Guided Factor Analysis on LUHMES CROP-seq Data
-- Deviance residual transformed + normal-mixture prior + by-stage estimation
Yifan Zhou (zhouyf@uchicago.edu)
2021-11-15
1 Single cell expression data
Source:
High-throughput single-cell functional elucidation of neurodevelopmental disease-associated genes reveals convergent mechanisms altering neuronal differentiation, GEO accession: GSE142078.
Perturbations:
CRISPR knock-down of 14 autism spectrum disorder (ASD)–associated genes (3 gRNAs per gene) + 5 non-targeting gRNAs.
Cells:
Lund human mesencephalic (LUHMES) neural progenitor cell line.
Cells from 3 batches were merged together into 1 analysis. All cells have only a single type of gRNA readout. Quality control resulted in 8708 cells.
Genes:
Top 6000 genes ranked by the multinomial deviance statistics were kept.
Normalization:
Deviance residual transformation.
Batch effect, unique UMI count, library size, and mitochondria percentage were all corrected for. The corrected and scaled expression data were used as input for subsequent factor analysis.
Here, our "guide", \(G\) matrix, consists of 15 types (14 genes + NTC) of gene-level knock-down conditions across cells.
In each case, Gibbs sampling was conducted for 3000 iterations, and the posterior mean estimates were averaged over the last 1000 iterations. SVD Initialization.
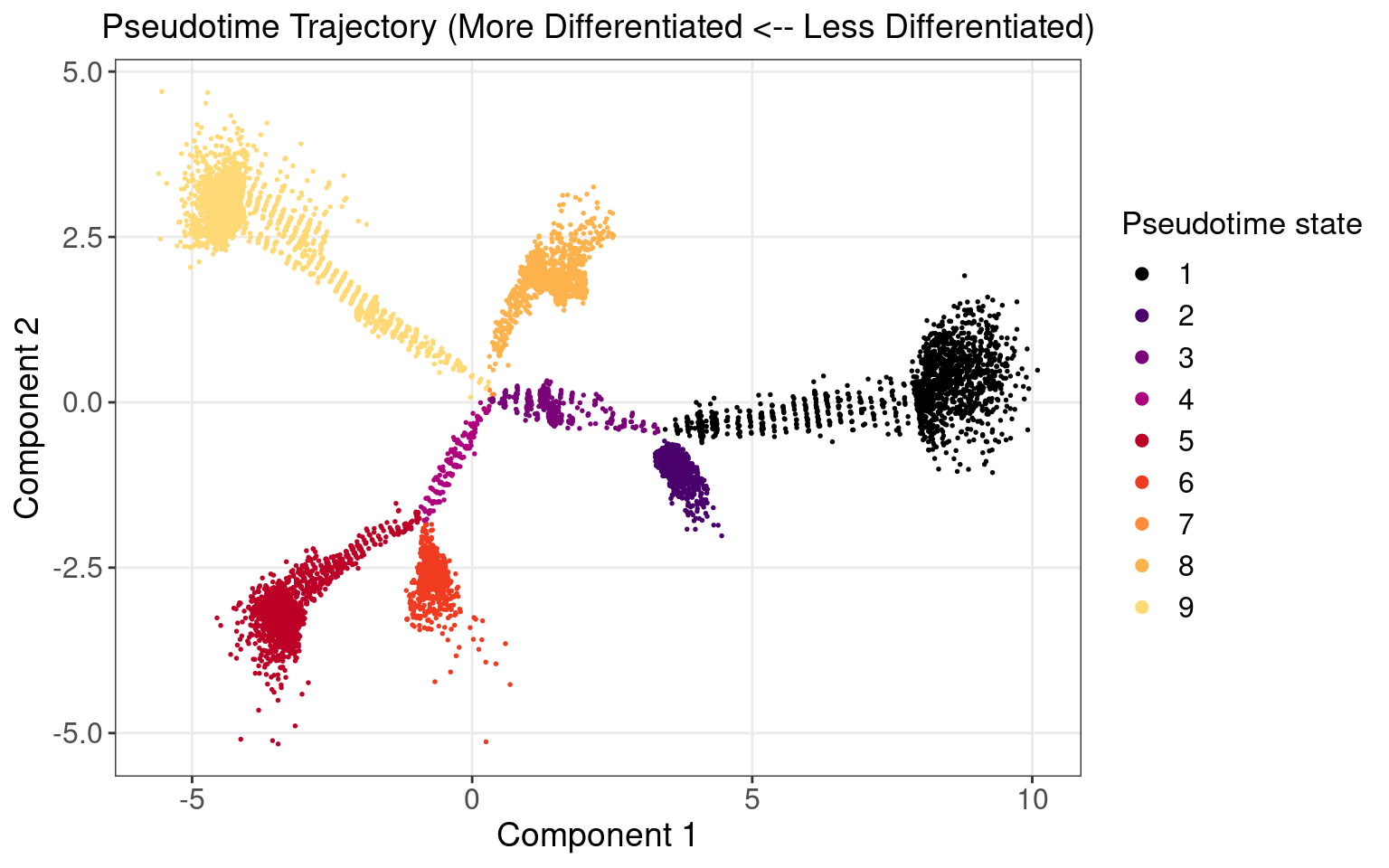
1.1 Monocle Pseudotime Trajectory
We attempted to reproduce the pseudotime trajectory of cells as reported in the original paper.
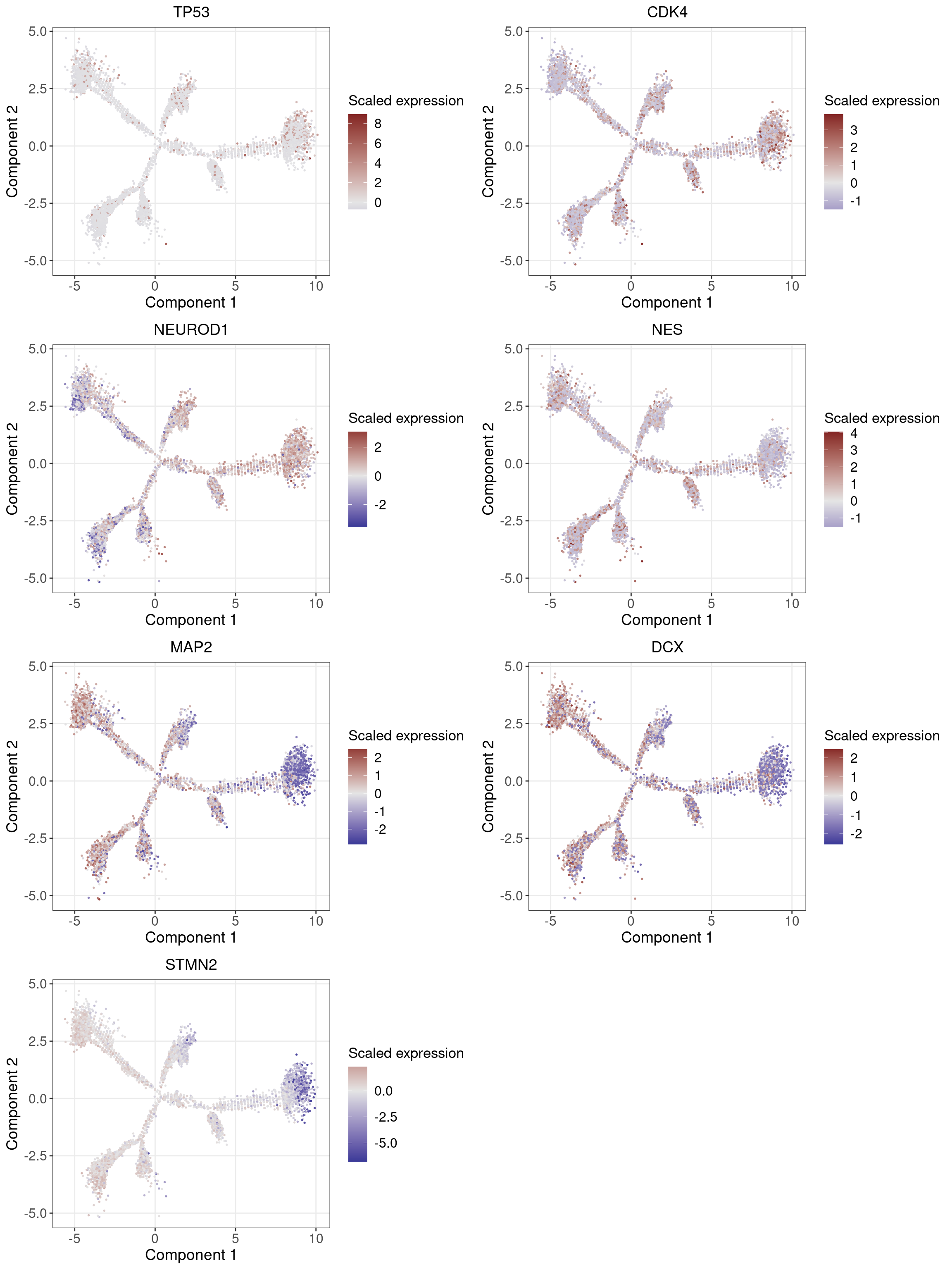
1.2 Marker Gene Expression in Pseudotime
Neuronal markers (MAP2 and DCX) increase along the pseudotime trajectory, whereas progenitor markers (TP53 and CDK4) decrease, consistent with Fig 3B of original paper.
Neuron progenitor (NPC): TP53, CDK4, NEUROD1, NES?
Post-mitotic neuron: MAP2, DCX, STMN2
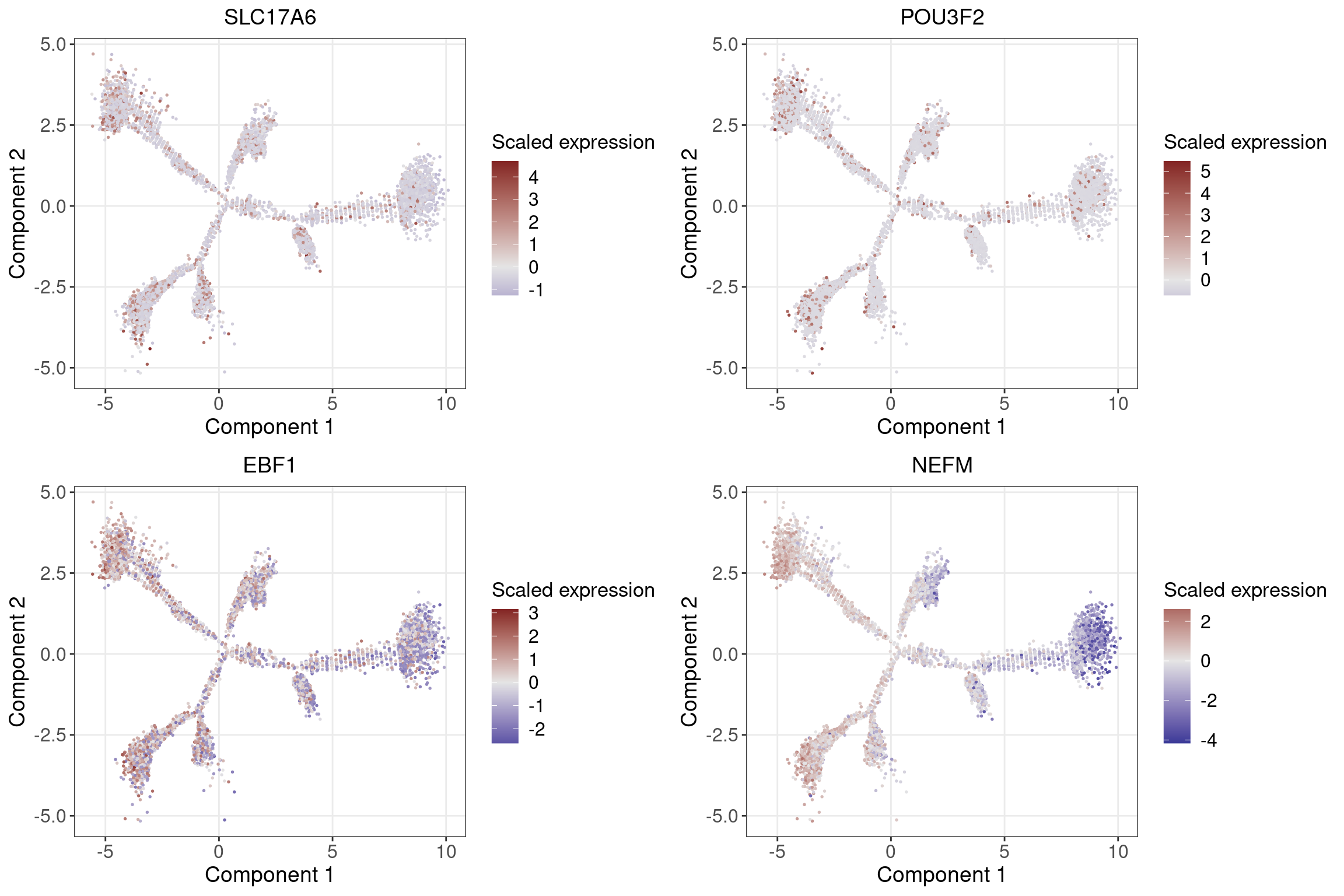
Glutamatergic neuron (excitatory): SLC17A6
Cortical neuron: BRN2 (POU3F2)
Developing striatal neurons: EBF1
Subtypes of inhibitory neurons: SST
Subtypes of extitatory neurons: NEFM
We separate the cells into 2 groups:
Early stage: 2523 cells in pseudotime state 1 - 3;
Late stage: 6185 cells in pseudotime satte 4 - 9.
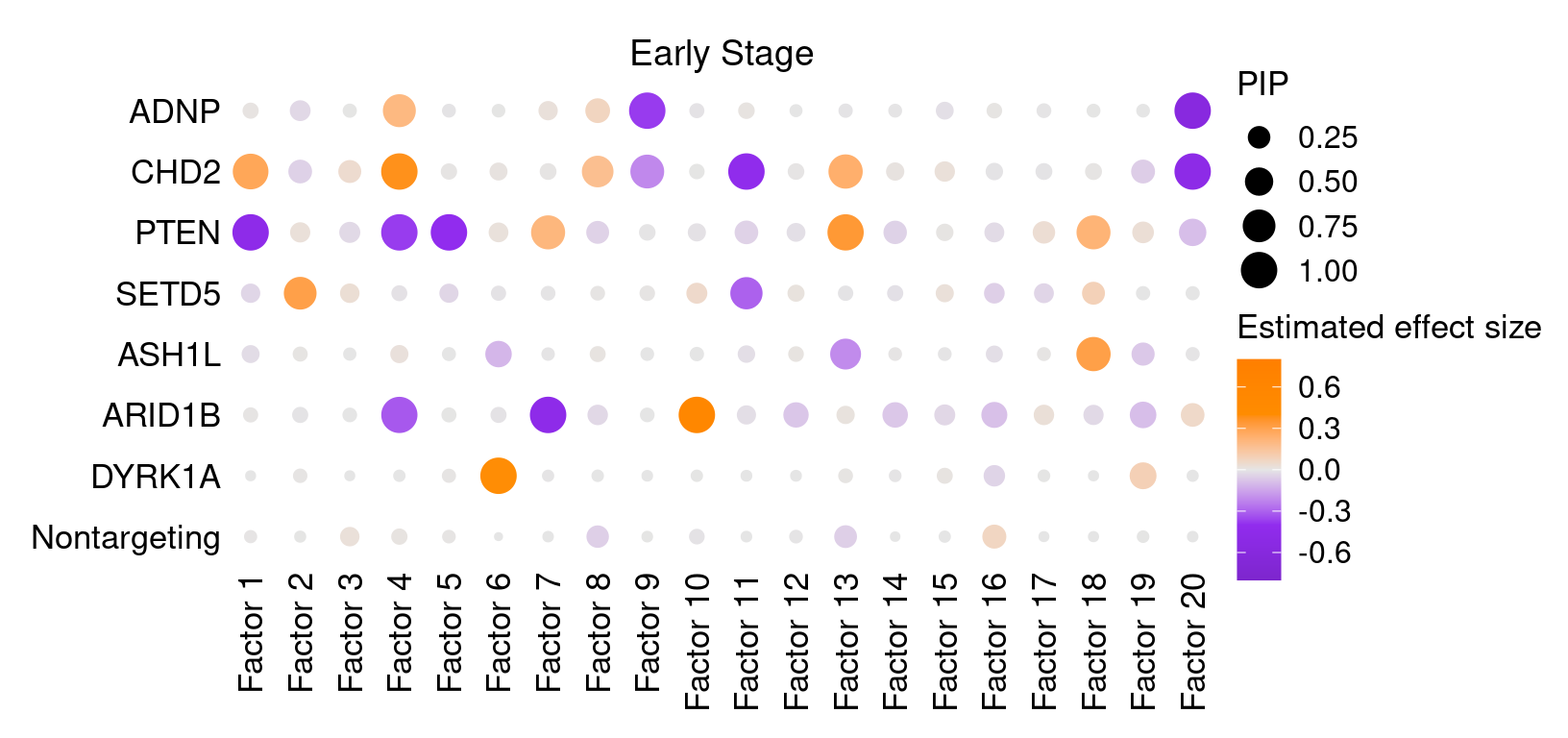
We modified GSFA so that the associations (\(\beta\)) between factors and perturbations are estimated for early and late stage cells separately.
