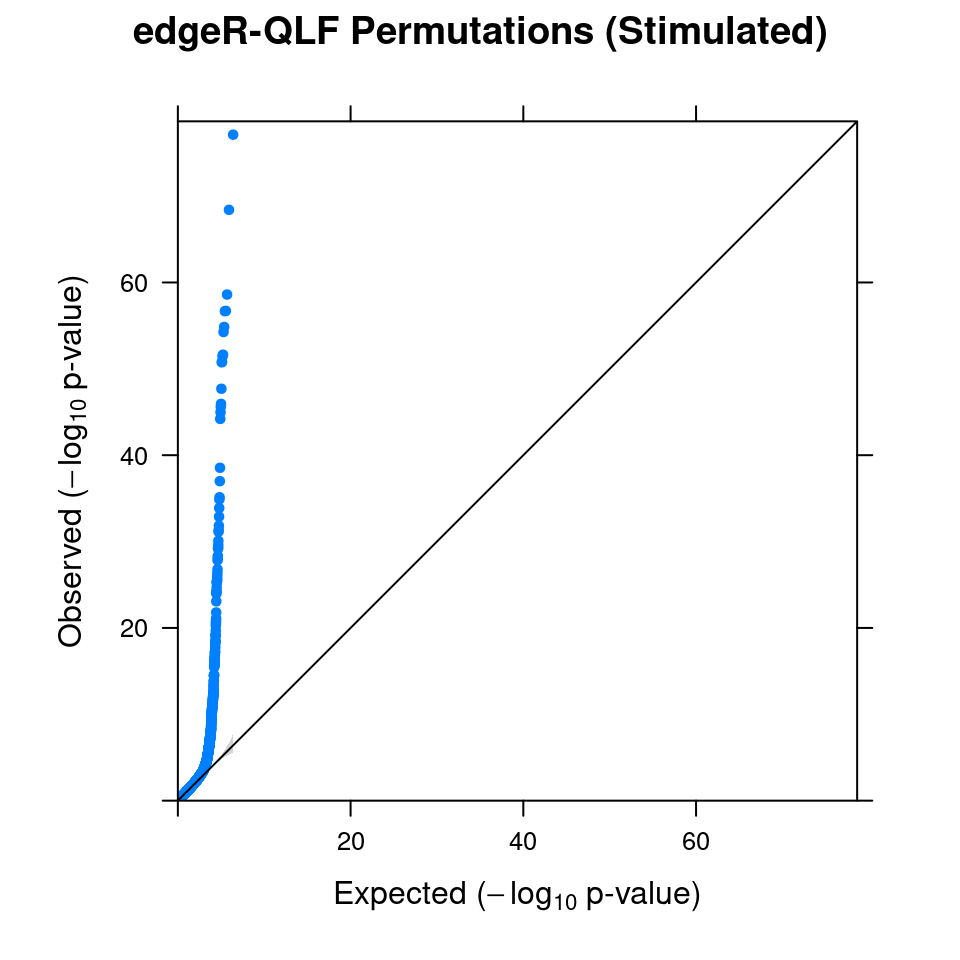
5.2 Original edgeR DGE result