2 Factor ~ Perturbation Association
2.1 Early stage cells
2.2 Late stage cells
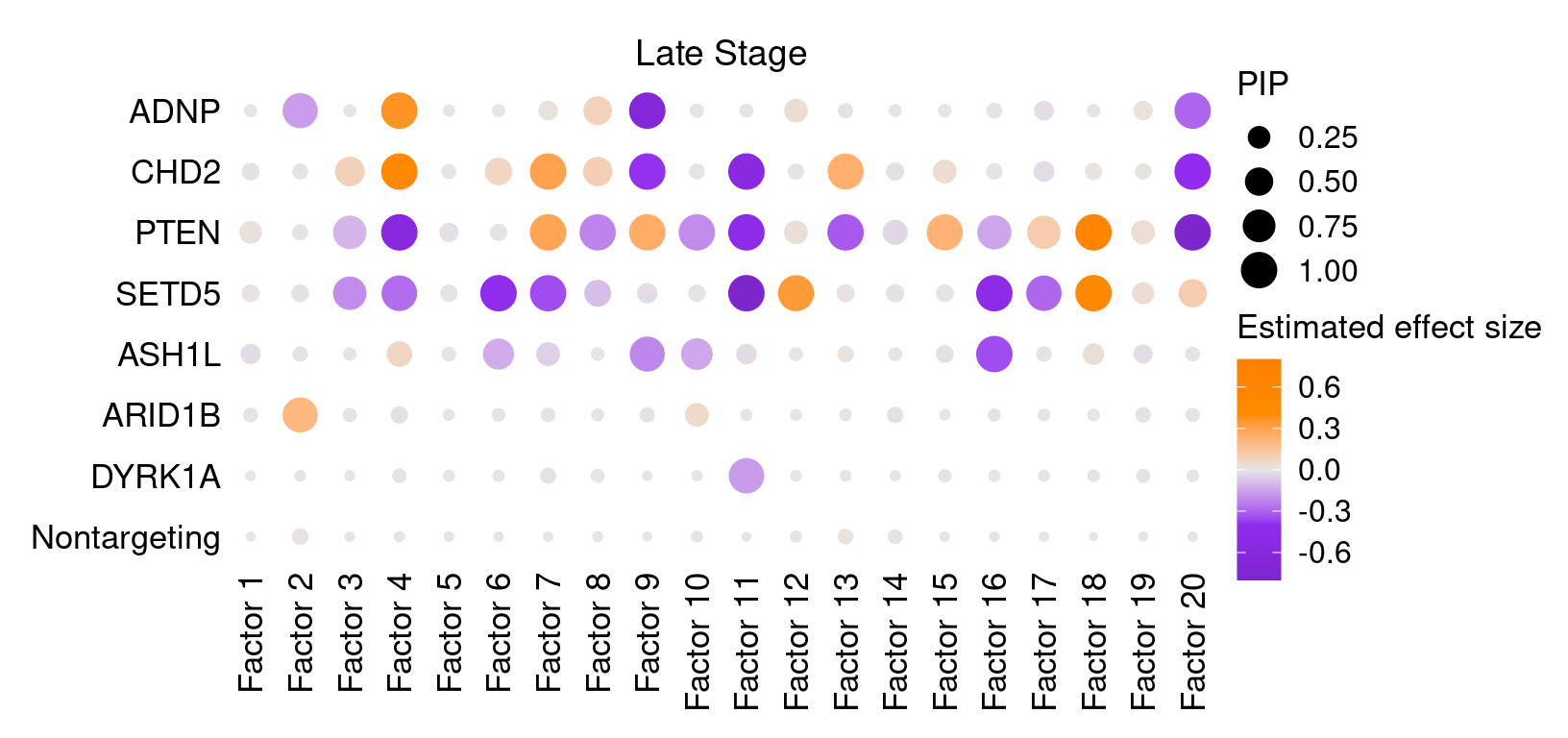
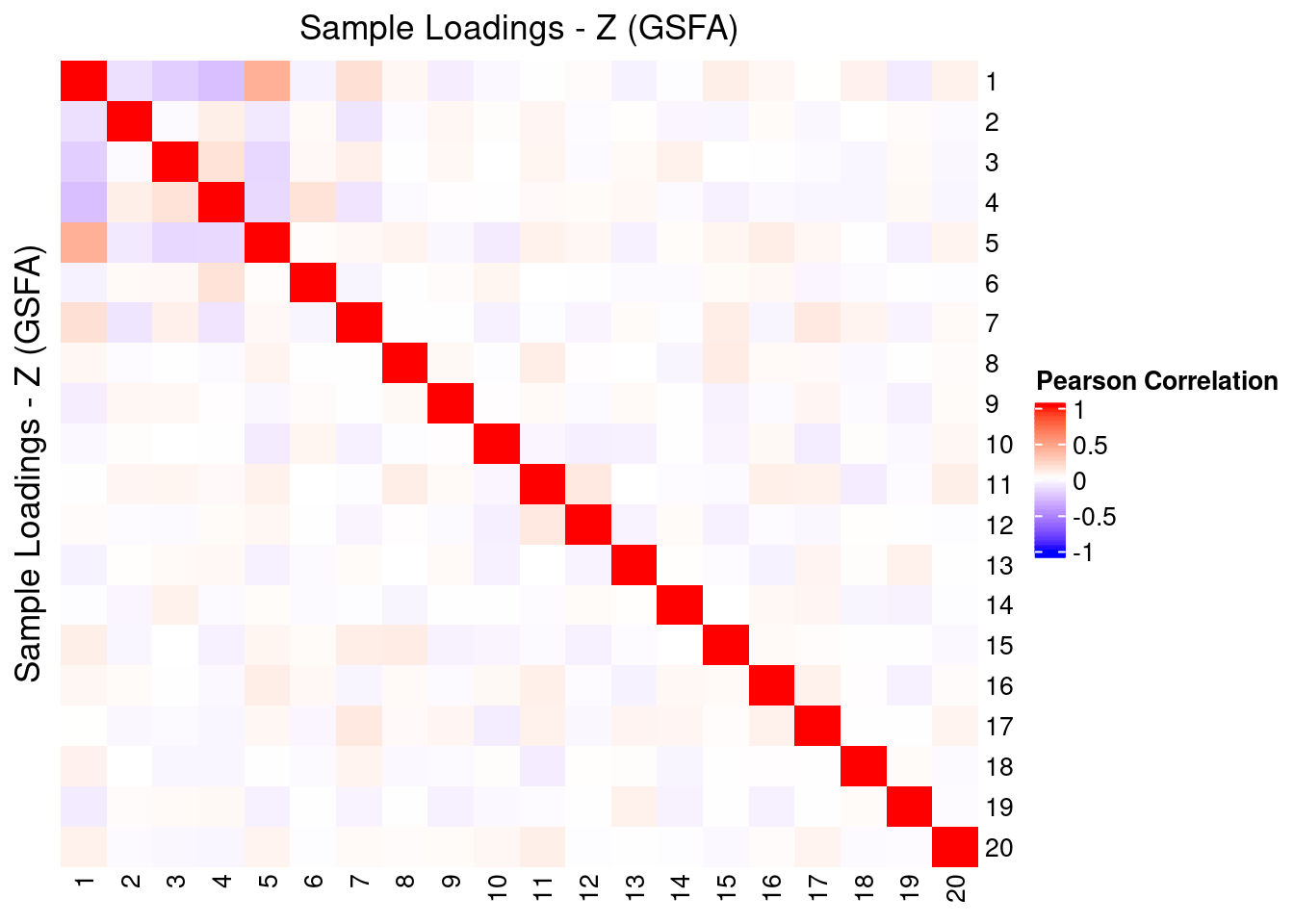
2.3 Correlation within Factors
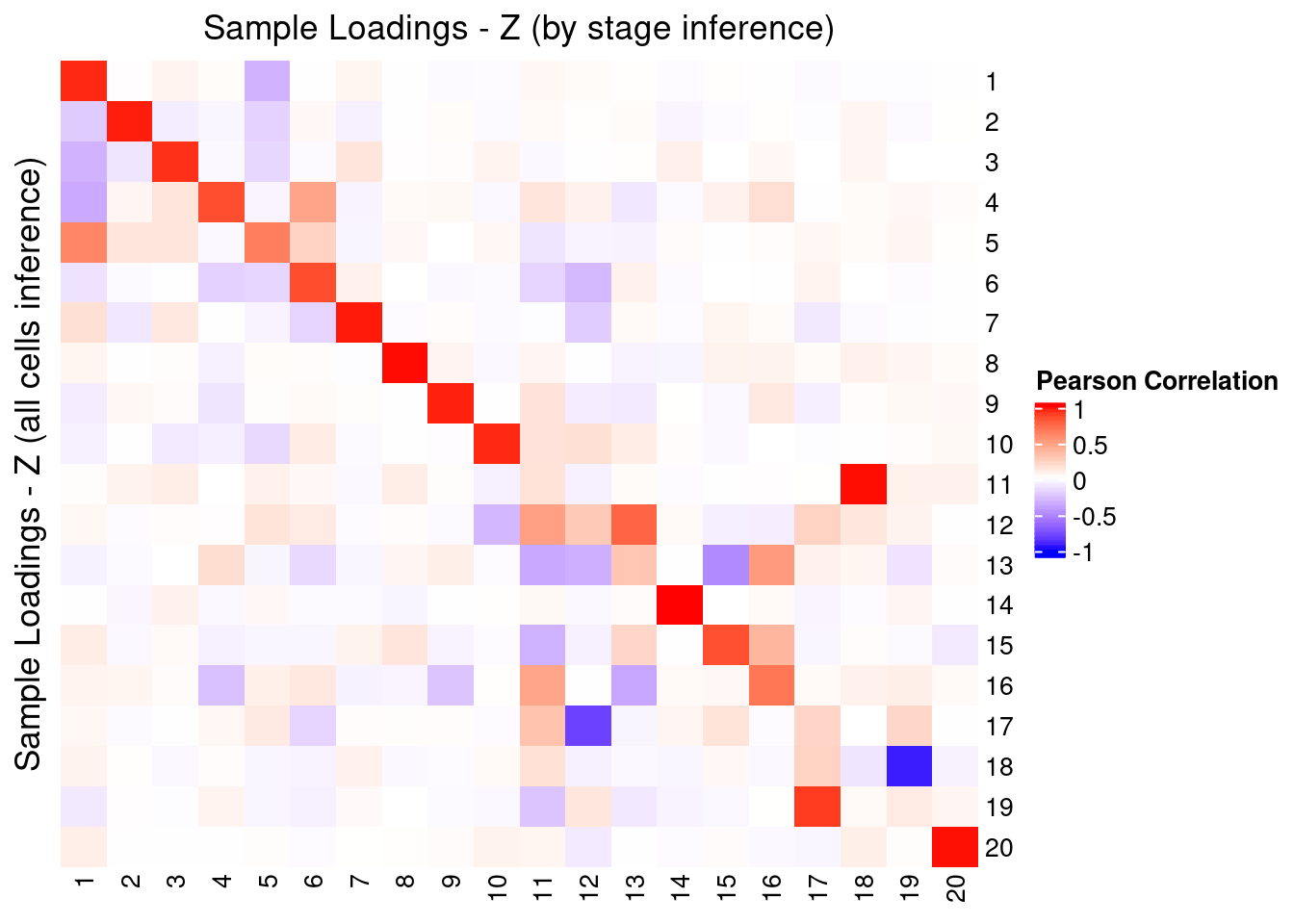
2.3.1 Correlation with factors inferred using all cells
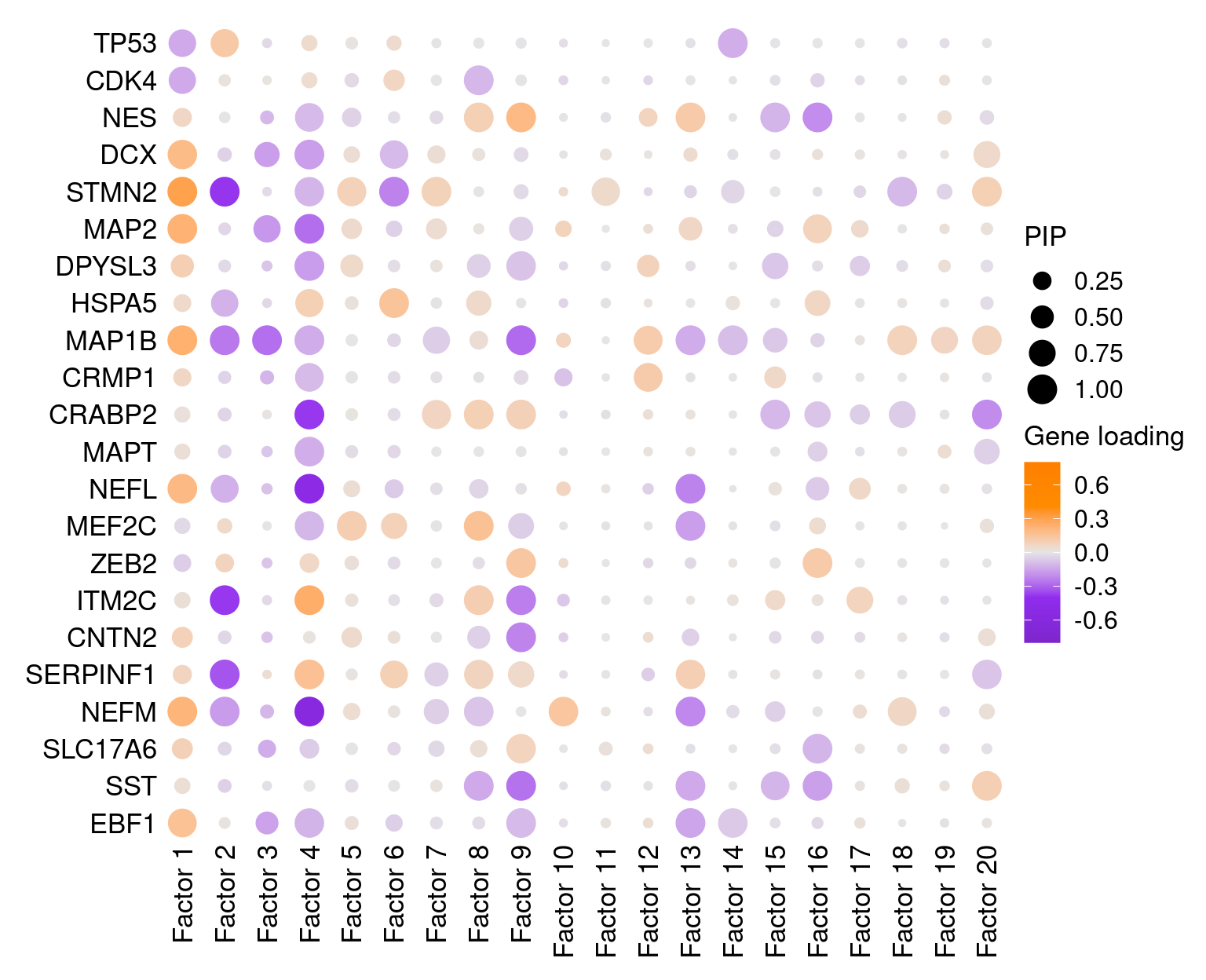
3 Factor Interpretation
3.1 Factor by gene loading
Cell differentiation markers and neuronal cell type markers:
| protein_name | gene_name | type |
|---|---|---|
| TP53 | TP53 | Cell proliferation |
| CDK4 | CDK4 | Cell proliferation |
| Nestin | NES | NPC |
| Doublecortin | DCX | Post-mitotic neuron |
| STMN2 | STMN2 | Post-mitotic neuron |
| MAP2 | MAP2 | Post-mitotic neuron |
| DPYSL3 | DPYSL3 | Mature neuron |
| HSPA5 | HSPA5 | Mature neuron |
| MAP1B | MAP1B | Mature neuron |
| CRMP1 | CRMP1 | Mature neuron |
| CRABP2 | CRABP2 | Mature neuron |
| MAPT | MAPT | Mature neuron |
| NEFL | NEFL | Mature neuron |
| MEF2C | MEF2C | Mature neuron |
| ZEB2 | ZEB2 | Mature neuron |
| ITM2C | ITM2C | Negative regulation of neuron projection |
| CNTN2 | CNTN2 | Negative regulation of neuron projection |
| SERPINF1 | SERPINF1 | Negative regulation of neuron projection |
| Neurofilament-M | NEFM | Excitatory neuron |
| VGLUT2 | SLC17A6 | Excitatory neuron |
| Somatostatin | SST | Inhibitory neuron |
| EBF1 | EBF1 | Developing striatal neuron |
3.2 Selected factor GO enrichment
4 DEG Interpretation
4.1 Number of DEGs detected under different methods
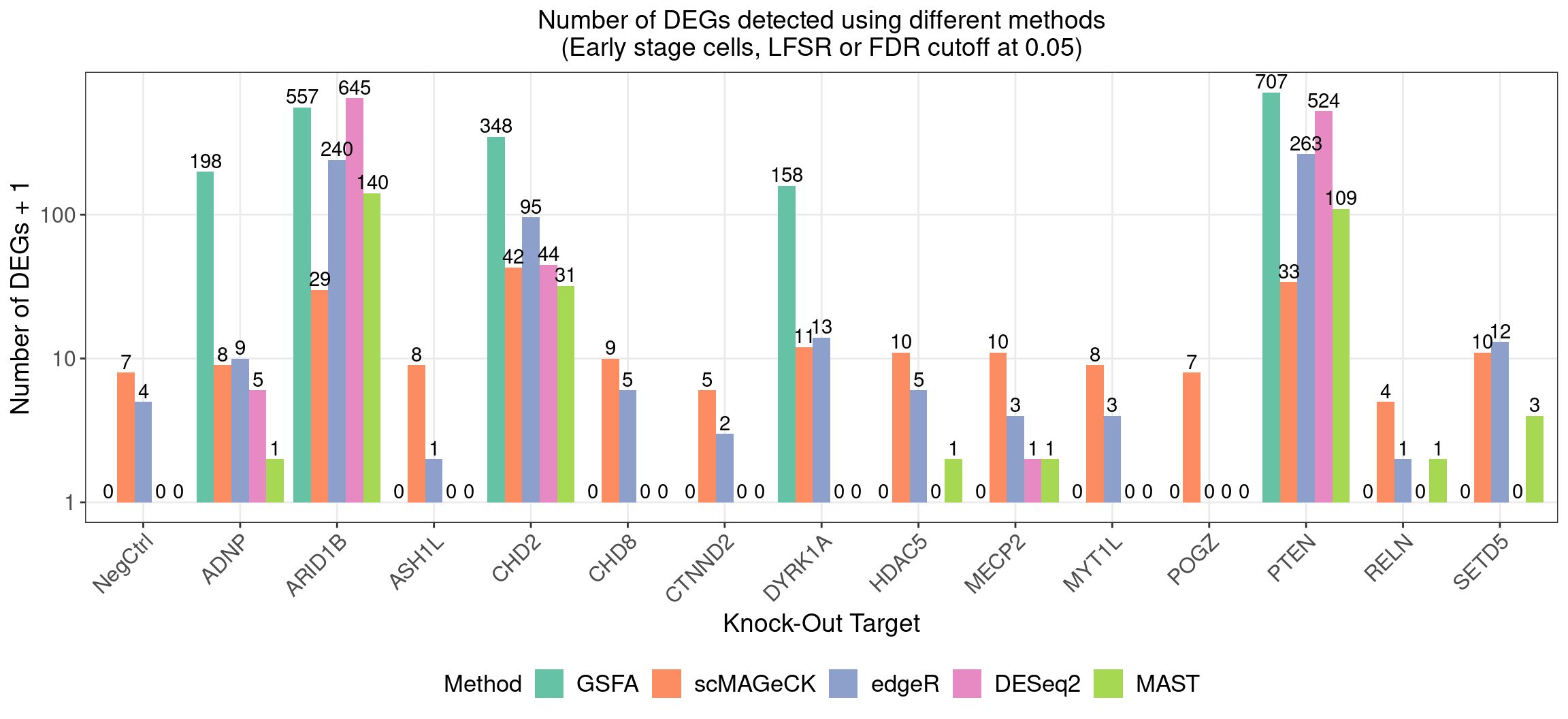
4.1.1 Early stage cells
Number of genes that passed GSFA LFSR < 0.05 under each perturbation:
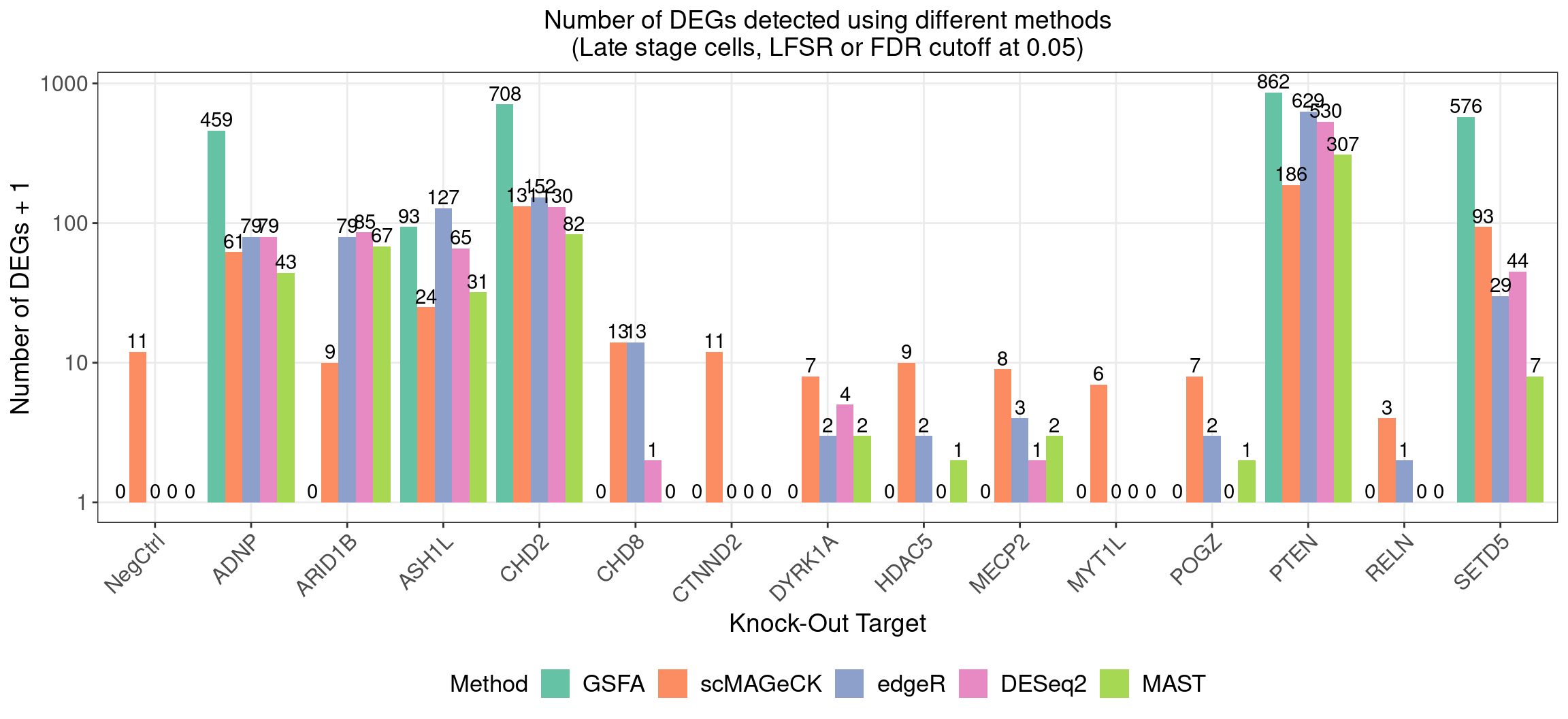
4.1.2 Late stage cells
Number of genes that passed GSFA LFSR < 0.05 under each perturbation:
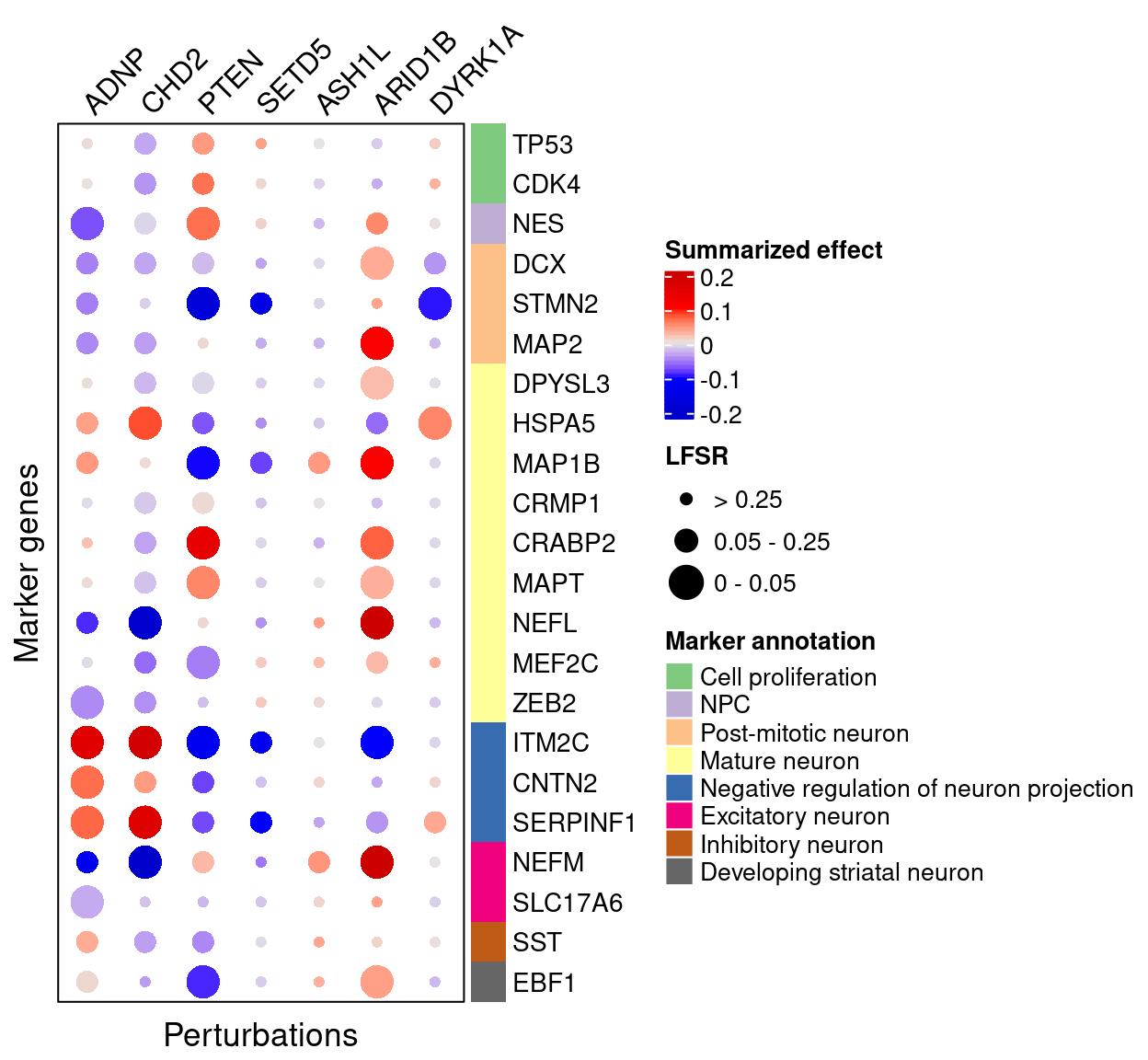
4.2 Perturbation effect on gene markers
Cell differentiation markers and neuronal cell type markers:
| protein_name | gene_name | type |
|---|---|---|
| TP53 | TP53 | Cell proliferation |
| CDK4 | CDK4 | Cell proliferation |
| Nestin | NES | NPC |
| Doublecortin | DCX | Post-mitotic neuron |
| STMN2 | STMN2 | Post-mitotic neuron |
| MAP2 | MAP2 | Post-mitotic neuron |
| DPYSL3 | DPYSL3 | Mature neuron |
| HSPA5 | HSPA5 | Mature neuron |
| MAP1B | MAP1B | Mature neuron |
| CRMP1 | CRMP1 | Mature neuron |
| CRABP2 | CRABP2 | Mature neuron |
| MAPT | MAPT | Mature neuron |
| NEFL | NEFL | Mature neuron |
| MEF2C | MEF2C | Mature neuron |
| ZEB2 | ZEB2 | Mature neuron |
| ITM2C | ITM2C | Negative regulation of neuron projection |
| CNTN2 | CNTN2 | Negative regulation of neuron projection |
| SERPINF1 | SERPINF1 | Negative regulation of neuron projection |
| Neurofilament-M | NEFM | Excitatory neuron |
| VGLUT2 | SLC17A6 | Excitatory neuron |
| Somatostatin | SST | Inhibitory neuron |
| EBF1 | EBF1 | Developing striatal neuron |
4.2.1 Early stage cells
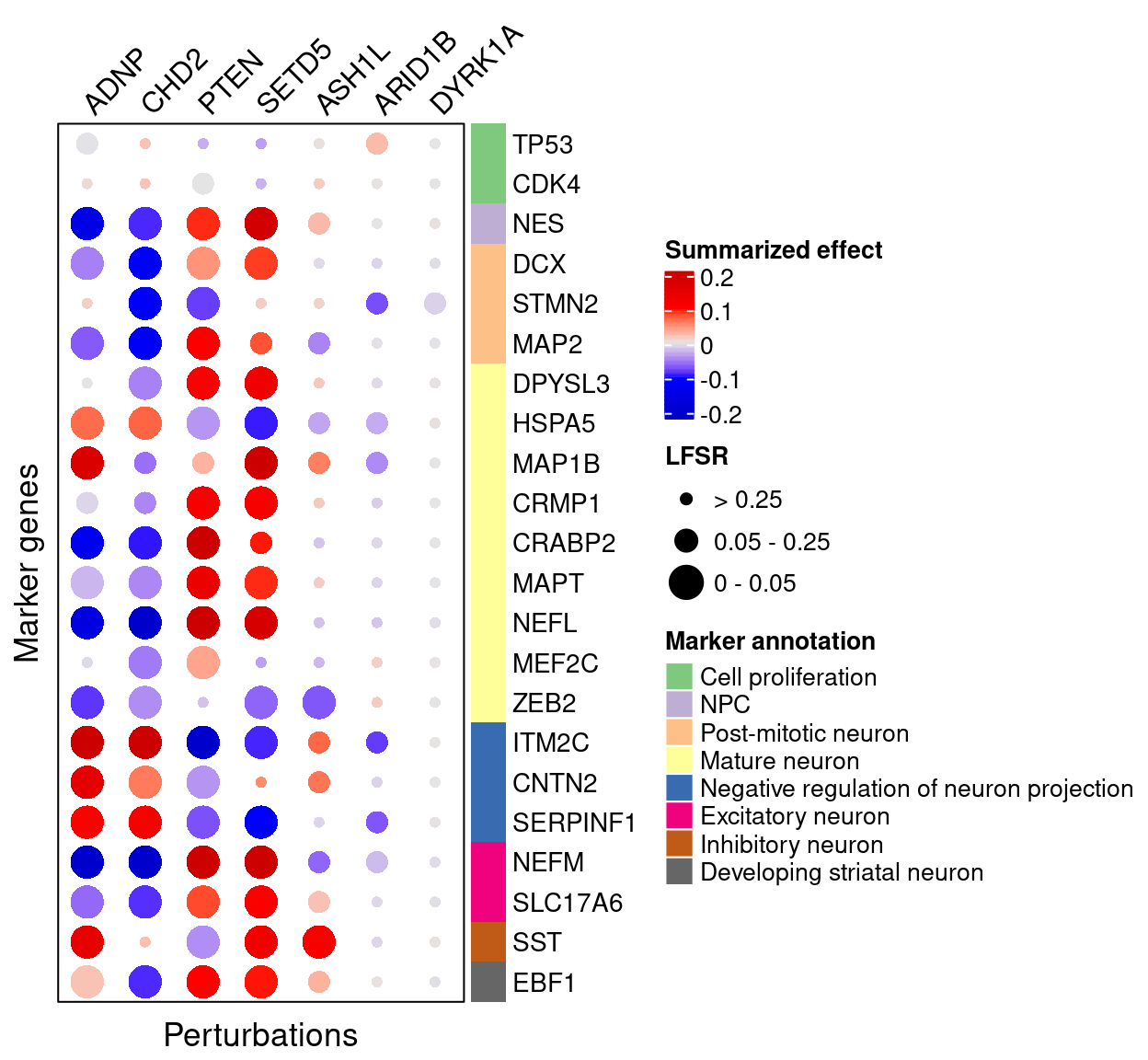
4.2.2 Late stage cells
4.3 DEG GO enrichment
4.3.1 Early stage cells
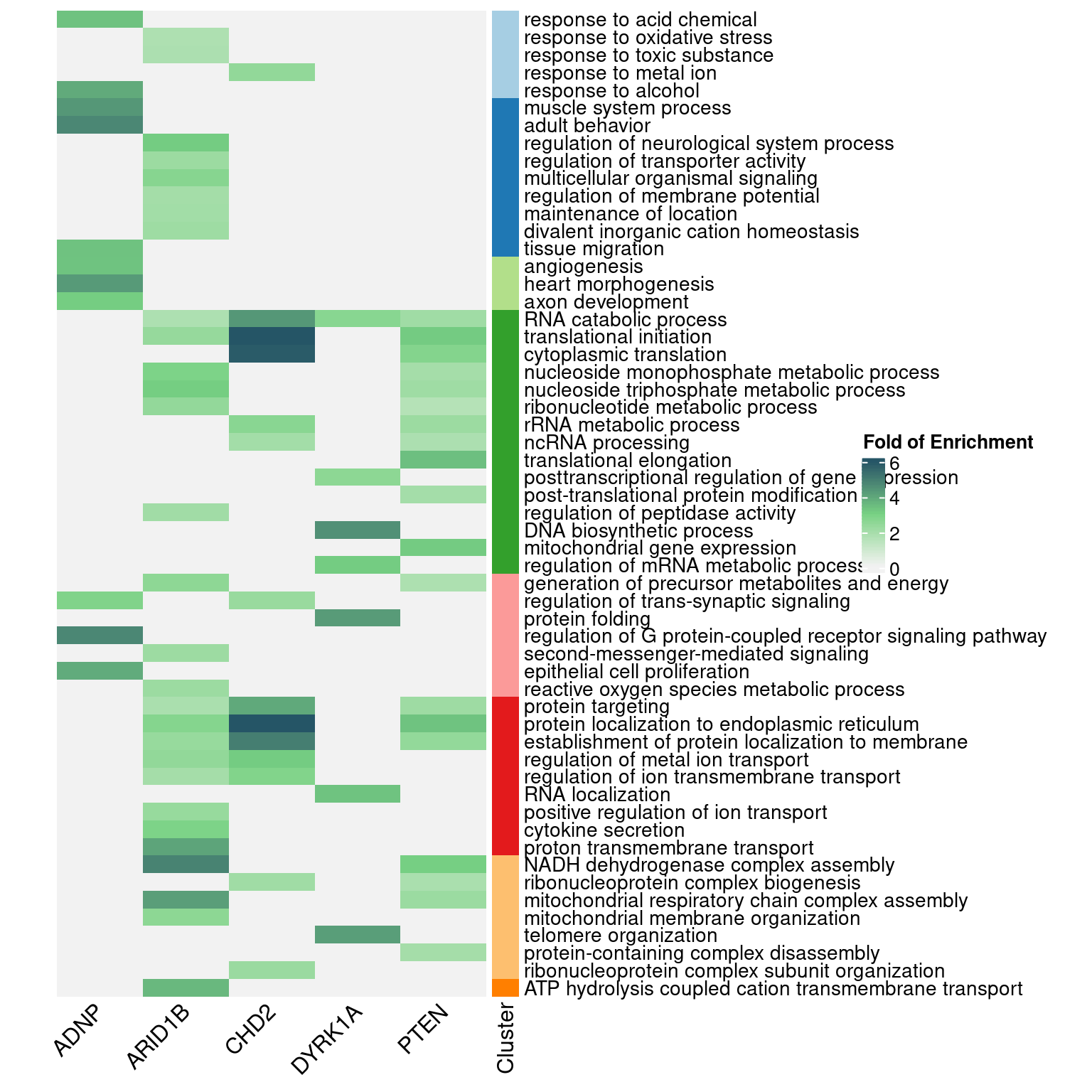
4.3.2 Late stage cells
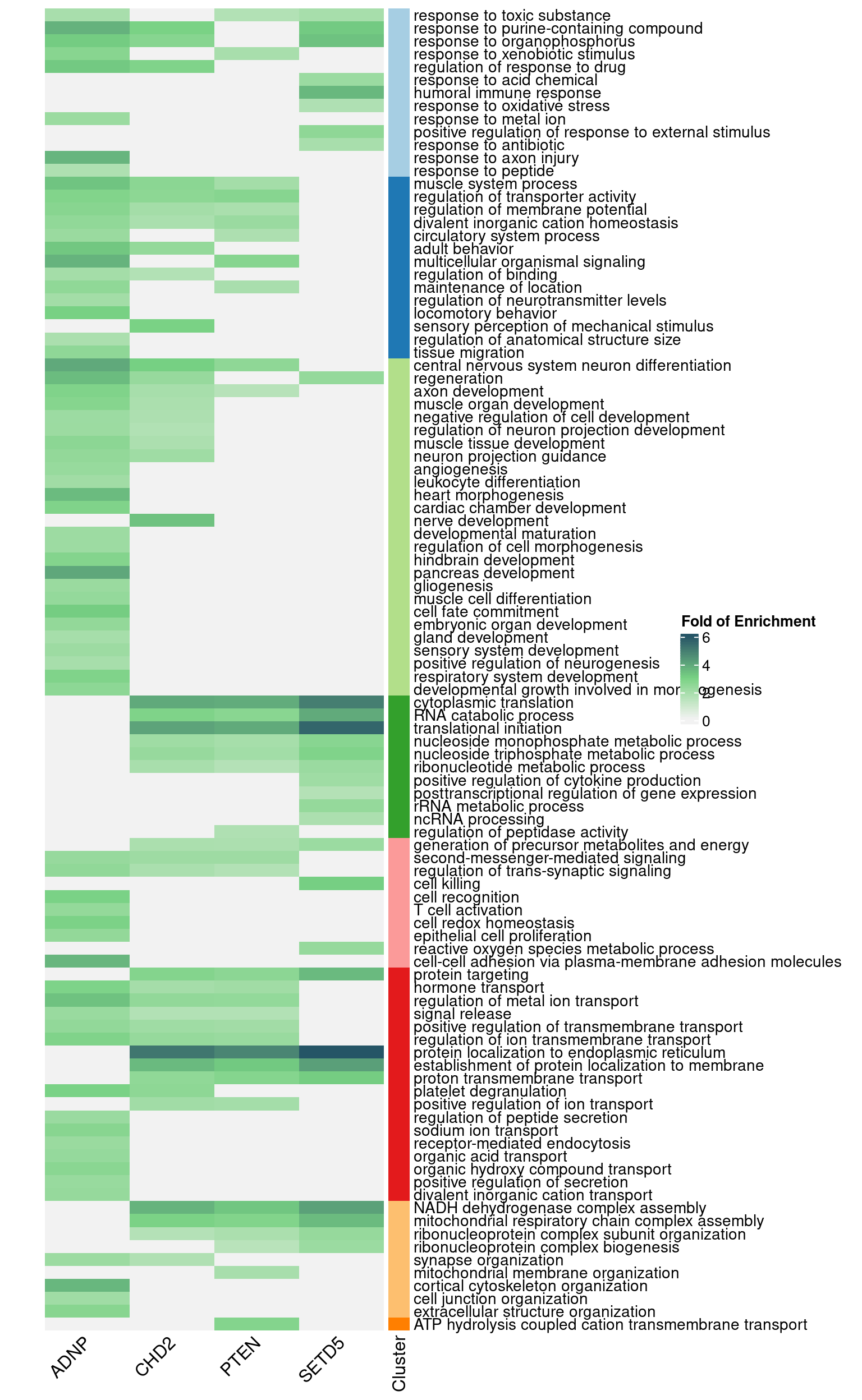
5 Gene Set Enrichment Analysis by Factor
Target genes: Genes w/ non-zero loadings in each factor (PIP cutoff at 0.95);
Backgroud genes: all 6000 genes used in factor analysis;
Statistical test: hypergeometric test (over-representation test);
Only GO terms/pathways that satisfy fold change \(\geq\) 2 and test FDR \(<\) 0.05 are shown below.
5.1 GO Slim Over-Representation Analysis
Gene sets: Gene ontology "Biological Process" (non-redundant).
Factor 1 : 7 significant GO terms| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0006414 | translational elongation | 4.53 | 3.25e-09 | 2.39e-06 | 21/109 | MRPL37; MRPL9; MRPS9; MRPL44; MRPS22; MRPL47; MRPS30; MRPL22; MRPS18B; MRPS10; EEF1A1; MRPL50; MRPS2; MRPS35; MRPL42; MRPS34; ERAL1; MRPL10; EIF4A3; GADD45GIP1; MRPL39 |
| GO:0140053 | mitochondrial gene expression | 4.06 | 5.70e-08 | 2.09e-05 | 20/116 | MRPL37; MRPL9; MRPS9; MRPL44; MRPS22; MRPL47; MRPS30; MRPL22; MRPS18B; MRPS10; TBRG4; MRPL50; MRPS2; MRPS35; MRPL42; MRPS34; ERAL1; MRPL10; GADD45GIP1; MRPL39 |
| GO:0016072 | rRNA metabolic process | 3.03 | 2.40e-06 | 5.88e-04 | 22/171 | RPF1; WDR43; MRPS9; NIFK; RPSA; RRP9; ABT1; BYSL; RPL7L1; DDX56; DCAF13; WDR74; GTPBP4; NGDN; RPS27L; RPS2; RSL1D1; NOB1; EIF4A3; GLTSCR2; RRP1B; RRP1 |
| GO:0032984 | protein-containing complex disassembly | 2.57 | 2.36e-05 | 2.89e-03 | 23/211 | PEX14; STMN1; MRPL37; MRPL9; MRPS9; MRPL44; MRPS22; MRPL47; MRPS30; MRPL22; MRPS18B; MRPS10; STMN4; STMN2; MRPL50; MRPS2; MRPS35; MRPL42; MRPS34; ERAL1; MRPL10; GADD45GIP1; MRPL39 |
| GO:0034470 | ncRNA processing | 2.53 | 1.92e-05 | 2.82e-03 | 24/223 | RPF1; WDR43; MRPS9; MRPL44; RPSA; RRP9; ABT1; BYSL; RPL7L1; DDX56; DCAF13; WDR74; GTPBP4; NGDN; RPS27L; RPS2; RSL1D1; NOB1; EIF4A3; TP53RK; GLTSCR2; TRMU; RRP1B; RRP1 |
| GO:0006091 | generation of precursor metabolites and energy | 2.37 | 3.94e-05 | 4.13e-03 | 25/248 | SDHB; HMGCL; NDUFS2; NDUFB5; ADH5; SDHA; PRELID1; NDUFA4; TBRG4; PMPCB; COX6C; CYC1; SDHD; GLRX3; NDUFA9; ATP5B; TRAP1; COQ9; NCOR1; IDH3B; SIRT6; UQCRFS1; GLTSCR2; MT-CO3; MT-ND4 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 2.28 | 6.65e-06 | 1.22e-03 | 32/330 | EIF3I; RPF1; WDR43; MRPS9; RPSA; RRP9; BRIX1; MRPL22; ABT1; BYSL; RPL7L1; DDX56; DCAF13; EIF3E; MRPS2; WDR74; GTPBP4; PRMT5; NGDN; RPS27L; RPS2; RSL1D1; NOB1; RPL23A; ERAL1; MRPL10; EIF4A3; GLTSCR2; EIF3D; RRP1B; RRP1 |
Factor 2 : 28 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 9.33 | 0.00e+00 | 0.00e+00 | 84/119 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RTN4; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; BCAP31; RPL10; RPS20; RPL7; RPL30; GPAA1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; SPCS2; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPS27L; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; VAPA; DDRGK1; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006413 | translational initiation | 6.90 | 0.00e+00 | 0.00e+00 | 82/157 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; MCTS1; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; PPP1CA; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPS27L; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; EIF4A1; RPL26; RPL23A; RPL23; RPL19; EIF1; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0090150 | establishment of protein localization to membrane | 5.70 | 0.00e+00 | 0.00e+00 | 88/204 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; YWHAQ; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; NSG1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; GDI1; RPS20; SDCBP; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; CALM1; HSP90AA1; SRP14; RPS27L; RPL4; RPLP1; RPS17; RPS2; RPS15A; ARL6IP1; RPL13; YWHAE; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; YWHAB; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3; WRB |
| GO:0002181 | cytoplasmic translation | 5.43 | 1.11e-15 | 1.36e-13 | 30/73 | RPL11; RPL31; RPL32; RPL15; TMA7; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; MCTS1; RPL30; RPL8; RPL41; RPL6; RPLP0; RPS29; RPLP1; RPL26; RPL19; RPL38; RPS21; RPL36; RPS28; RPL18A; RPL18; RPL13A |
| GO:0006401 | RNA catabolic process | 4.76 | 0.00e+00 | 0.00e+00 | 85/236 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; SSB; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; SET; RPL7A; RNH1; RPLP2; RPL27A; RPS13; RPS3; RPS25; HSPA8; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; SLIRP; RPS27L; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; YWHAB; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006605 | protein targeting | 4.56 | 0.00e+00 | 0.00e+00 | 90/261 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RAB7A; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; TOMM7; RPS4X; RPL36A; RPL39; RPL10; GDI1; RPS20; SDCBP; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; SPCS2; RPS3; RPS25; HSPA8; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; HSP90AA1; SRP14; RPS27L; RPL4; RPLP1; RPS17; RPS2; RPS15A; ARL6IP1; UQCRC2; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; YWHAB; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; ECH1; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0002576 | platelet degranulation | 4.41 | 6.95e-06 | 5.10e-04 | 12/36 | TAGLN2; SCCPDH; TUBA4A; MAGED2; APLP2; PSAP; CD63; CALM1; SCG3; ALDOA; LGALS3BP; SOD1 |
| GO:0051187 | cofactor catabolic process | 4.41 | 1.53e-03 | 4.00e-02 | 6/18 | ACOT7; PRDX1; BLVRA; PRDX4; PRDX5; PRDX2 |
| GO:0006081 | cellular aldehyde metabolic process | 3.92 | 6.13e-04 | 2.25e-02 | 8/27 | PARK7; TKT; TALDO1; AKR1C1; PGAM1; TPI1; ESD; IDH2 |
| GO:0006090 | pyruvate metabolic process | 3.78 | 9.19e-06 | 5.62e-04 | 14/49 | ENO1; PDHB; VDAC1; PDHA1; PGK1; LDHA; PGAM1; GAPDH; TPI1; LDHB; PKM; ALDOA; ENO3; BSG |
| GO:0001906 | cell killing | 3.78 | 8.03e-04 | 2.56e-02 | 8/28 | PRDX1; TUBB; HSP90AB1; RPL30; TUBB4B; GAPDH; RPS19; PVRL2 |
| GO:0070671 | response to interleukin-12 | 3.65 | 1.04e-03 | 3.17e-02 | 8/29 | CDC42; HNRNPA2B1; PPIA; TALDO1; CFL1; RPLP0; PSME2; SOD1 |
| GO:0009566 | fertilization | 3.57 | 2.96e-04 | 1.14e-02 | 10/37 | PARK7; H3F3A; CCT7; CCT5; VDAC2; CCT2; REC8; SLIRP; ALDOA; PVRL2 |
| GO:0016999 | antibiotic metabolic process | 3.42 | 1.67e-05 | 9.44e-04 | 15/58 | PARK7; PRDX1; MDH1; PDHB; MDH2; AKR1B1; PDHA1; PRDX4; PRDX5; AKR1C1; ALDH2; ESD; IDH2; PRDX2; SOD1 |
| GO:0072524 | pyridine-containing compound metabolic process | 3.41 | 9.00e-06 | 5.62e-04 | 16/62 | ENO1; MDH1; TKT; PDHB; MDH2; PDHA1; PGK1; TALDO1; LDHA; PGAM1; GAPDH; TPI1; PKM; IDH2; ALDOA; ENO3 |
| GO:0048857 | neural nucleus development | 3.41 | 1.67e-03 | 4.11e-02 | 8/31 | CDC42; ATP5F1; YWHAQ; ACTB; DYNLL1; CALM1; CKB; YWHAE |
| GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | 3.31 | 2.08e-03 | 4.49e-02 | 8/32 | ATP6V0B; ATP5F1; ATP5G3; ATP6AP1; ATP5C1; ATP5B; ATP6V0D1; ATP5A1 |
| GO:0045861 | negative regulation of proteolysis | 3.02 | 5.89e-07 | 4.80e-05 | 24/105 | PARK7; RPL11; TMEM59; RPL5; AGT; RPS7; SERPINI1; HSP90AB1; NGFRAP1; SDCBP; PRDX5; APLP2; GAPDH; PEBP1; MAP1A; ARL6IP1; HERPUD1; GABARAPL2; YWHAE; SERPINF1; RPL23; DDRGK1; CST3; CDKN2D |
| GO:0005996 | monosaccharide metabolic process | 2.74 | 6.50e-05 | 2.99e-03 | 18/87 | ENO1; MDH1; TKT; PDHB; MDH2; AKR1B1; PDHA1; PGK1; TALDO1; PPP1CA; PGAM1; GAPDH; TPI1; PKM; ALDOA; ENO3; G6PC3; CYB5R3 |
| GO:0016051 | carbohydrate biosynthetic process | 2.61 | 7.04e-04 | 2.35e-02 | 14/71 | ENO1; MDH1; TKT; MDH2; AKR1B1; PGK1; TALDO1; PPP1CA; PGAM1; GAPDH; TPI1; ALDOA; ENO3; G6PC3 |
| GO:0036230 | granulocyte activation | 2.49 | 1.70e-07 | 1.79e-05 | 36/191 | DDOST; ILF2; ARPC5; PSMD6; RAB7A; TUBB; CSNK2B; HSP90AB1; DYNLT1; PSMB1; PPIA; PRDX4; ATP6AP2; SDCBP; GGH; TUBB4B; PSMD13; GSTP1; NDUFC2; HSPA8; PSAP; PGAM1; CD63; CCT2; DYNLL1; ACTR10; HSP90AA1; SRP14; PKM; ALDOA; PSMD7; RAB5C; VAPA; CST3; FTL; CYB5R3 |
| GO:0002446 | neutrophil mediated immunity | 2.47 | 2.24e-07 | 2.05e-05 | 36/193 | DDOST; ILF2; ARPC5; PSMD6; RAB7A; TUBB; CSNK2B; HSP90AB1; DYNLT1; PSMB1; PPIA; PRDX4; ATP6AP2; SDCBP; GGH; TUBB4B; PSMD13; GSTP1; NDUFC2; HSPA8; PSAP; PGAM1; CD63; CCT2; DYNLL1; ACTR10; HSP90AA1; SRP14; PKM; ALDOA; PSMD7; RAB5C; VAPA; CST3; FTL; CYB5R3 |
| GO:0071897 | DNA biosynthetic process | 2.36 | 1.35e-03 | 3.71e-02 | 15/84 | RPS27A; CCT7; CTNNB1; CCT5; HSP90AB1; HNRNPA2B1; PPIA; PTGES3; CCT2; UBC; HSP90AA1; TERF2IP; CDKN2D; UBA52; UFD1L |
| GO:0052547 | regulation of peptidase activity | 2.31 | 1.54e-04 | 6.28e-03 | 22/126 | PARK7; AGT; ARL6IP5; SERPINI1; MTCH1; CYCS; NGFRAP1; BCAP31; PRDX5; RPS3; APLP2; GAPDH; PEBP1; PSME2; RPS27L; ARL6IP1; HERPUD1; YWHAE; SERPINF1; DDRGK1; CST3; CDKN2D |
| GO:0031098 | stress-activated protein kinase signaling cascade | 2.23 | 1.31e-03 | 3.71e-02 | 17/101 | CDC42; PRDX1; GADD45A; AGT; RPS27A; ARL6IP5; SKP1; EZR; AKR1B1; SDCBP; DNAJA1; GSTP1; RPS3; UBC; HACD3; UBA52; PRMT1 |
| GO:0019058 | viral life cycle | 2.22 | 7.02e-04 | 2.35e-02 | 19/113 | CDC42; RPS27A; RAB1A; RPSA; RAB7A; HSP90AB1; DYNLT1; PPIA; PABPC1; CHMP5; TSG101; HSPA8; UBC; PPIB; HACD3; VAPA; UBA52; PVRL2; CHMP2A |
| GO:0016072 | rRNA metabolic process | 2.16 | 6.52e-05 | 2.99e-03 | 28/171 | RPL11; RPS8; RPL5; RPS27; RPS7; RPSA; RPL14; RPL35A; H2AFY; RPS14; RPL10A; RPL7; RPS6; RPL35; RPL7A; RPS24; NPM3; RPS27L; RPS17; RPS2; RPL26; RPL27; EIF4A3; RPS21; RPS15; RPS28; RPS16; RPS19; RPS9 |
| GO:0016053 | organic acid biosynthetic process | 2.02 | 1.77e-03 | 4.11e-02 | 20/131 | ACOT7; PARK7; ENO1; PHGDH; MGST3; CDO1; ASNS; PGK1; PTGDS; LDHA; PGAM1; GAPDH; TPI1; PTGES3; HACD3; PKM; ALDOA; ENO3; TECR; XBP1 |
Factor 3 : 10 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 39.50 | 0.00e+00 | 0.00e+00 | 66/119 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS9; RPL28; RPS5; RPL3 |
| GO:0006413 | translational initiation | 30.00 | 0.00e+00 | 0.00e+00 | 66/157 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS9; RPL28; RPS5; RPL3 |
| GO:0002181 | cytoplasmic translation | 25.40 | 0.00e+00 | 0.00e+00 | 26/73 | RPL11; RPL31; RPL32; RPL15; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; RPL30; RPL8; RPL41; RPL6; RPLP0; RPS29; RPLP1; RPL26; RPL19; RPL36; RPS28; RPL18A; RPL18; RPL13A |
| GO:0090150 | establishment of protein localization to membrane | 23.40 | 0.00e+00 | 0.00e+00 | 67/204 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS9; RPL28; RPS5; RPL3 |
| GO:0006401 | RNA catabolic process | 19.90 | 0.00e+00 | 0.00e+00 | 66/236 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS9; RPL28; RPS5; RPL3 |
| GO:0006605 | protein targeting | 18.00 | 0.00e+00 | 0.00e+00 | 66/261 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS9; RPL28; RPS5; RPL3 |
| GO:0016072 | rRNA metabolic process | 8.75 | 4.33e-15 | 3.97e-13 | 21/171 | RPL11; RPS8; RPL5; RPS27; RPS7; RPL14; RPL35A; RPS14; RPL10A; RPL7; RPS6; RPL35; RPL7A; RPS24; RPS17; RPL26; RPS15; RPS28; RPS16; RPS19; RPS9 |
| GO:0034470 | ncRNA processing | 6.71 | 9.44e-13 | 7.70e-11 | 21/223 | RPL11; RPS8; RPL5; RPS27; RPS7; RPL14; RPL35A; RPS14; RPL10A; RPL7; RPS6; RPL35; RPL7A; RPS24; RPS17; RPL26; RPS15; RPS28; RPS16; RPS19; RPS9 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 6.70 | 0.00e+00 | 0.00e+00 | 31/330 | RPL11; RPS8; RPL5; RPS27; RPS7; RPL14; RPL24; RPL35A; RPS23; RPS14; RPL10A; RPL10; RPL7; RPS6; RPL35; RPL12; RPL7A; RPS24; RPL6; RPLP0; RPS17; RPL26; RPL23A; RPS15; RPS28; RPS16; RPS19; RPL13A; RPS9; RPS5; RPL3 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 6.55 | 3.13e-10 | 2.30e-08 | 17/185 | RPL11; RPL5; RPS27; RPL24; RPS23; RPS14; RPL10; RPL12; RPL6; RPLP0; RPL23A; RPS15; RPS28; RPS19; RPL13A; RPS5; RPL3 |
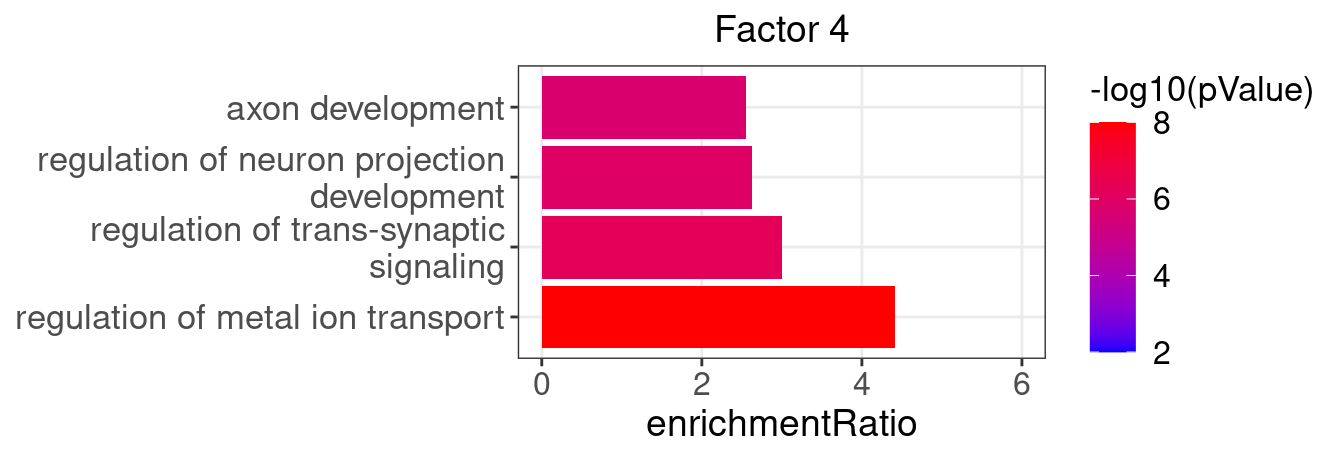
Factor 4 : 40 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0010737 | protein kinase A signaling | 8.28 | 1.69e-04 | 6.08e-03 | 5/11 | RAB13; AKAP12; EZR; GAL; AKAP6 |
| GO:0007218 | neuropeptide signaling pathway | 7.29 | 1.43e-03 | 2.84e-02 | 4/10 | PCSK1N; GAL; SSTR2; PTH2 |
| GO:0010644 | cell communication by electrical coupling | 7.01 | 4.30e-04 | 1.26e-02 | 5/13 | ATP1B1; FKBP1B; SRI; ANK3; CALM1 |
| GO:0035637 | multicellular organismal signaling | 4.66 | 1.33e-05 | 8.13e-04 | 11/43 | ATP1B1; AGT; FKBP1B; SCN3A; SCN9A; CHRNA1; SRI; AKAP9; ANK3; ATP2B1; CALM1 |
| GO:0010959 | regulation of metal ion transport | 4.42 | 3.03e-10 | 2.23e-07 | 24/99 | SEPN1; ATP1B1; AGT; FKBP1B; GNAI2; SNCA; CAMK2B; SRI; AKAP9; CHD7; GAL; FXYD6; ANK3; KCNIP2; CD63; JPH4; AKAP6; CALM1; B2M; VAMP2; NEDD4L; NKAIN4; HOMER3; FXYD7 |
| GO:0032409 | regulation of transporter activity | 4.27 | 4.17e-08 | 1.02e-05 | 19/81 | SEPN1; ATP1B1; FKBP1B; SNCA; MEF2C; ACTB; SRI; AKAP9; TMSB4X; GAL; FXYD6; ANK3; KCNIP2; JPH4; AKAP6; CALM1; VAMP2; NEDD4L; FXYD7 |
| GO:0031644 | regulation of neurological system process | 4.25 | 9.50e-04 | 2.17e-02 | 7/30 | AGT; NRXN1; AKAP9; CBLN1; EIF4A3; BAIAP2; CELF4 |
| GO:0099565 | chemical synaptic transmission, postsynaptic | 4.16 | 4.78e-04 | 1.35e-02 | 8/35 | NRXN1; CHRNA1; SNCA; MEF2C; CBLN1; EIF4A3; BAIAP2; CELF4 |
| GO:0034765 | regulation of ion transmembrane transport | 3.74 | 1.13e-08 | 4.14e-06 | 24/117 | SEPN1; ATP1B1; AGT; FKBP1B; SCN3A; SCN9A; SNCA; MEF2C; SRI; AKAP9; TMSB4X; CHD7; GAL; FXYD6; ANK3; KCNIP2; CD63; JPH4; AKAP6; CALM1; VAMP2; NEDD4L; CACNA1A; FXYD7 |
| GO:0006813 | potassium ion transport | 3.49 | 8.55e-04 | 2.09e-02 | 9/47 | ATP1B1; AKAP9; GAL; ANK3; KCNIP2; CD63; AKAP6; VAMP2; NEDD4L |
| GO:0003012 | muscle system process | 3.42 | 4.86e-07 | 7.81e-05 | 21/112 | LMNA; ATP1B1; AGT; FKBP1B; CHRNA1; MYL1; IGFBP5; MEF2C; HDAC2; CAMK2B; SRI; CALD1; KCNIP2; MYL6; AKAP6; CALM1; ACTC1; SSTR2; MYL12A; NEDD4L; PDE9A |
| GO:0034764 | positive regulation of transmembrane transport | 3.31 | 2.01e-04 | 6.71e-03 | 12/66 | ATP1B1; AGT; CAPN10; SNCA; SRI; AKAP9; TMSB4X; GAL; ANK3; KCNIP2; AKAP6; CALM1 |
| GO:0042391 | regulation of membrane potential | 3.19 | 1.60e-06 | 1.67e-04 | 21/120 | MLLT11; ATP1B1; FKBP1B; NRXN1; SCN3A; SCN9A; CHRNA1; SNCA; MEF2C; SRI; AKAP9; ANK3; KCNIP2; AKAP6; CBLN1; MAPT; EIF4A3; BAIAP2; CELF4; NEDD4L; CACNA1A |
| GO:0072507 | divalent inorganic cation homeostasis | 3.18 | 9.29e-06 | 6.20e-04 | 18/103 | SEPN1; SV2A; ATP1B1; AGT; FKBP1B; SNCA; SRI; CHD7; GNG3; ANK3; ATP2B1; HSP90B1; JPH4; AKAP6; CALM1; CALR; CACNA1A; SMDT1 |
| GO:0001764 | neuron migration | 3.18 | 5.33e-04 | 1.40e-02 | 11/63 | DNER; IGSF10; GPM6A; MAP1B; MEF2C; TUBB2B; MDGA1; CAMK2B; AUTS2; DCX; NRG1 |
| GO:0019932 | second-messenger-mediated signaling | 3.17 | 3.14e-05 | 1.77e-03 | 16/92 | SEPN1; ATP1B1; AGT; FKBP1B; NEUROD1; GNAI2; SRI; NRG1; GAL; JPH4; AKAP6; CALM1; MAPT; HOMER3; APLP1; PDE9A |
| GO:0070972 | protein localization to endoplasmic reticulum | 3.06 | 5.45e-06 | 4.00e-04 | 20/119 | RPS27; RPS7; RPL37A; RPL29; RPS3A; RPL37; RPS14; RPS18; RPL10A; RPL39; RPS20; RPL12; RPLP2; RPL41; RPLP1; RPS15A; RPL23; RPS21; RPS11; RPL28 |
| GO:0021953 | central nervous system neuron differentiation | 3.04 | 2.38e-03 | 4.26e-02 | 9/54 | DRAXIN; LMO4; MAP2; SOX4; MDGA1; FZD1; MYCBP2; CBLN1; MAPT |
| GO:0031345 | negative regulation of cell projection organization | 3.04 | 8.02e-04 | 2.03e-02 | 11/66 | DRAXIN; MAP2; ITM2C; PTPRG; NR2F1; DPYSL3; HDAC2; STMN2; B2M; EFNB3; NGFR |
| GO:0099177 | regulation of trans-synaptic signaling | 3.00 | 5.32e-07 | 7.81e-05 | 25/152 | AGT; NRXN1; GNAI2; NSG1; SNCA; MEF2C; CPLX2; TUBB2B; GRM4; AKAP12; CAMK2B; AKAP9; PCDH17; JPH4; CALM1; RAB26; CBLN1; EFNB3; VAMP2; MAPT; EIF4A3; BAIAP2; CELF4; CACNA1A; RAB3A |
| GO:0043270 | positive regulation of ion transport | 2.95 | 1.04e-03 | 2.18e-02 | 11/68 | ATP1B1; AGT; SNCA; SRI; AKAP9; TMSB4X; GAL; ANK3; KCNIP2; AKAP6; CALM1 |
| GO:0060537 | muscle tissue development | 2.88 | 3.91e-05 | 1.91e-03 | 18/114 | SEPN1; LMNA; AGT; ZBTB18; CHRNA1; IGFBP5; DNER; GPX1; MEF2C; FAM65B; UQCC2; MEOX2; NRG1; CHD7; MYL6; AKAP6; ACTC1; CALR |
| GO:0003013 | circulatory system process | 2.88 | 1.74e-04 | 6.08e-03 | 15/95 | ATP1B1; AGT; FKBP1B; MYL1; GPX1; GNAI2; MEOX2; SRI; AKAP9; CHD7; KCNIP2; ATP2B1; CALM1; ACTC1; NEDD4L |
| GO:0007517 | muscle organ development | 2.87 | 6.70e-05 | 2.89e-03 | 17/108 | SEPN1; ZBTB18; CHRNA1; DNER; GPX1; MEF2C; FAM65B; UQCC2; MEOX2; SRI; FZD1; NRG1; CHD7; MYL6; AKAP6; ACTC1; TCF12 |
| GO:0072511 | divalent inorganic cation transport | 2.72 | 1.34e-04 | 5.18e-03 | 17/114 | SEPN1; ATP1B1; AGT; FKBP1B; GNAI2; SNCA; GPM6A; CAMK2B; SRI; CHD7; ATP2B1; JPH4; AKAP6; CALM1; CACNA1A; HOMER3; SMDT1 |
| GO:0097485 | neuron projection guidance | 2.71 | 5.31e-04 | 1.40e-02 | 14/94 | DRAXIN; LHX9; NRXN1; ALCAM; GAP43; SPON2; PDLIM7; TUBB2B; EZR; DPYSL2; SPTAN1; FEZ1; MYCBP2; EFNB3 |
| GO:0051235 | maintenance of location | 2.70 | 2.28e-04 | 7.28e-03 | 16/108 | SEPN1; FKBP1B; SNCA; MDFI; EZR; SRI; AKAP9; TMSB4X; CHD7; FTH1; ANK3; HSP90B1; JPH4; AKAP6; CALM1; CALR |
| GO:0009914 | hormone transport | 2.69 | 9.04e-04 | 2.14e-02 | 13/88 | MPC2; AGT; FKBP1B; NEUROD1; CAPN10; SOX4; UQCC2; SRI; CHD7; GAL; VAMP2; CACNA1A; SELM |
| GO:0010975 | regulation of neuron projection development | 2.63 | 1.10e-06 | 1.35e-04 | 29/201 | DRAXIN; PTPRF; CRABP2; AGT; FKBP1B; MAP2; ITM2C; PTPRG; TNIK; MAP1B; MEF2C; NR2F1; DPYSL3; TUBB2B; HDAC2; CAMK2B; FZD1; NEFL; DPYSL2; STMN2; MYCBP2; B2M; SERPINF1; EFNB3; MAPT; NGFR; BAIAP2; NEDD4L; PREX1 |
| GO:0050808 | synapse organization | 2.62 | 3.78e-05 | 1.91e-03 | 21/146 | PTPRF; NRXN1; CHRNA1; DNER; GAP43; SNCA; GPM6A; MEF2C; TUBB; MDGA1; ACTB; CAMK2B; FZD1; NEFL; NRG1; ANK3; PCDH17; CBLN1; MAPT; BAIAP2; RAB3A |
| GO:0090130 | tissue migration | 2.60 | 1.25e-03 | 2.54e-02 | 13/91 | RAB13; AGT; IQSEC1; GPX1; PTPRG; MEF2C; MEOX2; TMSB4X; GLIPR2; ATP5B; ACTC1; SERPINF1; CALR |
| GO:0061564 | axon development | 2.55 | 2.04e-06 | 1.88e-04 | 29/207 | DRAXIN; STMN1; PTPRF; CRABP2; LHX9; FKBP1B; NRXN1; MAP2; ALCAM; GAP43; SPON2; MAP1B; NREP; PDLIM7; TUBB2B; EZR; AUTS2; NEFM; NEFL; DPYSL2; SPTAN1; FEZ1; ANK3; MYCBP2; EFNB3; MAPT; NGFR; BAIAP2; RAB3A |
| GO:0023061 | signal release | 2.52 | 4.54e-05 | 2.08e-03 | 22/159 | WLS; SV2A; MPC2; AGT; FKBP1B; NRXN1; NEUROD1; CAPN10; SNCA; MEF2C; CPLX2; SOX4; UQCC2; GRM4; SRI; CHD7; GAL; CALM1; VAMP2; CACNA1A; RAB3A; SELM |
| GO:0090150 | establishment of protein localization to membrane | 2.50 | 4.68e-06 | 3.81e-04 | 28/204 | RPS27; ATP1B1; RPS7; REEP1; RPL37A; RPL29; NSG1; RPS3A; RPL37; RPS14; RPS18; RPL10A; RPL39; RPS20; RPL12; RPLP2; ANK3; RPL41; CALM1; RPLP1; RAB26; RPS15A; VAMP2; RPL23; RAB31; RPS21; RPS11; RPL28 |
| GO:0006413 | translational initiation | 2.44 | 1.12e-04 | 4.57e-03 | 21/157 | RPS27; RPS7; RPL37A; EIF1B; RPL29; RPS3A; RPL37; RPS14; RPS18; RPL10A; RPL39; RPS20; RPL12; RPLP2; RPL41; RPLP1; RPS15A; RPL23; RPS21; RPS11; RPL28 |
| GO:0051961 | negative regulation of nervous system development | 2.38 | 1.95e-03 | 3.67e-02 | 14/107 | DRAXIN; PBX1; MAP2; ITM2C; PTPRG; NR2F1; DPYSL3; HDAC2; NRG1; STMN2; B2M; EFNB3; NGFR; CALR |
| GO:0010721 | negative regulation of cell development | 2.36 | 1.53e-03 | 2.96e-02 | 15/116 | DRAXIN; PBX1; MAP2; ITM2C; PTPRG; NR2F1; DPYSL3; HDAC2; NRG1; STMN2; GAL; B2M; EFNB3; NGFR; CALR |
| GO:0050769 | positive regulation of neurogenesis | 2.29 | 2.71e-04 | 8.29e-03 | 21/167 | CRABP2; AGT; FKBP1B; NEUROD1; MAP1B; MEF2C; DPYSL3; TUBB2B; HDAC2; CAMK2B; FZD1; NEFL; STMN2; TCF12; SERPINF1; MAPT; NGFR; BAIAP2; TCF4; NEDD4L; PCP4 |
| GO:0031346 | positive regulation of cell projection organization | 2.18 | 1.00e-03 | 2.17e-02 | 19/159 | CRABP2; AGT; FKBP1B; CDC42EP3; GPM6A; MAP1B; DPYSL3; TUBB2B; FAM65B; CAMK2B; AUTS2; FZD1; NEFL; STMN2; SERPINF1; MAPT; NGFR; BAIAP2; NEDD4L |
| GO:0022604 | regulation of cell morphogenesis | 2.08 | 1.01e-03 | 2.17e-02 | 21/184 | DRAXIN; EPB41; RHOC; CRABP2; CDC42EP3; MAP2; TNIK; MAP1B; TUBB2B; EZR; CAMK2B; NEFL; DPYSL2; MYCBP2; EFNB3; MAPT; NGFR; BAIAP2; NEDD4L; PREX1; CALR |
Factor 5 : 18 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 6.14 | 0.00e+00 | 0.00e+00 | 44/119 | RPS8; RPL5; RPS27; RPL31; RPL32; RPL15; RPL14; RPL24; SEC62; RPL35A; RPL34; RPS3A; RPS23; RPS14; RPS18; RPL10A; RPS4X; RPL36A; RPL10; RPS20; RPL7; RPL30; RPL12; RPL7A; RPLP2; SPCS2; RPS3; RPS25; RPS24; RPL6; RPLP0; RPS29; RPS27L; RPL4; RPLP1; RPS15A; RPL26; RPL23; RPS21; RPS28; RPL18; RPS9; RPL28; RPS5; RPL3 |
| GO:0006413 | translational initiation | 5.61 | 0.00e+00 | 0.00e+00 | 53/157 | RPS8; RPL5; RPS27; DDX1; RPL31; RPL32; RPL15; RPL14; RPL24; EIF4A2; RPL35A; RPL34; RPS3A; RPS23; RPS14; RPS18; RPL10A; HSPB1; RPS4X; RPL36A; RPL10; RPS20; RPL7; RPL30; PABPC1; EIF3E; RPL12; RPL7A; RPLP2; POLR2G; PPP1CA; RPS3; RPS25; RPS24; RPL6; RPLP0; RPS29; EIF5; EIF3J; RPS27L; RPL4; RPLP1; RPS15A; EIF4A1; RPL26; RPL23; EIF6; RPS21; RPS28; RPL18; RPS9; RPL28; RPS5; RPL3 |
| GO:0002181 | cytoplasmic translation | 4.78 | 7.27e-10 | 8.89e-08 | 21/73 | RPL31; RPL32; RPL15; RPL24; RPL35A; RPS23; RPL10A; RPL36A; RPL30; EIF3E; RPL6; RPLP0; RPS29; EIF5; EIF3J; RPLP1; RPL26; RPS21; EEF2; RPS28; RPL18 |
| GO:0010257 | NADH dehydrogenase complex assembly | 4.71 | 4.13e-08 | 3.03e-06 | 17/60 | NDUFB4; NDUFC1; NDUFS6; NDUFS4; NDUFB11; NDUFB9; NDUFA8; NDUFS3; NDUFS8; NDUFC2; NDUFB8; NDUFA12; NDUFB10; NDUFAB1; NDUFV2; NDUFS7; NDUFA6 |
| GO:0090150 | establishment of protein localization to membrane | 4.15 | 0.00e+00 | 0.00e+00 | 51/204 | RPS8; RPL5; RPS27; RPL31; RPL32; RPL15; RPL14; RPL24; SEC62; RPL35A; RPL34; RPS3A; RPS23; RPS14; GNB2L1; RPS18; RPL10A; CD24; GOPC; RPS4X; RPL36A; RPL10; RPS20; RPL7; RPL30; RPL12; RPL7A; RPLP2; RPS3; RPS25; RPS24; RPL6; RPLP0; RPS29; HSP90AA1; RPS27L; RPL4; RPLP1; RAB26; RPS15A; VPS35; RPL26; RPL23; NMT1; RPS21; RPS28; PDCD5; RPL18; RPS9; RPL28; RPS5; RPL3 |
| GO:0006401 | RNA catabolic process | 3.94 | 0.00e+00 | 0.00e+00 | 56/236 | RPS8; MAGOH; RPL5; RBM8A; RPS27; RPL31; RPL32; LSM3; RPL15; RPL14; RPL24; RPL35A; RPL34; RPS3A; RPS23; HNRNPA0; RPS14; RPS18; RPL10A; HSPB1; RPS4X; RPL36A; UPF3B; RPL10; RPS20; RPL7; RPL30; PABPC1; EIF3E; EXOSC4; RPL12; RPL7A; RPLP2; POLR2G; RPS3; RPS25; RPS24; EXOSC1; RPL6; RPLP0; RPS29; SLIRP; RPS27L; RPL4; RPLP1; RPS15A; RPL26; RPL23; RPS21; RPS28; HNRNPM; C19orf43; RPL18; RPS9; RPL28; RPS5; RPL3 |
| GO:0006414 | translational elongation | 3.81 | 3.20e-09 | 3.35e-07 | 25/109 | AURKAIP1; MRPS5; MRPL47; MRPS18C; MRPL22; GNB2L1; MRPS18A; EEF1A1; MRPL18; MRPL32; MRPL13; RPLP2; MRPL21; MRPL51; RPLP1; MRPL46; MRPL28; MRPL27; MRPS23; ICT1; MRPS7; MRPS26; EEF2; GADD45GIP1; MRPL40 |
| GO:1902600 | proton transmembrane transport | 3.67 | 2.05e-06 | 1.16e-04 | 17/77 | ATP6V0B; COX5B; ATP5G3; COX7C; COX7A2; TMSB4X; COX7B; COX6C; COX8A; ATP5B; COX6A1; ATP5E; NDUFS7; COX6B1; ATP5O; MT-CO1; MT-CYB |
| GO:0006605 | protein targeting | 3.37 | 4.44e-16 | 6.52e-14 | 53/261 | ATPIF1; RPS8; RPL5; RPS27; RPL31; RPL32; RPL15; RPL14; RPL24; SEC62; RPL35A; RPL34; RPS3A; RPS23; RPS14; RPS18; RPL10A; SLC25A6; RPS4X; RPL36A; RPL10; RPS20; RPL7; RPL30; RPL12; RPL7A; PMPCA; RPLP2; AIP; SPCS2; RPS3; RPS25; RPS24; RPL6; RPLP0; RPS29; HSP90AA1; RPS27L; RPL4; RPLP1; RPS15A; RPL26; RPL23; RPS21; RPS28; UBL5; PDCD5; ECH1; RPL18; RPS9; RPL28; RPS5; UBE2L3; RPL3 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 3.25 | 1.19e-05 | 5.58e-04 | 17/87 | NDUFB4; NDUFC1; NDUFS6; NDUFS4; NDUFB11; NDUFB9; NDUFA8; NDUFS3; NDUFS8; NDUFC2; NDUFB8; NDUFA12; NDUFB10; NDUFAB1; NDUFV2; NDUFS7; NDUFA6 |
| GO:0140053 | mitochondrial gene expression | 3.01 | 4.00e-06 | 2.09e-04 | 21/116 | AURKAIP1; MRPS5; MRPL47; MRPS18C; MRPL22; MRPS18A; MRPL18; MALSU1; MRPL32; MRPL13; MRPL21; MRPL51; MRPL46; MRPL28; MRPL27; MRPS23; ICT1; MRPS7; MRPS26; GADD45GIP1; MRPL40 |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.90 | 1.53e-08 | 1.40e-06 | 33/189 | ATPIF1; PARP1; COX5B; ATP5G3; NDUFB4; NDUFC1; NDUFS6; NDUFS4; COX7C; COX7A2; TMSB4X; NDUFB9; NDUFA8; NDUFS3; NDUFS8; NDUFC2; NDUFB8; TPI1; ATP5B; NDUFA12; COX6A1; RAN; NDUFB10; NDUFAB1; NDUFV2; EIF6; ATP5E; NDUFS7; UQCRFS1; NDUFA6; ATP5O; MT-CO1; MT-CYB |
| GO:0009123 | nucleoside monophosphate metabolic process | 2.73 | 1.18e-07 | 7.86e-06 | 32/195 | ATPIF1; PARP1; COX5B; ATP5G3; NDUFB4; NDUFC1; NDUFS6; NDUFS4; COX7C; COX7A2; TMSB4X; NDUFB9; NDUFA8; NDUFS3; NDUFS8; NDUFC2; NDUFB8; TPI1; ATP5B; NDUFA12; COX6A1; NDUFB10; NDUFAB1; NDUFV2; EIF6; ATP5E; NDUFS7; UQCRFS1; NDUFA6; ATP5O; MT-CO1; MT-CYB |
| GO:0071826 | ribonucleoprotein complex subunit organization | 2.42 | 1.22e-05 | 5.58e-04 | 27/185 | RPL5; RPS27; DDX1; LSM3; RPL24; YTHDC1; RPS23; RPS14; MRPL22; SNRPC; HSP90AB1; RPL10; EIF3E; RPL12; RPL6; RPLP0; HSP90AA1; EIF5; EIF3J; RPS27L; MRPS7; SNRPB; EIF6; PRPF6; RPS28; RPS5; RPL3 |
| GO:0006091 | generation of precursor metabolites and energy | 2.41 | 4.42e-07 | 2.70e-05 | 36/248 | SDHB; ATPIF1; COX5B; NDUFB4; NDUFC1; NDUFS6; NDUFS4; COX7C; COX7A2; COX7B; COX6C; NDUFB9; NDUFA8; NDUFS3; COX8A; PPP1CA; NDUFS8; NDUFC2; NDUFB8; TPI1; ATP5B; NDUFA12; COX6A1; VIMP; NDUFB10; NDUFAB1; NDUFV2; EIF6; ATP5E; NDUFS7; UQCRFS1; COX6B1; NDUFA6; ATP5O; MT-CO1; MT-CYB |
| GO:0022613 | ribonucleoprotein complex biogenesis | 2.32 | 3.24e-08 | 2.65e-06 | 46/330 | RPS8; RPL5; RPS27; DDX1; CEBPZ; LSM3; RPL14; RPL24; RPL35A; YTHDC1; RPS23; RPS14; MRPL22; SNRPC; RPL10A; HSP90AB1; MALSU1; RPL10; RPL7; DCAF13; EIF3E; EXOSC4; RPL12; RPL7A; TRMT112; RPS24; EXOSC1; NPM3; RPL6; RPLP0; RAN; HSP90AA1; EIF5; EIF3J; RPS27L; RPL26; DDX52; MRPS7; SNRPB; EIF6; RPS21; PRPF6; RPS28; RPS9; RPS5; RPL3 |
| GO:0009259 | ribonucleotide metabolic process | 2.13 | 2.09e-05 | 9.01e-04 | 33/257 | ATPIF1; PARP1; COX5B; ATP5G3; NDUFB4; NDUFC1; NDUFS6; NDUFS4; COX7C; COX7A2; TMSB4X; NDUFB9; NDUFA8; NDUFS3; NDUFS8; NDUFC2; NDUFB8; TPI1; ATP5B; NDUFA12; COX6A1; RAN; NDUFB10; NDUFAB1; NDUFV2; EIF6; ATP5E; NDUFS7; UQCRFS1; NDUFA6; ATP5O; MT-CO1; MT-CYB |
| GO:0016072 | rRNA metabolic process | 2.04 | 1.26e-03 | 4.64e-02 | 21/171 | RPS8; RPL5; RPS27; RPL14; RPL35A; RPS14; RPL10A; RPL7; DCAF13; EXOSC4; RPL7A; TRMT112; RPS24; EXOSC1; NPM3; RPS27L; RPL26; DDX52; RPS21; RPS28; C19orf43; RPS9 |
Factor 6 : 12 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0051131 | chaperone-mediated protein complex assembly | 12.30 | 2.04e-04 | 0.018682 | 4/11 | HSPD1; HSP90AB1; CCT2; HSP90AA1 |
| GO:0070670 | response to interleukin-4 | 12.30 | 2.04e-04 | 0.018682 | 4/11 | HSP90AB1; HSPA5; TUBA1B; XBP1 |
| GO:0001818 | negative regulation of cytokine production | 5.05 | 3.92e-04 | 0.028773 | 7/47 | HSP90AB1; TMSB4X; LAPTM4B; UBE2L6; HMGB1; C1QBP; PIN1 |
| GO:0071897 | DNA biosynthetic process | 4.44 | 2.93e-05 | 0.009816 | 11/84 | CENPF; HNRNPD; HSP90AB1; HNRNPA2B1; PPIA; DKC1; UBE2L6; HNRNPA1; CCT2; HNRNPC; HSP90AA1 |
| GO:0006457 | protein folding | 4.18 | 4.28e-07 | 0.000314 | 17/138 | PDIA6; FKBP1B; HSPD1; HSPE1; CANX; HSP90AB1; PPIA; PRDX4; HSPA5; HSPA8; PFDN5; CCT2; HSP90B1; HSP90AA1; PDIA3; PPIB; CALR |
| GO:0032200 | telomere organization | 4.13 | 2.83e-04 | 0.023113 | 9/74 | GNL3; HNRNPD; HSP90AB1; HNRNPA2B1; DKC1; HNRNPA1; CCT2; HNRNPC; HSP90AA1 |
| GO:0006338 | chromatin remodeling | 3.59 | 8.02e-04 | 0.049030 | 9/85 | NASP; ANP32E; PTMA; HMGB2; NPM1; HELLS; NPM3; HMGB1; HNRNPC |
| GO:0006403 | RNA localization | 3.26 | 4.53e-05 | 0.009816 | 15/156 | SFPQ; SRSF7; SSB; HNRNPA3; NOP58; NPM1; SRSF3; HNRNPA2B1; DKC1; HNRNPA1; PA2G4; CCT2; RAN; SRSF2; FBL |
| GO:1903311 | regulation of mRNA metabolic process | 3.07 | 5.35e-05 | 0.009816 | 16/177 | SERBP1; PRDX6; SRSF7; PCBP4; HNRNPD; NPM1; SRSF3; HNRNPA2B1; PABPC1; HSPA8; HNRNPA1; HNRNPC; RBM25; C1QBP; SRSF2; HNRNPM |
| GO:0060249 | anatomical structure homeostasis | 2.95 | 6.83e-04 | 0.045598 | 12/138 | PRDX1; NEUROD1; GNL3; HNRNPD; HSP90AB1; HNRNPA2B1; DKC1; HNRNPA1; CCT2; HNRNPC; HSP90AA1; ACTG1 |
| GO:0006401 | RNA catabolic process | 2.59 | 1.67e-04 | 0.018682 | 18/236 | SERBP1; SSB; PCBP4; HNRNPD; RPL37; NPM1; RPS4X; RPL39; RPL10; DKC1; PABPC1; RPS13; HSPA8; HNRNPC; HNRNPM; C19orf43; RPS9; RPL28 |
| GO:0010608 | posttranscriptional regulation of gene expression | 2.49 | 7.97e-05 | 0.011700 | 21/286 | SERBP1; EPRS; EIF5B; NCL; PCBP4; HNRNPD; NPM1; SOX4; HNRNPA2B1; RPS4X; RPL10; DKC1; PABPC1; HSPA8; PA2G4; RAN; HNRNPC; C1QBP; HNRNPM; CALR; RPS9 |
Factor 7 : 19 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0010257 | NADH dehydrogenase complex assembly | 6.83 | 0.00e+00 | 0.00e+00 | 30/60 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFC1; NDUFS6; NDUFA2; NDUFB2; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFA8; NDUFS8; NDUFC2; NDUFB8; NDUFA12; NDUFB1; NDUFB10; NDUFAB1; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND3; MT-ND4 |
| GO:0070972 | protein localization to endoplasmic reticulum | 6.54 | 0.00e+00 | 0.00e+00 | 57/119 | RPL11; RPS8; RPL5; RPS27; RTN4; RPS27A; RPL37A; RPL32; RPL15; RPSA; SPCS1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SEC61G; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; SEC61B; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 5.97 | 0.00e+00 | 0.00e+00 | 38/87 | NDUFS5; COX20; NDUFB3; NDUFAF3; COX17; NDUFB4; NDUFC1; NDUFS6; NDUFA2; UQCC2; NDUFB2; NDUFB11; NDUFA1; UQCRB; NDUFB9; TMEM261; NDUFB6; NDUFA8; NDUFS8; NDUFC2; NDUFB8; COX14; NDUFA12; NDUFB1; NDUFB10; NDUFAB1; COA3; NDUFS7; NDUFA11; PET100; NDUFA7; NDUFB7; NDUFA13; NDUFA3; UQCR10; NDUFA6; MT-ND3; MT-ND4 |
| GO:1902600 | proton transmembrane transport | 5.68 | 0.00e+00 | 0.00e+00 | 32/77 | PARK7; ATP6V0B; COX5B; ATP5G3; COX17; ATP5I; COX7C; COX7A2; NDUFA4; ATP5J2; ATP6V1F; TMSB4X; COX7B; COX6C; ATP6V1G1; COX8A; ATP5L; ATP5G2; SLC25A3; COX6A1; COX5A; COX4I1; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; COX6B1; ATP5J; ATP5O; MT-CO1; MT-ND4 |
| GO:0006413 | translational initiation | 5.48 | 0.00e+00 | 0.00e+00 | 63/157 | RPL11; RPS8; RPL5; RPS27; DDX1; RPS27A; RPL37A; RPL32; RPL15; RPSA; EIF4A2; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; MCTS1; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL35; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; EIF3J; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; EIF1; RPL18A; EIF3K; RPS16; RPS19; RPL18; RPL13A; RPS11; EIF3L; RPL3 |
| GO:0002181 | cytoplasmic translation | 5.24 | 3.40e-14 | 1.92e-12 | 28/73 | RPL11; RPL32; RPL15; TMA7; RPL9; RPS23; RPL10A; RPL39; MCTS1; EIF4EBP1; RPL30; EIF3E; EIF3H; EIF3F; RPS26; RPL41; RPL6; RPLP0; RPS29; EIF3J; RPL26; RPL19; EEF2; RPL18A; EIF3K; RPL18; RPL13A; EIF3L |
| GO:0090150 | establishment of protein localization to membrane | 4.35 | 0.00e+00 | 0.00e+00 | 65/204 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPL10A; CD24; RPS12; FIS1; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPS6; SEC61B; RPL35; RPL7A; RPL27A; RPS13; TIMM10; RPS3; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; CALM1; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; VAMP2; RPL26; RPL23A; RPL23; RPL19; ROMO1; TIMM13; RPL18A; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | 4.27 | 5.84e-05 | 2.38e-03 | 10/32 | ATP6V0B; ATP5G3; ATP6V1F; ATP5L; ATP5G2; ATP5G1; ATP5H; ATP5A1; ATP5E; ATP5O |
| GO:0009141 | nucleoside triphosphate metabolic process | 4.19 | 0.00e+00 | 0.00e+00 | 58/189 | PARK7; ATPIF1; NDUFS5; UQCRH; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; UQCRB; NDUFB9; NDUFB6; NDUFA8; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5G2; NDUFA12; COX6A1; RAN; NDUFB1; COX5A; NDUFB10; NDUFAB1; DCTPP1; COX4I1; ATP5G1; NME1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| GO:0009123 | nucleoside monophosphate metabolic process | 3.92 | 0.00e+00 | 0.00e+00 | 56/195 | PARK7; ATPIF1; NDUFS5; UQCRH; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; HDDC2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; UQCRB; NDUFB9; NDUFB6; NDUFA8; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5G2; NDUFA12; COX6A1; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| GO:0017004 | cytochrome complex assembly | 3.90 | 6.45e-04 | 2.49e-02 | 8/28 | COX20; COX17; UQCC2; UQCRB; COX14; COA3; PET100; UQCR10 |
| GO:0006605 | protein targeting | 3.72 | 0.00e+00 | 0.00e+00 | 71/261 | RPL11; ATPIF1; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; SPCS1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; TOMM7; SEC61G; FIS1; SLC25A6; TIMM17B; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPS6; SEC61B; RPL35; RPL7A; RPL27A; RPS13; IMMP1L; TIMM10; RPS3; TIMM8B; RPS26; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; ROMO1; TIMM13; UBL5; RPL18A; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| GO:0006401 | RNA catabolic process | 3.59 | 0.00e+00 | 0.00e+00 | 62/236 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; LSM3; RPL15; RPSA; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; LSM5; LINC01420; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; EIF3E; RPS6; RPL35; RPL7A; RPL27A; RPS13; RNASEH2C; RPS3; RPS26; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; SLIRP; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; LSM7; RPL18A; LSM4; RPS16; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| GO:0006091 | generation of precursor metabolites and energy | 3.47 | 0.00e+00 | 0.00e+00 | 63/248 | PARK7; SH3BGRL3; ATPIF1; NDUFS5; UQCRH; COX20; COX5B; NDUFB3; COX17; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; GLRX; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; COX7B; NDUFA1; UQCRB; COX6C; NDUFB9; NDUFB6; NDUFA8; COX8A; NDUFS8; NDUFC2; ATP5L; CISD1; NDUFB8; TPI1; BLOC1S1; NDUFA12; COX6A1; HMGB1; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5H; ATP5A1; ATP5E; NDUFS7; UQCR11; NDUFA7; NDUFB7; COX6B1; ETFB; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| GO:0007006 | mitochondrial membrane organization | 3.34 | 5.54e-07 | 2.90e-05 | 21/86 | ATPIF1; ATP5G3; ATP5I; TMEM14A; DYNLT1; ATP5J2; YWHAZ; TIMM10; ATP5L; ATP5G2; CALM1; ATP5G1; ATP5H; ATP5A1; ROMO1; ATP5E; TIMM13; NDUFA13; PDCD5; ATP5J; ATP5O |
| GO:0009259 | ribonucleotide metabolic process | 3.14 | 2.22e-16 | 1.36e-14 | 59/257 | PARK7; ATPIF1; NDUFS5; UQCRH; COX5B; DBI; ATP5G3; NDUFB3; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; HINT1; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; UQCRB; NDUFB9; NDUFB6; NDUFA8; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5G2; NDUFA12; COX6A1; RAN; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5G1; NME1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| GO:0006839 | mitochondrial transport | 2.42 | 1.73e-05 | 7.46e-04 | 26/147 | ATPIF1; COX5B; ATP5I; TMEM14A; DYNLT1; TOMM7; ATP5J2; FIS1; SLC25A6; TIMM17B; YWHAZ; IMMP1L; TIMM10; TIMM8B; ATP5L; ATP5H; ATP5A1; ROMO1; ATP5E; TIMM13; UBL5; NDUFA13; PDCD5; SMDT1; ATP5J; ATP5O |
| GO:0070585 | protein localization to mitochondrion | 2.35 | 1.00e-03 | 3.68e-02 | 16/93 | ATPIF1; TOMM7; FIS1; SLC25A6; TIMM17B; YWHAZ; IMMP1L; TIMM10; TIMM8B; DDIT3; CALM1; ROMO1; TIMM13; UBL5; NDUFA13; PDCD5 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 2.29 | 8.39e-06 | 4.11e-04 | 31/185 | MRPL20; RPL11; RPL5; RPS27; SNRPE; DDX1; SNRPG; LSM3; RPSA; RPS23; RPS14; LINC01420; MCTS1; RPL10; EIF3E; EIF3H; EIF3F; SNRPF; RPL6; RPLP0; EIF3J; RPL23A; SNRPD1; LSM4; EIF3K; RPS19; SNRPD2; RPL13A; EIF3L; RPL3; SNU13 |
Factor 8 : 4 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0050954 | sensory perception of mechanical stimulus | 4.63 | 2.45e-04 | 0.04498 | 8/39 | CASP3; FAM65B; POU3F4; NAV2; COL2A1; CACNB3; SRRM4; SIX1 |
| GO:0070972 | protein localization to endoplasmic reticulum | 3.41 | 3.72e-06 | 0.00273 | 18/119 | RPL32; RPSA; RPL34; RPS18; RPL36A; RPL10; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; OS9; RPL6; RPLP0; RPS17; RPL26; RPL23 |
| GO:0006413 | translational initiation | 2.59 | 1.70e-04 | 0.04158 | 18/157 | RPL32; RPSA; RPL34; RPS18; RPL36A; RPL10; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; DENR; RPS17; RPL26; RPL23 |
| GO:0090150 | establishment of protein localization to membrane | 2.43 | 8.08e-05 | 0.02965 | 22/204 | RPL32; RPSA; NSG1; RPL34; GNB2L1; RPS18; RPL36A; RPL10; RPL12; RPL27A; RPS13; RPS3; CACNB3; RPS26; RPL41; RPL6; RPLP0; CALM1; RPS17; VAMP2; RPL26; RPL23 |
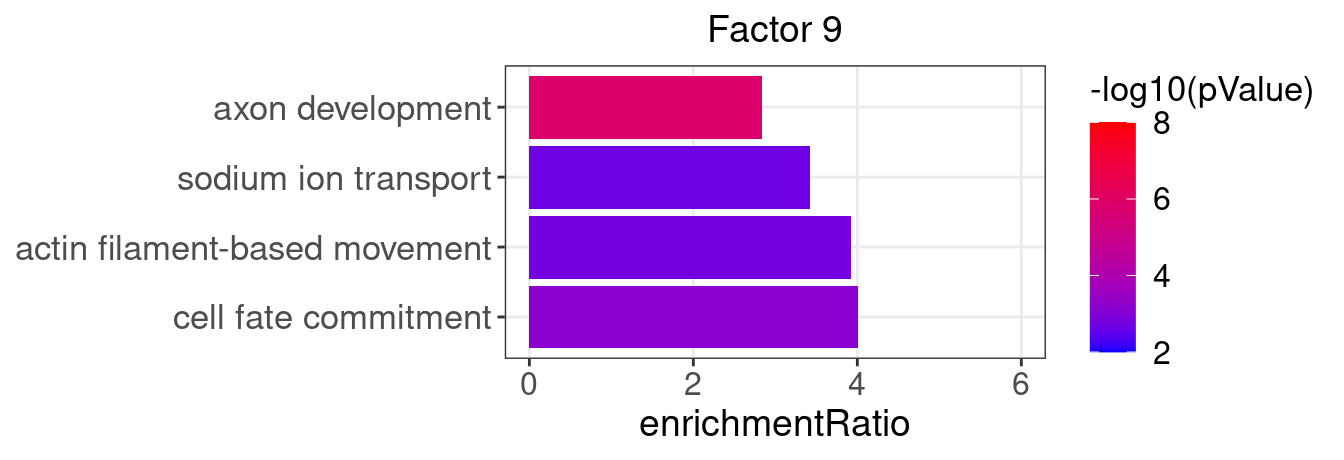
Factor 9 : 35 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0014075 | response to amine | 9.41 | 5.44e-04 | 0.028630 | 4/10 | HDAC2; ASIC1; CALM1; NME1 |
| GO:0048645 | animal organ formation | 8.56 | 8.26e-04 | 0.031152 | 4/11 | GNG5; CTNNB1; ROBO1; SIX1 |
| GO:0007586 | digestion | 7.24 | 1.67e-03 | 0.043405 | 4/13 | NEUROD1; SST; NEUROG1; LIMA1 |
| GO:0010644 | cell communication by electrical coupling | 7.24 | 1.67e-03 | 0.043405 | 4/13 | RYR2; CAMK2D; ANK3; CALM1 |
| GO:0035270 | endocrine system development | 4.41 | 1.96e-03 | 0.044935 | 6/32 | PBX1; SIX3; NEUROD1; CDKN1C; SIX1; ONECUT2 |
| GO:0035637 | multicellular organismal signaling | 4.38 | 3.73e-04 | 0.028630 | 8/43 | RYR2; SCN3A; CHRNA1; ANK2; CAMK2D; ANK3; ATP2B1; CALM1 |
| GO:0014074 | response to purine-containing compound | 4.22 | 1.09e-03 | 0.036375 | 7/39 | JUN; RYR2; IGFBP5; SPARC; HDAC2; AKAP6; NME1 |
| GO:0003007 | heart morphogenesis | 4.04 | 6.41e-05 | 0.015684 | 11/64 | JUN; GNG5; RYR2; ID2; CTNNB1; ROBO1; IFT57; COL4A3BP; OLFM1; SIX1; ACTC1 |
| GO:0045165 | cell fate commitment | 4.01 | 6.99e-04 | 0.030163 | 8/47 | ID2; NEUROD1; CTNNB1; GAP43; CASP3; NEUROG1; SIX1; ONECUT2 |
| GO:0008544 | epidermis development | 3.92 | 8.59e-05 | 0.015766 | 11/66 | POU3F1; CRABP2; PKP4; IGFBP5; SATB1; CTNNB1; CASP3; HDAC2; AGPAT2; GAL; TNFRSF19 |
| GO:0030048 | actin filament-based movement | 3.92 | 1.72e-03 | 0.043405 | 7/42 | RYR2; ANK2; CAMK2D; TPM2; MYL6; ACTC1; TNNC2 |
| GO:0003012 | muscle system process | 3.78 | 8.09e-07 | 0.000572 | 18/112 | RYR2; CHRNA1; IGFBP5; ANK2; CAMK2D; SORBS2; COL4A3BP; NEUROG1; HDAC2; MTPN; TPM2; VCL; MYL6; AKAP6; CALM1; ACTC1; SSTR2; TNNC2 |
| GO:0043588 | skin development | 3.72 | 5.79e-04 | 0.028630 | 9/57 | STMN1; POU3F1; PKP4; IGFBP5; CTNNB1; CASP3; HDAC2; GAL; TNFRSF19 |
| GO:0006814 | sodium ion transport | 3.42 | 2.02e-03 | 0.044935 | 8/55 | SCN3A; CAMK2D; GLRX; SLC17A6; FXYD6; ANK3; ASIC1; NKAIN4 |
| GO:0097305 | response to alcohol | 3.36 | 6.57e-04 | 0.030140 | 10/70 | EEF1B2; CTNNB1; SPARC; HDAC2; LANCL2; TNC; ATP2B1; CALM1; ACTC1; MT-ND4 |
| GO:0032409 | regulation of transporter activity | 3.20 | 5.54e-04 | 0.028630 | 11/81 | RYR2; ANK2; CAMK2D; GLRX; ACTB; TMSB4X; GAL; FXYD6; ANK3; AKAP6; CALM1 |
| GO:0060560 | developmental growth involved in morphogenesis | 3.17 | 3.33e-04 | 0.028630 | 12/89 | DRAXIN; CRABP2; ZEB2; CTNNB1; ALCAM; MAP1B; TNC; OLFM1; GAL; VCL; SIX1; SYT4 |
| GO:0048568 | embryonic organ development | 3.14 | 1.20e-04 | 0.017667 | 14/105 | MFAP2; NES; PBX1; RYR2; ID2; NCOA1; SIX3; NEUROD1; CTNNB1; IFT57; NEUROG1; CTHRC1; CDKN1C; SIX1 |
| GO:0043270 | positive regulation of ion transport | 3.11 | 2.11e-03 | 0.045645 | 9/68 | RYR2; ANK2; GLRX; TMSB4X; GAL; ANK3; AKAP6; CALM1; SYT4 |
| GO:0050673 | epithelial cell proliferation | 3.00 | 5.56e-04 | 0.028630 | 12/94 | JUN; ID2; IGFBP5; SCG2; CTNNB1; ROBO1; IFT57; SPARC; CDKN1C; SIX1; PGF; NME1 |
| GO:0097485 | neuron projection guidance | 3.00 | 5.56e-04 | 0.028630 | 12/94 | DRAXIN; SHC1; EFNA3; LHX9; CNTN2; ROBO1; ALCAM; GAP43; SPON2; PDLIM7; ETV1; NCAM1 |
| GO:0048880 | sensory system development | 2.88 | 8.14e-04 | 0.031152 | 12/98 | MFAP2; JUN; NES; SIX3; ZEB2; NEUROD1; CTNNB1; HDAC2; CDKN1C; ATP2B1; MAB21L1; CELF4 |
| GO:0051961 | negative regulation of nervous system development | 2.86 | 5.34e-04 | 0.028630 | 13/107 | DRAXIN; PBX1; CNTN2; ID2; SIX3; ITM2C; CTNNB1; NR2F1; DPYSL3; HDAC2; B2M; LINGO1; SYT4 |
| GO:0010959 | regulation of metal ion transport | 2.85 | 8.92e-04 | 0.031169 | 12/99 | RYR2; CTNNB1; ANK2; CAMK2D; GLRX; GAL; FXYD6; ANK3; AKAP6; CALM1; B2M; NKAIN4 |
| GO:0090130 | tissue migration | 2.84 | 1.49e-03 | 0.043405 | 11/91 | JUN; ZEB2; SCG2; IQSEC1; ROBO1; SPARC; MEOX2; TMSB4X; ACTC1; PFN1; MAPRE2 |
| GO:0061564 | axon development | 2.84 | 1.56e-06 | 0.000572 | 25/207 | DRAXIN; STMN1; JUN; S100A6; SHC1; EFNA3; CRABP2; LHX9; CNTN2; ZEB2; ROBO1; ALCAM; GAP43; SPON2; CASP3; MAP1B; PDLIM7; ETV1; TNC; OLFM1; NCAM1; ANK3; VCL; LINGO1; ADARB1 |
| GO:0010721 | negative regulation of cell development | 2.84 | 3.51e-04 | 0.028630 | 14/116 | DRAXIN; PBX1; CNTN2; ID2; SIX3; ITM2C; CTNNB1; NR2F1; DPYSL3; HDAC2; GAL; B2M; LINGO1; SYT4 |
| GO:0007517 | muscle organ development | 2.83 | 5.85e-04 | 0.028630 | 13/108 | RYR2; CHRNA1; CTNNB1; NEUROG1; ETV1; MEOX2; MTPN; MYL6; AKAP6; SIX1; ACTC1; TCF12; ADARB1 |
| GO:0007389 | pattern specification process | 2.57 | 2.25e-03 | 0.047172 | 12/110 | PBX1; SIX3; ZEB2; NEUROD1; CTNNB1; ROBO1; IFT57; NEUROG1; MEOX2; PBX3; EMX2; SIX1 |
| GO:0007015 | actin filament organization | 2.50 | 8.49e-04 | 0.031152 | 15/141 | STMN1; ARPC5; CDC42EP3; CD47; WDR1; SORBS2; DPYSL3; CAP2; MTPN; TMSB4X; TPM2; CFL1; LIMA1; ACTC1; PFN1 |
| GO:0030900 | forebrain development | 2.44 | 1.62e-03 | 0.043405 | 14/135 | DRAXIN; POU3F1; ARPC5; CNTN2; ID2; SIX3; ZEB2; NEUROD1; CTNNB1; ROBO1; CASP3; EMX2; NME1; SSTR2 |
| GO:0002009 | morphogenesis of an epithelium | 2.42 | 1.73e-03 | 0.043405 | 14/136 | PBX1; RYR2; ZEB2; IGFBP5; CTNNB1; IFT57; WDR1; CTHRC1; TNC; CFL1; VCL; SIX1; PGF; PFN1 |
| GO:0051098 | regulation of binding | 2.27 | 1.67e-03 | 0.043405 | 16/166 | STMN1; EPB41; JUN; NES; ID2; NEUROD1; CTNNB1; HDAC2; ACTB; TMSB4X; CTHRC1; CALM1; B2M; CCPG1; WFIKKN1; NME1 |
| GO:0050769 | positive regulation of neurogenesis | 2.25 | 1.77e-03 | 0.043405 | 16/167 | CRABP2; ID2; NCOA1; ZEB2; NEUROD1; CTNNB1; ROBO1; MAP1B; NEUROG1; DPYSL3; HDAC2; TCF12; NME1; SYT4; TCF4; PCP4 |
| GO:0009636 | response to toxic substance | 2.23 | 2.01e-03 | 0.044935 | 16/169 | JUN; EPHX1; CHRNA1; EEF1B2; CASP3; SPARC; HDAC2; ACTB; PON2; TNC; RPS3; ASIC1; CALM1; ACTC1; NXN; MT-ND4 |
Factor 11 : 12 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 30.90 | 0.00e+00 | 0.00e+00 | 77/119 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006413 | translational initiation | 23.40 | 0.00e+00 | 0.00e+00 | 77/157 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0002181 | cytoplasmic translation | 19.60 | 0.00e+00 | 0.00e+00 | 30/73 | RPL11; RPL31; RPL32; RPL15; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; RPL30; RPL8; RPL41; RPL6; RPLP0; RPS29; RPLP1; RPL26; RPL19; RPL38; RPL17; RPS21; EEF2; RPL36; RPS28; RPL18A; RPL18; RPL13A |
| GO:0090150 | establishment of protein localization to membrane | 18.30 | 0.00e+00 | 0.00e+00 | 78/204 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006401 | RNA catabolic process | 15.60 | 0.00e+00 | 0.00e+00 | 77/236 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006605 | protein targeting | 14.30 | 0.00e+00 | 0.00e+00 | 78/261 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0016072 | rRNA metabolic process | 6.98 | 3.33e-15 | 3.06e-13 | 25/171 | RPL11; RPS8; RPL5; RPS27; RPS7; RPSA; RPL14; RPL35A; RPS14; RPL10A; RPL7; RPS6; RPL35; RPL7A; RPS24; RPS17; RPS2; RPL26; RPL27; RPS21; RPS15; RPS28; RPS16; RPS19; RPS9 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 5.35 | 0.00e+00 | 0.00e+00 | 37/330 | RPL11; RPS8; RPL5; RPS27; RPS7; RPSA; RPL14; RPL24; RPL35A; RPS23; RPS14; RPS10; RPL10A; RPL10; RPL7; RPS6; RPL35; RPL12; RPL7A; RPS24; RPL6; RPLP0; RPS17; RPS2; RPL26; RPL23A; RPL27; RPL38; RPS21; RPS15; RPS28; RPS16; RPS19; RPL13A; RPS9; RPS5; RPL3 |
| GO:0034470 | ncRNA processing | 5.35 | 1.71e-12 | 1.39e-10 | 25/223 | RPL11; RPS8; RPL5; RPS27; RPS7; RPSA; RPL14; RPL35A; RPS14; RPL10A; RPL7; RPS6; RPL35; RPL7A; RPS24; RPS17; RPS2; RPL26; RPL27; RPS21; RPS15; RPS28; RPS16; RPS19; RPS9 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 5.16 | 7.84e-10 | 5.76e-08 | 20/185 | RPL11; RPL5; RPS27; RPSA; RPL24; RPS23; RPS14; RPS10; RPL10; RPL12; RPL6; RPLP0; RPL23A; RPL38; RPS15; RPS28; RPS19; RPL13A; RPS5; RPL3 |
| GO:0034248 | regulation of cellular amide metabolic process | 3.16 | 6.27e-05 | 4.18e-03 | 15/227 | RPL5; IGFBP5; RPS14; GNB2L1; SOX4; EEF1A1; RPS4X; RPL10; RPS3; GAPDH; RPL26; RPL38; EEF2; RPL13A; RPS9 |
| GO:0010608 | posttranscriptional regulation of gene expression | 2.67 | 2.52e-04 | 1.54e-02 | 16/286 | RPL5; RPS27A; IGFBP5; RPS14; GNB2L1; SOX4; RPS4X; RPL10; RPS3; GAPDH; RPL26; RPL38; EEF2; UBA52; RPL13A; RPS9 |
Factor 12 : 5 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 12.00 | 2.71e-08 | 1.99e-05 | 9/119 | RPL31; RPL37A; RPL37; SEC61G; RPL39; RPLP2; RPS24; RPS2; RPL38 |
| GO:0006413 | translational initiation | 9.09 | 3.02e-07 | 7.40e-05 | 9/157 | RPL31; RPL37A; RPL37; RPL39; RPLP2; RPS24; RPS2; RPL38; EIF2S2 |
| GO:0090150 | establishment of protein localization to membrane | 7.00 | 2.78e-06 | 5.09e-04 | 9/204 | RPL31; RPL37A; RPL37; RPL39; RPLP2; RPS24; HSP90AA1; RPS2; RPL38 |
| GO:0006605 | protein targeting | 6.69 | 2.50e-07 | 7.40e-05 | 11/261 | RPL31; RPL37A; RPL37; SEC61G; RPL39; RPLP2; RPS24; UBC; HSP90AA1; RPS2; RPL38 |
| GO:0006401 | RNA catabolic process | 6.05 | 9.24e-06 | 1.36e-03 | 9/236 | RPL31; RPL37A; RPL37; RPL39; RPLP2; RPS24; UBC; RPS2; RPL38 |
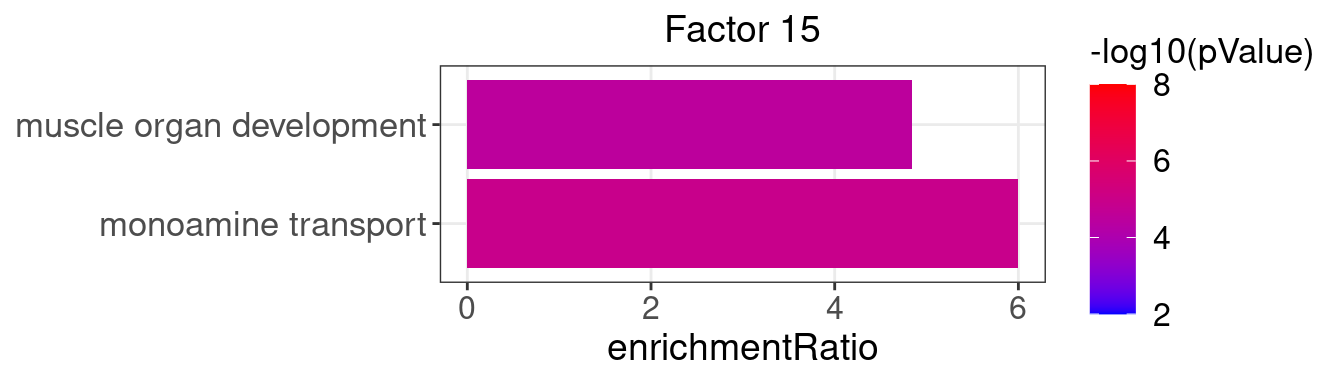
Factor 15 : 3 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0015844 | monoamine transport | 15.40 | 1.18e-05 | 0.00868 | 5/17 | GPM6B; SYT7; SNCG; SYT1; SYT4 |
| GO:0015696 | ammonium transport | 11.40 | 5.87e-05 | 0.01437 | 5/23 | GPM6B; SYT7; SNCG; SYT1; SYT4 |
| GO:0007517 | muscle organ development | 4.84 | 3.32e-05 | 0.01219 | 10/108 | TNNI1; CHRNA1; BASP1; NEUROG1; ETV1; G6PD; CNTFR; MYL6; EFNB2; SIX1 |
Factor 18 : 11 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 12.70 | 0.00e+00 | 0.00e+00 | 32/119 | RPL22; RPL5; RPS7; RTN4; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL37; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; KDELR1; RPL13A; RPS9; RPL3 |
| GO:0006413 | translational initiation | 12.30 | 0.00e+00 | 0.00e+00 | 41/157 | RPL22; RPL5; RPS7; RPL37A; RPL15; RPSA; EIF1B; RPL14; EIF4A2; RPS3A; RPL37; NPM1; RPL10A; RPS4X; RPL10; RPS20; RPL7; PABPC1; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS29; RPL4; RPS2; RPL23; EIF1; RPL36; RPS28; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| GO:0002181 | cytoplasmic translation | 11.70 | 3.33e-15 | 4.07e-13 | 18/73 | RPL15; TMA7; RPL10A; EIF3E; EIF3H; EIF3F; EIF3M; RPS26; RPL41; RPL6; RPLP0; RPS29; EEF2; RPL36; RPS28; RPL13A; EIF3D; EIF3L |
| GO:0006401 | RNA catabolic process | 7.21 | 0.00e+00 | 0.00e+00 | 36/236 | RPL22; YBX1; RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL37; NPM1; RPL10A; RPS4X; RPL10; RPS20; RPL7; PABPC1; EIF3E; RPL35; RPL7A; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS29; SLIRP; RPL4; RPS2; RPL23; CIRBP; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| GO:0090150 | establishment of protein localization to membrane | 7.19 | 0.00e+00 | 0.00e+00 | 31/204 | RPL22; RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL37; GNB2L1; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| GO:0006605 | protein targeting | 5.80 | 0.00e+00 | 0.00e+00 | 32/261 | RPL22; RPL5; TOMM20; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL37; RPL10A; SLC25A6; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 4.60 | 3.90e-08 | 3.58e-06 | 18/185 | RPL5; RPSA; NPM1; RBMX; RPL10; EIF3E; EIF3H; EIF3F; EIF3M; RPL6; RPLP0; CIRBP; RPS28; GLTSCR2; RPL13A; EIF3D; EIF3L; RPL3 |
| GO:0034248 | regulation of cellular amide metabolic process | 3.75 | 8.85e-07 | 7.22e-05 | 18/227 | RPL5; EIF1B; EIF4A2; NPM1; GNB2L1; EEF1A1; RPS4X; RPL10; PABPC1; EIF3E; EIF3H; RPS3; EIF1; CIRBP; EEF2; RPL13A; RPS9; EIF3D |
| GO:0022613 | ribonucleoprotein complex biogenesis | 3.73 | 2.18e-09 | 2.29e-07 | 26/330 | RPL5; RPS7; RPSA; RPL14; NPM1; RPL10A; RBMX; RPL10; RPL7; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPL6; RPLP0; RPS2; CIRBP; RPS28; GLTSCR2; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| GO:0016072 | rRNA metabolic process | 3.32 | 2.26e-04 | 1.51e-02 | 12/171 | RPL5; RPS7; RPSA; RPL14; RPL10A; RPL7; RPL35; RPL7A; RPS2; RPS28; GLTSCR2; RPS9 |
| GO:0010608 | posttranscriptional regulation of gene expression | 3.31 | 1.51e-06 | 1.11e-04 | 20/286 | YBX1; RPL5; H3F3A; EIF1B; EIF4A2; NPM1; GNB2L1; RPS4X; RPL10; PABPC1; EIF3E; EIF3H; POLR2L; RPS3; EIF1; CIRBP; EEF2; RPL13A; RPS9; EIF3D |
Factor 20 : 2 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 7.57 | 2.99e-05 | 0.0220 | 7/45 | STMN1; RGS16; RGS2; ATP2B4; ABHD6; RGS10; CALM1 |
| GO:0003012 | muscle system process | 4.35 | 8.47e-05 | 0.0311 | 10/112 | RGS2; ATP2B4; AGT; RYR2; MYL1; NEUROG1; CAMK2B; CALM1; SSTR2; PDE9A |
| Factor | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| Signif_GO_terms | 7 | 28 | 10 | 40 | 18 | 12 | 19 | 4 | 35 | 0 | 12 | 5 | 0 | 0 | 3 | 0 | 0 | 11 | 0 | 2 |
5.1.1 Factors of interest
4, 9, 15
6 Gene Set Enrichment Analysis by Perturbation
Target genes: Genes w/ LFSR < 0.05 under each perturbation marker gene;
Backgroud genes: all 6000 genes used in factor analysis;
Statistical test: hypergeometric test (over-representation test);
Only GO terms/pathways that satisfy fold change \(\geq\) 2 and test FDR \(<\) 0.05 are shown below.
6.1 GO Slim Over-Representation Analysis
Gene sets: Gene ontology "Biological Process" (non-redundant).
6.1.1 Early stage cells
ADNP : 12 significant GO terms| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0007218 | neuropeptide signaling pathway | 12.10 | 2.04e-04 | 0.028393 | 4/10 | GAL; NMB; SSTR2; PTH2 |
| GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 4.72 | 5.84e-04 | 0.038949 | 7/45 | STMN1; RGS16; RGS2; ATP2B4; CNTN2; ABHD6; DNM1 |
| GO:0030534 | adult behavior | 4.72 | 5.84e-04 | 0.038949 | 7/45 | CNTN2; ID2; HDAC2; PBX3; LGI4; SEZ6L; CSTB |
| GO:0003012 | muscle system process | 4.34 | 5.63e-07 | 0.000413 | 16/112 | RGS2; ATP2B4; AGT; IGFBP5; CAMK2D; SORBS2; COL4A3BP; HDAC2; CAMK2B; MTPN; TPM2; VCL; ACTC1; SSTR2; TNNC2; PDE9A |
| GO:0003007 | heart morphogenesis | 4.27 | 2.13e-04 | 0.028393 | 9/64 | JUN; ID2; NRP2; ROBO1; IFT57; COL4A3BP; OLFM1; SIX1; ACTC1 |
| GO:0097305 | response to alcohol | 3.90 | 4.24e-04 | 0.034555 | 9/70 | RGS2; EEF1B2; SPARC; HDAC2; LANCL2; TNC; GSTP1; ACTC1; MT-ND4 |
| GO:0050673 | epithelial cell proliferation | 3.88 | 4.98e-05 | 0.012178 | 12/94 | JUN; VASH2; ID2; NRP2; IGFBP5; SCG2; ROBO1; IFT57; SPARC; SIX1; PGF; SERPINF1 |
| GO:0001101 | response to acid chemical | 3.34 | 3.95e-04 | 0.034555 | 11/100 | FABP3; ATP2B4; CASP3; SPARC; HDAC2; LANCL2; TNC; GSTP1; SIX1; SERPINF1; BAIAP2 |
| GO:0090130 | tissue migration | 3.34 | 7.33e-04 | 0.044844 | 10/91 | JUN; AGT; ZEB2; NRP2; SCG2; IQSEC1; ROBO1; SPARC; ACTC1; SERPINF1 |
| GO:0001525 | angiogenesis | 3.31 | 2.32e-04 | 0.028393 | 12/110 | JUN; SHC1; EFNA3; VASH2; AGT; NRP2; SCG2; ROBO1; SPARC; VAV2; PGF; SERPINF1 |
| GO:0061564 | axon development | 3.08 | 3.13e-06 | 0.001149 | 21/207 | STMN1; JUN; S100A6; SHC1; EFNA3; CNTN2; ZEB2; NRP2; ROBO1; SPON2; CASP3; ETV1; TNC; OLFM1; NCAM1; ANK3; VCL; LINGO1; BAIAP2; DCC; ADARB1 |
| GO:0099177 | regulation of trans-synaptic signaling | 2.80 | 4.23e-04 | 0.034555 | 14/152 | PRKCZ; CNTN2; AGT; ABHD6; CPLX1; CPLX2; CAMK2B; DNM1; SYT7; NTF3; RAB26; BAIAP2; CELF4; DCC |
ARID1B : 24 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0010257 | NADH dehydrogenase complex assembly | 4.83 | 9.74e-14 | 2.38e-11 | 28/60 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFS6; NDUFA2; NDUFB2; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFA8; NDUFS8; NDUFC2; NDUFB8; NDUFB1; NDUFB10; NDUFAB1; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND3; MT-ND6 |
| GO:0010644 | cell communication by electrical coupling | 4.78 | 7.47e-04 | 3.05e-02 | 6/13 | ATP1B1; FKBP1B; DBN1; SRI; ANK3; CALM1 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 4.17 | 1.95e-14 | 7.17e-12 | 35/87 | NDUFS5; COX20; NDUFB3; NDUFAF3; COX17; NDUFB4; NDUFS6; NDUFA2; UQCC2; NDUFB2; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFA8; NDUFS8; NDUFC2; NDUFB8; COX14; NDUFB1; NDUFB10; NDUFAB1; COA3; NDUFS7; NDUFA11; PET100; NDUFA7; NDUFB7; NDUFA13; NDUFA3; UQCR10; NDUFA6; MT-ND3; MT-ND6 |
| GO:1902600 | proton transmembrane transport | 4.03 | 4.15e-12 | 5.30e-10 | 30/77 | PARK7; COX5B; ATP5G3; COX17; ATP5I; COX7C; COX7A2; NDUFA4; ATP5J2; ATP6V1F; ATP6V0E2; TMSB4X; COX7B; COX6C; COX8A; ATP5L; ATP5B; SLC25A3; COX6A1; COX5A; COX4I1; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; COX6B1; ATP5J; ATP5O; MT-CO1 |
| GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | 3.56 | 1.19e-04 | 6.25e-03 | 11/32 | ATP5G3; ATP6V1F; ATP6V0E2; ATP5L; ATP5B; ATP2B1; ATP5G1; ATP5H; ATP5A1; ATP5E; ATP5O |
| GO:0031644 | regulation of neurological system process | 3.11 | 1.52e-03 | 4.31e-02 | 9/30 | PRKCZ; AGT; NRXN1; AKAP9; FABP5; CBLN1; EIF4A3; BAIAP2; CELF4 |
| GO:0009141 | nucleoside triphosphate metabolic process | 3.07 | 2.22e-15 | 1.63e-12 | 56/189 | PARK7; ATPIF1; NDUFS5; UQCRH; ATP1B1; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; SNCA; NDUFS6; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; NDUFB9; NDUFB6; NDUFA8; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5B; COX6A1; RAN; NDUFB1; COX5A; NDUFB10; NDUFAB1; DCTPP1; COX4I1; ATP5G1; NME1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3 |
| GO:0050663 | cytokine secretion | 2.88 | 1.62e-03 | 4.31e-02 | 10/36 | PRKCZ; S100A13; AGT; SELK; BTN2A2; EZR; TMSB4X; CADM1; HMGB1; MIF |
| GO:0009123 | nucleoside monophosphate metabolic process | 2.87 | 1.65e-13 | 3.04e-11 | 54/195 | PARK7; ATPIF1; NDUFS5; UQCRH; ATP1B1; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; SNCA; NDUFS6; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; HDDC2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; NDUFB9; NDUFB6; NDUFA8; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5B; COX6A1; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3 |
| GO:0070972 | protein localization to endoplasmic reticulum | 2.70 | 1.47e-07 | 1.20e-05 | 31/119 | RPL5; RPS27A; RPL15; RPSA; SPCS1; RPL9; RPS3A; RPS12; SEC61G; RPS4X; RPL7; SEC61B; RPL35; RPL12; RPL7A; RPLP2; RPS3; RPS26; RPLP0; SRP14; RPL4; RPS2; RPL13; RPL23A; RPL19; RPS21; RPS19; RPL18; RPL13A; RPL28; RPL3 |
| GO:0035637 | multicellular organismal signaling | 2.65 | 1.99e-03 | 4.88e-02 | 11/43 | ATP1B1; AGT; FKBP1B; SCN3A; SCN9A; SRI; AKAP9; ANK3; ATP2B1; CALM1; SOD1 |
| GO:0007006 | mitochondrial membrane organization | 2.53 | 4.52e-05 | 2.62e-03 | 21/86 | ATPIF1; ATP5G3; ATP5I; SNCA; DYNLT1; ATP5J2; YWHAZ; TIMM10; ATP5L; ATP5B; CALM1; ATP5G1; ATP5H; ATP5A1; ROMO1; ATP5E; TIMM13; NDUFA13; PDCD5; ATP5J; ATP5O |
| GO:0006091 | generation of precursor metabolites and energy | 2.50 | 4.34e-12 | 5.30e-10 | 60/248 | PARK7; SH3BGRL3; ATPIF1; NDUFS5; UQCRH; COX20; COX5B; NDUFB3; COX17; NDUFB4; ATP5I; SNCA; NDUFS6; COX7C; GLRX; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; COX7B; NDUFA1; COX6C; NDUFB9; NDUFB6; NDUFA8; COX8A; NDUFS8; NDUFC2; ATP5L; CISD1; NDUFB8; TPI1; BLOC1S1; ATP5B; COX6A1; HMGB1; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5H; ATP5A1; ATP5E; NDUFS7; UQCR11; NDUFA7; NDUFB7; COX6B1; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3 |
| GO:0010959 | regulation of metal ion transport | 2.41 | 4.63e-05 | 2.62e-03 | 23/99 | SEPN1; ATP1B1; AGT; FKBP1B; GNAI2; SNCA; GLRX; CAMK2B; SRI; AKAP9; SARAF; CHD7; ANK3; KCNIP2; CD63; JPH4; AKAP6; CALM1; B2M; NKAIN4; HOMER3; FXYD7; ATF4 |
| GO:0009259 | ribonucleotide metabolic process | 2.38 | 6.85e-11 | 7.18e-09 | 59/257 | PARK7; ATPIF1; NDUFS5; UQCRH; ATP1B1; COX5B; DBI; ATP5G3; NDUFB3; NDUFB4; ATP5I; SCD5; SNCA; NDUFS6; COX7C; HINT1; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; NDUFB9; NDUFB6; NDUFA8; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5B; COX6A1; RAN; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5G1; NME1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; PDE9A; MT-CO1; MT-ND3 |
| GO:0006413 | translational initiation | 2.31 | 1.37e-06 | 1.01e-04 | 35/157 | RPL5; RPS27A; RPL15; RPSA; EIF1B; EIF4A2; RPL9; RPS3A; RPS12; RPS4X; RPL7; EIF3E; EIF3H; RPL35; RPL12; RPL7A; RPLP2; EIF3F; RPS3; RPS26; RPLP0; EIF3J; RPL4; RPS2; RPL13; RPL23A; RPL19; RPS21; RPS19; RPL18; RPL13A; RPL28; EIF3L; RPL3; ATF4 |
| GO:0043270 | positive regulation of ion transport | 2.28 | 1.70e-03 | 4.31e-02 | 15/68 | PARK7; ATP1B1; AGT; COX17; SNCA; GLRX; SRI; AKAP9; TMSB4X; ANK3; KCNIP2; AKAP6; CALM1; MIF; ATF4 |
| GO:0090150 | establishment of protein localization to membrane | 2.28 | 5.50e-08 | 5.05e-06 | 45/204 | RPL5; ATP1B1; RPS27A; REEP1; RPL15; RPSA; NSG1; RPL9; RPS3A; GNB2L1; CD24; RPS12; FIS1; RPS4X; RPL7; YWHAZ; SEC61B; RPL35; RPL12; RPL7A; RPLP2; TIMM10; RPS3; ANK3; RPS26; RPLP0; CALM1; SRP14; RPL4; RPS2; RAB26; RPL13; RPL23A; RPL19; RAB31; ROMO1; RPS21; TIMM13; NDUFA13; PDCD5; RPS19; RPL18; RPL13A; RPL28; RPL3 |
| GO:0032409 | regulation of transporter activity | 2.17 | 1.53e-03 | 4.31e-02 | 17/81 | PARK7; SEPN1; ATP1B1; FKBP1B; COX17; SNCA; GLRX; ACTB; SRI; AKAP9; TMSB4X; ANK3; KCNIP2; JPH4; AKAP6; CALM1; FXYD7 |
| GO:0072593 | reactive oxygen species metabolic process | 2.17 | 1.16e-03 | 4.07e-02 | 18/86 | PARK7; ATPIF1; PRDX1; GADD45A; AGT; GPX1; GNAI2; SNCA; HSP90AB1; PRDX5; GSTP1; SH3PXD2A; MAPT; ROMO1; PREX1; PRDX2; NDUFA13; SOD1 |
| GO:0019932 | second-messenger-mediated signaling | 2.14 | 1.02e-03 | 3.73e-02 | 19/92 | SEPN1; ATP1B1; AGT; FKBP1B; NEUROD1; GNAI2; SELK; HINT1; SRI; FIS1; NRG1; PEBP1; JPH4; AKAP6; CALM1; MAPT; HOMER3; APLP1; PDE9A |
| GO:0072507 | divalent inorganic cation homeostasis | 2.11 | 6.71e-04 | 2.90e-02 | 21/103 | SEPN1; ATP1B1; AGT; FKBP1B; SELK; SNCA; CD24; SRI; FIS1; CHD7; ANK3; DDIT3; ATP2B1; HSP90B1; HMGB1; TPT1; JPH4; AKAP6; CALM1; CACNA1A; ATF4 |
| GO:0052547 | regulation of peptidase activity | 2.05 | 3.29e-04 | 1.51e-02 | 25/126 | PARK7; NRDC; LAMTOR5; AGT; HSPE1; GPX1; SNCA; GNB2L1; CYCS; HIP1; FIS1; PCSK1N; NGFRAP1; ANP32B; PRDX5; RPS3; PEBP1; HMGB1; MAPT; NGFR; TIMP2; CST3; NDUFA13; PDCD5; CSTB |
| GO:0051235 | maintenance of location | 2.01 | 1.28e-03 | 4.27e-02 | 21/108 | PARK7; SEPN1; HNRNPU; FKBP1B; TMSB10; SNCA; SKP1; DBN1; MDFI; EZR; SRI; AKAP9; TMSB4X; CHD7; TXN; ANK3; DDIT3; HSP90B1; JPH4; AKAP6; CALM1 |
CHD2 : 13 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 8.32 | 0.00e+00 | 0.00e+00 | 58/119 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL39; RPL10; RPL7; RPL30; RPS6; RPL35; HSPA5; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPL3 |
| GO:0006413 | translational initiation | 6.85 | 0.00e+00 | 0.00e+00 | 63/157 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; EIF1B; RPL14; RPL24; RPL35A; RPL9; EIF4E; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL39; RPL10; COPS5; RPL7; RPL30; EIF3E; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS15A; RPL13; EIF4A1; RPL26; RPL23A; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPL3; ATF4 |
| GO:0002181 | cytoplasmic translation | 5.85 | 1.02e-13 | 1.24e-11 | 25/73 | RPL11; RPL31; RPL32; RPL15; TMA7; RPL24; RPL35A; RPL9; RPS23; RPL39; RPL30; EIF3E; RPL41; RPL6; RPLP0; RPS29; RPLP1; RPL26; RPL19; RPL38; RPL17; RPS28; RPL18A; RPL18; RPL13A |
| GO:0090150 | establishment of protein localization to membrane | 4.94 | 0.00e+00 | 0.00e+00 | 59/204 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL39; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; CALM1; RPL4; RPLP1; RPS17; RPS15A; RPL13; VAMP2; RPL26; RPL23A; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPL3 |
| GO:0006401 | RNA catabolic process | 4.34 | 0.00e+00 | 0.00e+00 | 60/236 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; PCBP4; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL39; RPL10; RPL7; RPL30; EIF3E; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SLIRP; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPL3 |
| GO:0006605 | protein targeting | 3.93 | 0.00e+00 | 0.00e+00 | 60/261 | RPL22; PEX14; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; PARL; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; AKAP12; RPS4X; RPL39; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPL3 |
| GO:0010959 | regulation of metal ion transport | 3.10 | 1.28e-05 | 1.05e-03 | 18/99 | SEPN1; AGT; GNAI2; SNCA; CAMK2B; AKAP9; CHD7; GAL; KCNIP2; CD63; JPH4; CALM1; B2M; VAMP2; NEDD4L; NKAIN4; FXYD7; ATF4 |
| GO:0034765 | regulation of ion transmembrane transport | 2.77 | 3.91e-05 | 2.87e-03 | 19/117 | EPHB2; SEPN1; RGS2; AGT; SCN3A; CRBN; SNCA; AKAP9; TMSB4X; CHD7; GAL; KCNIP2; CD63; JPH4; CALM1; VAMP2; NEDD4L; CACNA1A; FXYD7 |
| GO:0016072 | rRNA metabolic process | 2.60 | 5.05e-06 | 5.01e-04 | 26/171 | RPL11; RPS8; RPL5; RPS27; RPSA; RPL14; RPL35A; RPS14; RPL7L1; CHD7; RPL7; RPS6; RPL35; RPL7A; RPS24; RPS17; RSL1D1; NOB1; RPL26; ZNHIT3; RPL27; RPS15; RPS28; RPS16; RPS19; GLTSCR2 |
| GO:0010038 | response to metal ion | 2.40 | 8.73e-04 | 4.58e-02 | 16/114 | PEF1; S100A13; ID2; SNCA; HSPA5; SYT7; KCNIP2; CALM1; B2M; SERPINF1; NEDD4L; BSG; JUNB; CALR; ATF4; MT-CO1 |
| GO:0099177 | regulation of trans-synaptic signaling | 2.25 | 4.83e-04 | 2.83e-02 | 20/152 | PRKCZ; EPHB2; AGT; GNAI2; SNCA; EIF4E; CPLX2; GRM4; AKAP12; CAMK2B; AKAP9; SYT7; NTF3; JPH4; CALM1; CBLN1; VAMP2; BAIAP2; CELF4; CACNA1A |
| GO:0071826 | ribonucleoprotein complex subunit organization | 2.22 | 1.65e-04 | 1.10e-02 | 24/185 | RPL11; RPL5; RPS27; RPSA; RPL24; RPS23; RPS14; MRPL22; RPS10; RPL10; EIF3E; STRAP; RPL6; RPLP0; RPL23A; ZNHIT3; RPL38; CELF4; RPS15; RPS28; RPS19; GLTSCR2; RPL13A; RPL3 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 2.07 | 5.46e-06 | 5.01e-04 | 40/330 | RPL11; RPS8; RPL5; RPS27; RPSA; RPL14; RPL24; RPL35A; RPS23; RPS14; MRPL22; RPS10; RPL7L1; RPL10; CHD7; RPL7; EIF3E; RPS6; RPL35; RPL7A; RPS24; STRAP; RPL6; RPLP0; RPS17; RSL1D1; NOB1; RPL26; RPL23A; ZNHIT3; RPL27; RPL38; CELF4; RPS15; RPS28; RPS16; RPS19; GLTSCR2; RPL13A; RPL3 |
DYRK1A : 9 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0051131 | chaperone-mediated protein complex assembly | 12.50 | 1.93e-04 | 0.017703 | 4/11 | HSPD1; HSP90AB1; CCT2; HSP90AA1 |
| GO:0070670 | response to interleukin-4 | 12.50 | 1.93e-04 | 0.017703 | 4/11 | HSP90AB1; HSPA5; TUBA1B; XBP1 |
| GO:0071897 | DNA biosynthetic process | 4.50 | 2.57e-05 | 0.008269 | 11/84 | CENPF; HNRNPD; HSP90AB1; HNRNPA2B1; PPIA; DKC1; UBE2L6; HNRNPA1; CCT2; HNRNPC; HSP90AA1 |
| GO:0006457 | protein folding | 4.24 | 3.49e-07 | 0.000256 | 17/138 | PDIA6; FKBP1B; HSPD1; HSPE1; CANX; HSP90AB1; PPIA; PRDX4; HSPA5; HSPA8; PFDN5; CCT2; HSP90B1; HSP90AA1; PDIA3; PPIB; CALR |
| GO:0032200 | telomere organization | 4.18 | 2.55e-04 | 0.020819 | 9/74 | GNL3; HNRNPD; HSP90AB1; HNRNPA2B1; DKC1; HNRNPA1; CCT2; HNRNPC; HSP90AA1 |
| GO:0006403 | RNA localization | 3.31 | 3.85e-05 | 0.008269 | 15/156 | SFPQ; SRSF7; SSB; HNRNPA3; NOP58; NPM1; SRSF3; HNRNPA2B1; DKC1; HNRNPA1; PA2G4; CCT2; RAN; SRSF2; FBL |
| GO:1903311 | regulation of mRNA metabolic process | 3.11 | 4.51e-05 | 0.008269 | 16/177 | SERBP1; PRDX6; SRSF7; PCBP4; HNRNPD; NPM1; SRSF3; HNRNPA2B1; PABPC1; HSPA8; HNRNPA1; HNRNPC; RBM25; C1QBP; SRSF2; HNRNPM |
| GO:0006401 | RNA catabolic process | 2.62 | 1.40e-04 | 0.017109 | 18/236 | SERBP1; SSB; PCBP4; HNRNPD; RPL37; NPM1; RPS4X; RPL39; RPL10; DKC1; PABPC1; RPS13; HSPA8; HNRNPC; HNRNPM; C19orf43; RPS9; RPL28 |
| GO:0010608 | posttranscriptional regulation of gene expression | 2.53 | 6.48e-05 | 0.009508 | 21/286 | SERBP1; EPRS; EIF5B; NCL; PCBP4; HNRNPD; NPM1; SOX4; HNRNPA2B1; RPS4X; RPL10; DKC1; PABPC1; HSPA8; PA2G4; RAN; HNRNPC; C1QBP; HNRNPM; CALR; RPS9 |
PTEN : 12 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0006414 | translational elongation | 3.41 | 1.11e-15 | 2.72e-13 | 47/109 | AURKAIP1; MRPS15; MRPL37; MRPL9; MRPS5; MRPS9; MRPL44; MRPS22; MRPL47; MRPS18C; MRPS30; MRPL22; GNB2L1; MRPS18B; MRPS10; MRPL2; MRPS18A; EEF1A1; MRPL18; MRPL32; MRPL13; MRPL50; MRPS2; RPLP2; MRPL21; MRPL51; MRPS35; MRPL42; MRPL52; MRPL46; MRPS11; MRPL28; MRPS34; EIF5A; ERAL1; MRPL10; MRPL27; MRPS23; ICT1; MRPS7; MRPL12; MRPS26; EEF2; GADD45GIP1; MRPL40; MRPL39; MRPS6 |
| GO:0070972 | protein localization to endoplasmic reticulum | 3.33 | 4.44e-16 | 1.63e-13 | 50/119 | RPS8; RPL5; RPS27; RPS7; RPL31; RPL32; RPL15; RPL14; RPL29; RPL24; SEC62; RPL35A; SRP72; RPL34; ANK2; RPS23; RPS14; RPS18; RPL10A; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL12; RPL7A; RPLP2; SPCS2; RPS3; RPS25; RPS24; RPS26; RPL6; RPLP0; RPS27L; RPL4; RPS2; RPS15A; RPL26; RPL23; RPS21; ASNA1; KDELR1; RPL18; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0140053 | mitochondrial gene expression | 3.14 | 9.36e-14 | 1.72e-11 | 46/116 | AURKAIP1; MRPS15; MRPL37; MRPL9; MRPS5; MRPS9; MRPL44; MRPS22; MRPL47; MRPS18C; MRPS30; MRPL22; MRPS18B; MRPS10; MRPL2; MRPS18A; MRPL18; MALSU1; MRPL32; TBRG4; HSD17B10; MRPL13; MRPL50; MRPS2; MRPL21; MRPL51; MRPS35; MRPL42; MRPL52; MRPL46; MRPS11; MRPL28; MRPS34; ERAL1; MRPL10; MRPL27; MRPS23; ICT1; MRPS7; MRPL12; MRPS26; UQCC1; GADD45GIP1; MRPL40; MRPL39; MRPS6 |
| GO:0006413 | translational initiation | 3.13 | 0.00e+00 | 0.00e+00 | 62/157 | EIF3I; RPS8; RPL5; RPS27; RPS7; DDX1; RPL31; RPL32; RPL15; RPL14; RPL29; RPL24; EIF4A2; RPL35A; EIF4E; RPL34; RPS23; RPS14; RPS18; RPL10A; HSPB1; RPS4X; RPL36A; RPL39; RPL10; EIF4EBP1; RPS20; COPS5; RPL7; RPL30; PABPC1; RPL12; RPL7A; RPLP2; EIF3F; POLR2G; PPP1CA; RPS3; RPS25; CDC123; RPS24; RPS26; RPL6; RPLP0; EIF5; EIF3J; RPS27L; RPL4; RPS2; RPS15A; EIF4A1; RPL26; RPL23; EIF6; RPS21; RPL18; RPS11; RPS9; RPL28; RPS5; EIF3L; RPL3; ATF4 |
| GO:0010257 | NADH dehydrogenase complex assembly | 3.04 | 3.50e-07 | 2.33e-05 | 23/60 | NDUFS2; NDUFB4; NDUFB5; NDUFC1; NDUFS6; NDUFS4; NDUFA5; NDUFB11; NDUFB9; NDUFA8; NDUFS3; NDUFV1; NDUFS8; NDUFC2; TMEM126B; NDUFA9; NDUFA12; NDUFAB1; NDUFV2; NDUFS7; NDUFA13; NDUFA6; MT-ND4 |
| GO:0002181 | cytoplasmic translation | 2.71 | 1.31e-06 | 7.41e-05 | 25/73 | EIF3I; RPL31; RPL32; RPL15; TMA7; RPL29; RPL24; RPL35A; RPS23; RPL10A; RPL36A; RPL39; EIF4EBP1; RPL30; EIF3F; RPS26; RPL6; RPLP0; EIF5; EIF3J; RPL26; RPS21; EEF2; RPL18; EIF3L |
| GO:0090150 | establishment of protein localization to membrane | 2.37 | 1.91e-11 | 2.80e-09 | 61/204 | HPCA; RPS8; RPL5; RPS27; RPS7; RPL31; C2orf47; RPL32; RPL15; RPL14; RPL29; RPL24; SEC62; RPL35A; SRP72; RPL34; RPS23; RPS14; GNB2L1; RPS18; RPL10A; CD24; GOPC; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL12; RPL7A; RPLP2; BAD; RPS3; RPS25; RPS24; RPS26; RPL6; RPLP0; CALM1; RPS27L; RPL4; RPS2; RPS15A; VPS35; TIMM22; VAMP2; RPL26; RPL23; NMT1; RPS21; ASNA1; RAB8A; NDUFA13; PDCD5; RPL18; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 2.18 | 1.24e-04 | 5.07e-03 | 24/87 | NDUFS2; NDUFB4; NDUFB5; NDUFC1; NDUFS6; NDUFS4; NDUFA5; NDUFB11; NDUFB9; NDUFA8; NDUFS3; NDUFV1; NDUFS8; NDUFC2; TMEM126B; NDUFA9; NDUFA12; NDUFAB1; NDUFV2; UQCC1; NDUFS7; NDUFA13; NDUFA6; MT-ND4 |
| GO:0016072 | rRNA metabolic process | 2.18 | 8.45e-08 | 6.89e-06 | 47/171 | RPS8; RPF1; RPL5; RPS27; RPS7; WDR43; MRPS9; NIFK; RPL14; EXOSC7; RRP9; RPL35A; RPS14; ABT1; RPL10A; BYSL; RPL7L1; GTF3C6; DDX56; CHD7; RPL7; DCAF13; EXOSC4; RPL7A; WDR74; TRMT112; GTPBP4; RPS24; EXOSC1; NPM3; EXOSC8; NGDN; NOP10; RPS27L; MRPS11; RPS2; RSL1D1; NOB1; RPL26; DDX52; MAPT; UTP18; RPS21; C19orf43; GLTSCR2; RPS9; RRP1B; RRP1 |
| GO:0006605 | protein targeting | 2.12 | 1.49e-10 | 1.82e-08 | 70/261 | PEX14; HMGCL; ATPIF1; HPCA; RPS8; RPL5; RPS27; TIMM17A; RPS7; RPL31; RPL32; RPL15; RPL14; RPL29; RPL24; SEC62; PARL; RPL35A; SRP72; SCARB2; RPL34; RPS23; RPS14; RPS18; RPL10A; AKAP12; TOMM7; PMPCB; SLC25A6; TIMM17B; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL12; RPL7A; PMPCA; RPLP2; AIP; SPCS2; RPS3; RPS25; RPS24; RPS26; RPL6; RPLP0; DNAJC15; RPS27L; RPL4; RPS2; RPS15A; TIMM22; RPL26; RPL23; RPS21; TIMM44; ASNA1; NDUFA13; PDCD5; ECH1; TIMM50; RPL18; RPS11; RPS9; RPL28; RPS5; UBE2L3; RPL3 |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.10 | 1.22e-07 | 8.95e-06 | 50/189 | ATPIF1; NDUFS2; PARP1; GUK1; DGUOK; SUCLG1; COX5B; ATP5G3; DTYMK; NDUFB4; NDUFB5; ATP5I; SNCA; NDUFC1; SDHA; NDUFS6; NDUFS4; UQCRQ; PMPCB; NDUFA5; TMSB4X; NDUFB9; CYC1; NDUFA8; NDUFS3; BAD; NDUFV1; NDUFS8; NDUFC2; SDHD; NDUFA9; ATP5B; NDUFA12; COX6A1; DNAJC15; COX5A; NDUFAB1; COQ9; NCOR1; ATP5G1; NDUFV2; ITPA; EIF6; NDUFS7; SIRT6; UQCRFS1; NDUFA6; ATP5O; MT-CO3; MT-ND4 |
| GO:0006401 | RNA catabolic process | 2.08 | 4.49e-09 | 4.71e-07 | 62/236 | PPP1R8; RPS8; MAGOH; RPL5; RBM8A; RPS27; RPS7; RPL31; RPL32; LSM3; RPL15; RPL14; EXOSC7; PCBP4; RPL29; RPL24; RPL35A; RPL34; RPS23; RPS14; RPS18; RPL10A; TBRG4; HSPB1; RPS4X; RPL36A; RPL39; UPF3B; RPL10; RPS20; RPL7; RPL30; PABPC1; EXOSC4; RPL12; RPL7A; RPLP2; POLR2G; RPS3; RPS25; RPS24; EXOSC1; RPS26; RPL6; RPLP0; EXOSC8; SLIRP; RPS27L; RPL4; RPS2; RNPS1; RPS15A; RPL26; RPL23; RPS21; HNRNPM; C19orf43; RPL18; RPS11; RPS9; RPL28; RPS5; RPL3 |
| Marker | ADNP | ARID1B | CHD2 | DYRK1A | PTEN |
| Signif_GO_terms | 12 | 24 | 13 | 9 | 12 |
6.1.2 Late stage cells
ADNP : 69 significant GO terms| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0007218 | neuropeptide signaling pathway | 6.59 | 4.47e-04 | 1.03e-02 | 5/10 | PCSK1N; GAL; NMB; SSTR2; PTH2 |
| GO:0010644 | cell communication by electrical coupling | 6.09 | 1.98e-04 | 6.55e-03 | 6/13 | ATP1B1; RYR2; FKBP1B; CAMK2D; SRI; ANK3 |
| GO:0002448 | mast cell mediated immunity | 5.27 | 4.73e-03 | 4.88e-02 | 4/10 | S100A13; SPON2; CPLX2; VAMP2 |
| GO:0014075 | response to amine | 5.27 | 4.73e-03 | 4.88e-02 | 4/10 | RGS2; HDAC2; RGS10; ASIC1 |
| GO:0045576 | mast cell activation | 5.27 | 4.73e-03 | 4.88e-02 | 4/10 | S100A13; CPLX2; VAMP2; PVRL2 |
| GO:0031016 | pancreas development | 3.96 | 2.84e-03 | 3.72e-02 | 6/20 | WLS; NEUROD1; CTNNB1; SOX4; ONECUT2; XBP1 |
| GO:0021953 | central nervous system neuron differentiation | 3.91 | 1.26e-06 | 2.03e-04 | 16/54 | LMO4; CNTN2; ZEB2; MAP2; CTNNB1; ROBO1; SOX4; MDGA1; FZD1; MTPN; MYCBP2; POU4F1; HSP90AA1; CBLN1; MAPT; ADARB1 |
| GO:0014074 | response to purine-containing compound | 3.72 | 9.92e-05 | 4.05e-03 | 11/39 | SEPN1; JUN; RYR2; IGFBP5; SPARC; HDAC2; EZR; AKAP9; HSPA5; HSP90B1; BSG |
| GO:0035637 | multicellular organismal signaling | 3.68 | 5.37e-05 | 2.47e-03 | 12/43 | ATP1B1; AGT; RYR2; FKBP1B; SCN3A; SCN9A; CHRNA1; ANK2; CAMK2D; SRI; AKAP9; ANK3 |
| GO:0048678 | response to axon injury | 3.64 | 1.05e-03 | 1.80e-02 | 8/29 | PTPRF; JUN; FKBP1B; GAP43; NREP; NEFL; TNC; RTN4R |
| GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 3.63 | 1.28e-04 | 4.93e-03 | 11/40 | PTPRF; CNTN2; PTPRG; ROBO1; ALCAM; MDGA1; CADM1; PCDH17; CBLN1; BSG; PVRL2 |
| GO:0030865 | cortical cytoskeleton organization | 3.60 | 4.80e-03 | 4.88e-02 | 6/22 | EPB41; RAB13; WDR1; EZR; EPB41L3; CALR |
| GO:0003007 | heart morphogenesis | 3.50 | 3.17e-06 | 3.88e-04 | 17/64 | JUN; GNG5; RYR2; ID2; CTNNB1; ROBO1; IFT57; COL4A3BP; SOX4; FZD1; NRG1; CHD7; OLFM1; POU4F1; SIX1; ACTC1; TEAD2 |
| GO:0031099 | regeneration | 3.47 | 1.37e-05 | 9.54e-04 | 15/57 | SEPN1; PTPRF; JUN; FKBP1B; GPX1; GAP43; NREP; MTPN; NEFL; TNC; GSTP1; PGF; PKM; ENO3; RTN4R |
| GO:0010959 | regulation of metal ion transport | 3.33 | 4.41e-08 | 1.08e-05 | 25/99 | SEPN1; ATP1B1; AGT; RYR2; FKBP1B; CTNNB1; GNAI2; SNCA; ANK2; CAMK2D; GLRX; CAMK2B; SRI; AKAP9; CHD7; GAL; FXYD6; ANK3; KCNIP2; CD63; JPH4; B2M; VAMP2; NKAIN4; FXYD7 |
| GO:0003012 | muscle system process | 3.30 | 8.44e-09 | 3.10e-06 | 28/112 | ENO1; LMNA; ATP1B1; RGS2; AGT; RYR2; FKBP1B; CHRNA1; MYL1; IGFBP5; ANK2; CAMK2D; SORBS2; COL4A3BP; HDAC2; CAMK2B; SRI; CALD1; MTPN; TPM2; VCL; KCNIP2; ACTC1; SSTR2; MYL12A; MYL12B; TNNC2; PDE9A |
| GO:0030534 | adult behavior | 3.22 | 3.97e-04 | 9.49e-03 | 11/45 | CNTN2; ID2; SNCA; HDAC2; CHD7; PBX3; PCDH17; EFNB3; LGI4; SEZ6L; CSTB |
| GO:2001023 | regulation of response to drug | 3.18 | 4.93e-03 | 4.88e-02 | 7/29 | RGS2; AGT; NCOA1; SNCA; TXN; SYT7; GAL |
| GO:0046683 | response to organophosphorus | 3.10 | 3.19e-03 | 3.90e-02 | 8/34 | JUN; IGFBP5; SPARC; EZR; AKAP9; HSPA5; HSP90B1; BSG |
| GO:0045165 | cell fate commitment | 3.09 | 5.93e-04 | 1.21e-02 | 11/47 | LMO4; ID2; NEUROD1; CTNNB1; GAP43; CASP3; EBF2; NRG1; POU4F1; SIX1; ONECUT2 |
| GO:0007626 | locomotory behavior | 2.98 | 1.64e-04 | 5.73e-03 | 14/62 | CNTN2; ID2; SNCA; SOBP; NRG1; CHD7; PBX3; ZNF385A; NOVA1; EFNB3; SNAP25; LGI4; SEZ6L; CSTB |
| GO:0002576 | platelet degranulation | 2.93 | 4.65e-03 | 4.88e-02 | 8/36 | TAGLN2; TUBA4A; MANF; WDR1; SPARC; VCL; CD63; LGALS3BP |
| GO:0008037 | cell recognition | 2.93 | 4.65e-03 | 4.88e-02 | 8/36 | CNTN2; ROBO1; GAP43; CASP3; CCT5; CADM1; EFNB3; BSG |
| GO:0045454 | cell redox homeostasis | 2.89 | 2.98e-03 | 3.84e-02 | 9/41 | SH3BGRL3; PRDX1; PDIA6; GPX1; GLRX; PRDX4; TXN; PDIA3; NXN |
| GO:0009914 | hormone transport | 2.85 | 2.25e-05 | 1.38e-03 | 19/88 | MPC2; AGT; FKBP1B; NEUROD1; CAPN10; CPLX1; SOX4; UQCC2; SRI; CHD7; SYT7; GAL; DYNLL1; NMB; DOC2B; VAMP2; SNAP25; CACNA1A; SELM |
| GO:0034765 | regulation of ion transmembrane transport | 2.82 | 1.39e-06 | 2.03e-04 | 25/117 | SEPN1; ATP1B1; RGS2; AGT; RYR2; FKBP1B; SCN3A; SCN9A; WDR1; SNCA; ANK2; CAMK2D; GLRX; SRI; AKAP9; CHD7; GAL; FXYD6; ANK3; KCNIP2; CD63; JPH4; VAMP2; CACNA1A; FXYD7 |
| GO:0003205 | cardiac chamber development | 2.81 | 2.27e-03 | 3.21e-02 | 10/47 | LMO4; RYR2; ID2; ROBO1; ANK2; SOX4; FZD1; NRG1; CHD7; POU4F1 |
| GO:0060541 | respiratory system development | 2.81 | 2.27e-03 | 3.21e-02 | 10/47 | SEPN1; IGFBP5; CTNNB1; SPARC; CHD7; TNC; PPP1CA; SIX1; RAB3A; GNG8 |
| GO:0061564 | axon development | 2.80 | 1.24e-10 | 9.08e-08 | 44/207 | STMN1; PTPRF; JUN; S100A6; SHC1; EFNA3; CRABP2; LHX9; CNTN2; FKBP1B; ZEB2; MAP2; TWF2; ROBO1; ALCAM; GAP43; SPON2; CASP3; MAP1B; NREP; EZR; ETV1; NEFM; NEFL; DPYSL2; TNC; SPTAN1; OLFM1; NCAM1; FEZ1; ANK3; VCL; MYCBP2; POU4F1; HSP90AA1; LINGO1; EFNB3; MAPT; NGFR; BAIAP2; BSG; RAB3A; RTN4R; ADARB1 |
| GO:0032409 | regulation of transporter activity | 2.77 | 8.88e-05 | 3.83e-03 | 17/81 | SEPN1; ATP1B1; RYR2; FKBP1B; SNCA; ANK2; CAMK2D; GLRX; SRI; AKAP9; GAL; FXYD6; ANK3; KCNIP2; JPH4; VAMP2; FXYD7 |
| GO:0030902 | hindbrain development | 2.73 | 1.10e-03 | 1.84e-02 | 12/58 | RERE; WLS; NEUROD1; CTNNB1; MTPN; HSPA5; POU4F1; CBLN1; SSTR2; SEZ6L; MT-CO1; MT-ND4 |
| GO:0007517 | muscle organ development | 2.69 | 1.37e-05 | 9.54e-04 | 22/108 | SEPN1; RGS2; RYR2; ZBTB18; CHRNA1; DNER; CTNNB1; GPX1; FAM65B; UQCC2; ETV1; SRI; FZD1; MTPN; NRG1; CHD7; POU4F1; SIX1; ACTC1; TCF12; XBP1; ADARB1 |
| GO:0042391 | regulation of membrane potential | 2.64 | 7.70e-06 | 7.06e-04 | 24/120 | PRKCZ; JUN; ATP1B1; RYR2; FKBP1B; SCN3A; SCN9A; CHRNA1; WDR1; SNCA; ANK2; CAMK2D; GLRX; SRI; AKAP9; ANK3; KCNIP2; ASIC1; CBLN1; MAPT; EIF4A3; BAIAP2; CELF4; CACNA1A |
| GO:0009410 | response to xenobiotic stimulus | 2.64 | 4.01e-04 | 9.49e-03 | 15/75 | SEPN1; PHGDH; RGS2; EPHX1; RYR2; GPX1; SNCA; HDAC2; EIF4EBP1; FBXO32; HSPA5; GSTP1; RGS10; ASIC1; SERPINF1 |
| GO:0043062 | extracellular structure organization | 2.64 | 1.51e-03 | 2.41e-02 | 12/60 | MFAP2; AGT; CD47; SPARC; GPM6B; PRDX4; TNC; ITGAE; MFAP4; CST3; PLTP; BSG |
| GO:0006814 | sodium ion transport | 2.64 | 2.35e-03 | 3.26e-02 | 11/55 | ATP1B1; SCN3A; SCN9A; CAMK2D; GLRX; SLC17A6; FXYD6; ANK3; ASIC1; NKAIN4; FXYD7 |
| GO:0015850 | organic hydroxy compound transport | 2.59 | 4.27e-03 | 4.82e-02 | 10/51 | AGT; NCOA1; MFSD10; SNCA; GPM6B; SYT7; GAL; LIMA1; PLTP; SELM |
| GO:0060537 | muscle tissue development | 2.54 | 3.33e-05 | 1.88e-03 | 22/114 | SEPN1; LMNA; RGS2; AGT; RYR2; ZBTB18; ID2; CHRNA1; IGFBP5; DNER; CTNNB1; GPX1; SORBS2; FAM65B; UQCC2; MTPN; NRG1; CHD7; POU4F1; SIX1; ACTC1; CALR |
| GO:0060560 | developmental growth involved in morphogenesis | 2.52 | 2.98e-04 | 7.81e-03 | 17/89 | PRKCZ; CRABP2; ZEB2; MAP2; CTNNB1; TWF2; ALCAM; MAP1B; DPYSL2; TNC; OLFM1; GAL; VCL; SIX1; HSP90AA1; MAPT; RTN4R |
| GO:0090130 | tissue migration | 2.46 | 3.92e-04 | 9.49e-03 | 17/91 | JUN; RAB13; AGT; ZEB2; SCG2; IQSEC1; GPX1; PTPRG; ROBO1; SPARC; GLIPR2; ATP5B; CORO1C; ACTC1; SERPINF1; MAPRE2; CALR |
| GO:0051235 | maintenance of location | 2.44 | 1.39e-04 | 5.09e-03 | 20/108 | SEPN1; RYR2; FKBP1B; TWF2; SNCA; ANK2; CAMK2D; MDFI; EZR; SRI; AKAP9; CHD7; TXN; HSPA5; ANK3; HSP90B1; JPH4; EPB41L3; CALR; KDELR1 |
| GO:0072507 | divalent inorganic cation homeostasis | 2.43 | 2.14e-04 | 6.55e-03 | 19/103 | SEPN1; SV2A; ATP1B1; AGT; RYR2; FKBP1B; SNCA; ANK2; CAMK2D; SRI; CHD7; ANK3; HSP90B1; TPT1; JPH4; NMB; CALR; CACNA1A; SMDT1 |
| GO:0099177 | regulation of trans-synaptic signaling | 2.43 | 7.16e-06 | 7.06e-04 | 28/152 | PRKCZ; CNTN2; AGT; GNAI2; CPLX1; NSG1; SNCA; CPLX2; GRM4; AKAP12; CAMK2B; AKAP9; DNM1; SYT7; ASIC1; PCDH17; JPH4; RAB26; CBLN1; EFNB3; VAMP2; MAPT; EIF4A3; BAIAP2; CELF4; SNAP25; CACNA1A; RAB3A |
| GO:0034764 | positive regulation of transmembrane transport | 2.40 | 3.51e-03 | 4.09e-02 | 12/66 | ATP1B1; AGT; RYR2; CAPN10; SNCA; ANK2; GLRX; SRI; AKAP9; GAL; ANK3; KCNIP2 |
| GO:0048568 | embryonic organ development | 2.39 | 2.78e-04 | 7.81e-03 | 19/105 | MFAP2; NES; VASH2; RYR2; ID2; NCOA1; SIX3; NEUROD1; CTNNB1; IFT57; FAM65B; MDFI; SOBP; CHD7; CTHRC1; CDKN1C; SIX1; EIF4A3; TEAD2 |
| GO:0050673 | epithelial cell proliferation | 2.38 | 5.82e-04 | 1.21e-02 | 17/94 | JUN; VASH2; ID2; IGFBP5; SCG2; CTNNB1; GPX1; ROBO1; IFT57; SPARC; EGFL7; CDKN1C; SIX1; PGF; SERPINF1; NGFR; XBP1 |
| GO:0097485 | neuron projection guidance | 2.38 | 5.82e-04 | 1.21e-02 | 17/94 | SHC1; EFNA3; LHX9; CNTN2; ROBO1; ALCAM; GAP43; SPON2; EZR; ETV1; DPYSL2; SPTAN1; NCAM1; FEZ1; MYCBP2; EFNB3; BSG |
| GO:0042110 | T cell activation | 2.36 | 6.61e-04 | 1.28e-02 | 17/95 | PRKCZ; FKBP1B; SATB1; CTNNB1; CD47; CASP3; SOX4; FAM65B; BTN2A2; CHD7; CD151; CADM1; B2M; EFNB3; PREX1; KDELR1; XBP1 |
| GO:0042692 | muscle cell differentiation | 2.35 | 2.32e-04 | 6.81e-03 | 20/112 | SEPN1; LMNA; RGS2; AGT; IGFBP5; DNER; CTNNB1; GPX1; WDR1; ANK2; CASP3; SORBS2; FAM65B; MTPN; NRG1; SIX1; ACTC1; WFIKKN1; CALR; XBP1 |
| GO:0015849 | organic acid transport | 2.33 | 4.52e-03 | 4.88e-02 | 12/68 | FABP3; SV2A; MPC2; RGS2; AGT; NCOA1; SNCA; SLC17A6; NMB; VAMP2; BSG; RAB3A |
| GO:0072511 | divalent inorganic cation transport | 2.31 | 2.97e-04 | 7.81e-03 | 20/114 | SEPN1; ATP1B1; AGT; RYR2; FKBP1B; TMEM163; CTNNB1; GNAI2; SNCA; ANK2; CAMK2D; GPM6A; CAMK2B; SRI; CHD7; ASIC1; TPT1; JPH4; CACNA1A; SMDT1 |
| GO:0019932 | second-messenger-mediated signaling | 2.29 | 1.30e-03 | 2.12e-02 | 16/92 | SEPN1; ATP1B1; RGS2; AGT; RYR2; FKBP1B; NEUROD1; GNAI2; ANK2; CAMK2D; SRI; NRG1; GAL; JPH4; MAPT; PDE9A |
| GO:0001525 | angiogenesis | 2.28 | 5.14e-04 | 1.14e-02 | 19/110 | JUN; SHC1; EFNA3; VASH2; AGT; SCG2; CTNNB1; GPX1; ROBO1; SPARC; VAV2; EGFL7; ATP5B; PGF; PKM; SERPINF1; NGFR; RNF213; XBP1 |
| GO:0051047 | positive regulation of secretion | 2.26 | 7.87e-04 | 1.44e-02 | 18/105 | PRKCZ; WLS; MPC2; AGT; CAPN10; GNAI2; SNCA; SOX4; EZR; SRI; SYT7; GAL; CADM1; DYNLL1; NMB; DOC2B; RAB3A; XBP1 |
| GO:0023061 | signal release | 2.24 | 4.82e-05 | 2.36e-03 | 27/159 | WLS; SV2A; MPC2; AGT; FKBP1B; NEUROD1; CAPN10; CPLX1; SNCA; CPLX2; SOX4; UQCC2; GRM4; SRI; CHD7; SYT7; GAL; ASIC1; DYNLL1; TRIM9; NMB; DOC2B; VAMP2; SNAP25; CACNA1A; RAB3A; SELM |
| GO:0042063 | gliogenesis | 2.22 | 2.52e-03 | 3.43e-02 | 15/89 | MXRA8; POU3F1; PHGDH; CNTN2; ID2; DNER; CTNNB1; GAP43; WDR1; SOX4; HDAC2; NRG1; GSTP1; MAPT; LGI4 |
| GO:0003013 | circulatory system process | 2.22 | 1.85e-03 | 2.73e-02 | 16/95 | ATP1B1; RGS2; AGT; RYR2; ID2; FKBP1B; MYL1; GPX1; GNAI2; ANK2; CAMK2D; SRI; AKAP9; CHD7; KCNIP2; ACTC1 |
| GO:0006898 | receptor-mediated endocytosis | 2.22 | 1.85e-03 | 2.73e-02 | 16/95 | CNTN2; SNCA; SPARC; EZR; HIP1; DNM1; CALY; BICD1; CD63; HSP90B1; HSP90AA1; B2M; LGALS3BP; RAB31; SNAP25; CALR |
| GO:0002791 | regulation of peptide secretion | 2.22 | 7.26e-04 | 1.37e-02 | 19/113 | PRKCZ; WLS; MPC2; FKBP1B; NEUROD1; CAPN10; SOX4; BTN2A2; UQCC2; EZR; SRI; CHD7; SYT7; CADM1; DYNLL1; DOC2B; SNAP25; CACNA1A; XBP1 |
| GO:0010038 | response to metal ion | 2.20 | 8.12e-04 | 1.45e-02 | 19/114 | JUN; S100A13; RYR2; ID2; SNCA; CAMK2D; CASP3; SPARC; HSPA5; SYT7; PPP1CA; ANK3; KCNIP2; B2M; SERPINF1; MAPT; BSG; CALR; MT-CO1 |
| GO:0050808 | synapse organization | 2.17 | 2.14e-04 | 6.55e-03 | 24/146 | PTPRF; CNTN2; CHRNA1; DNER; CTNNB1; GAP43; SNCA; GPM6A; SPARC; MDGA1; CAMK2B; FZD1; NEFL; NRG1; TNC; ANK3; PCDH17; POU4F1; SIX1; CBLN1; MAPT; BAIAP2; RAB3A; SEZ6L |
| GO:0010975 | regulation of neuron projection development | 2.16 | 1.43e-05 | 9.54e-04 | 33/201 | PTPRF; CRABP2; RGS2; CNTN2; AGT; FKBP1B; ZEB2; MAP2; ITM2C; TWF2; PTPRG; ROBO1; TNIK; MAP1B; NR2F1; HDAC2; CAMK2B; FZD1; NEFL; DPYSL2; HSPA5; OLFM1; MYCBP2; B2M; LINGO1; SERPINF1; EFNB3; MAPT; NGFR; BAIAP2; SNAP25; PREX1; RTN4R |
| GO:0010721 | negative regulation of cell development | 2.16 | 1.01e-03 | 1.76e-02 | 19/116 | RGS2; CNTN2; ID2; SIX3; MAP2; ITM2C; CTNNB1; PTPRG; NR2F1; HDAC2; NRG1; GAL; CORO1C; B2M; LINGO1; EFNB3; NGFR; CALR; RTN4R |
| GO:0048880 | sensory system development | 2.15 | 2.57e-03 | 3.43e-02 | 16/98 | MFAP2; JUN; NES; SIX3; ZEB2; NEUROD1; CTNNB1; MAB21L2; GPM6A; HDAC2; DCX; CHD7; CDKN1C; POU4F1; SERPINF1; CELF4 |
| GO:0022604 | regulation of cell morphogenesis | 2.15 | 4.08e-05 | 2.14e-03 | 30/184 | EPB41; S100A13; CRABP2; CNTN2; ZEB2; MAP2; ARPC2; TWF2; ROBO1; TNIK; WDR1; MAP1B; SPARC; EZR; CAMK2B; NEFL; DPYSL2; OLFM1; CORO1C; MYCBP2; LINGO1; EFNB3; MAPT; NGFR; BAIAP2; MYL12B; EPB41L3; PREX1; CALR; RTN4R |
| GO:0021700 | developmental maturation | 2.15 | 3.51e-03 | 4.09e-02 | 15/92 | RERE; CNTN2; ID2; SIX3; CTNNB1; CAMK2B; NEFL; CDKN1C; GAL; KCNIP2; CD63; RAB3A; LGI4; SEZ6L; XBP1 |
| GO:0034330 | cell junction organization | 2.07 | 3.89e-03 | 4.46e-02 | 16/102 | RAB13; AGT; PKP4; IQSEC1; CTNNB1; WDR1; ANK2; GPM6B; CD151; CADM1; VCL; SLK; CORO1C; EPB41L3; MAPRE2; PVRL2 |
| GO:0002521 | leukocyte differentiation | 2.06 | 3.15e-03 | 3.90e-02 | 17/109 | PRKCZ; JUN; ID2; SATB1; CMTM7; CTNNB1; SOX4; BTN2A2; CHD7; TPD52; FAM213A; POU4F1; B2M; PREX1; KDELR1; XBP1; MFNG |
| GO:0001505 | regulation of neurotransmitter levels | 2.01 | 3.09e-03 | 3.90e-02 | 18/118 | PHGDH; SV2A; RGS2; AGT; CD47; CPLX1; SNCA; CPLX2; GRM4; GPM6B; SYT7; ASIC1; TRIM9; HSP90AA1; DOC2B; VAMP2; SNAP25; RAB3A |
CHD2 : 32 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 5.15 | 0.00e+00 | 0.00e+00 | 75/119 | RPL22; RPL11; RPS8; RPL5; RPS27; RYR2; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; SPCS1; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SEC61G; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; HSPA5; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS9; RPS5; RPL3 |
| GO:0014075 | response to amine | 4.90 | 4.46e-04 | 1.26e-02 | 6/10 | RGS2; HDAC2; RGS10; CALM1; NME1; SOD1 |
| GO:0010737 | protein kinase A signaling | 4.46 | 8.80e-04 | 2.15e-02 | 6/11 | RAB13; AKAP12; EZR; GAL; AKAP6; MIF |
| GO:0051339 | regulation of lyase activity | 4.08 | 6.28e-04 | 1.65e-02 | 7/14 | PARK7; HPCA; GNAI2; CAP2; AKAP9; GABBR2; PHPT1 |
| GO:0006413 | translational initiation | 4.06 | 0.00e+00 | 0.00e+00 | 78/157 | RPL22; RPL11; RPS8; RPL5; RPS27; DDX1; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; EIF1B; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; EIF3J; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS9; RPS5; EIF3L; RPL3 |
| GO:0002181 | cytoplasmic translation | 3.92 | 4.22e-14 | 5.16e-12 | 35/73 | RPL11; RPL31; RPL32; RPL15; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; EIF4EBP1; RPL30; EIF3E; EIF3H; RPL8; EIF3F; RPL41; RPL6; RPLP0; RPS29; EIF3J; RPLP1; RPL26; RPL19; RPL38; RPL17; EEF2; RPL36; RPS28; RPL18A; RPL18; RPL13A; EIF3L |
| GO:0010644 | cell communication by electrical coupling | 3.77 | 2.63e-03 | 4.54e-02 | 6/13 | ATP1B1; RYR2; FKBP1B; CAMK2D; SRI; CALM1 |
| GO:0010257 | NADH dehydrogenase complex assembly | 3.68 | 2.31e-10 | 2.43e-08 | 27/60 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFS6; NDUFA2; NDUFA5; NDUFB2; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFS8; NDUFC2; NDUFB8; NDUFB10; NDUFAB1; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND1; MT-ND3 |
| GO:0090150 | establishment of protein localization to membrane | 3.52 | 0.00e+00 | 0.00e+00 | 88/204 | RPL22; RPL11; HPCA; RPS8; RPL5; RPS27; ATP1B1; RPS27A; REEP1; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; CPLX1; NSG1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPL10A; RPS12; FIS1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPL8; RPS6; SEC61B; RPL7A; RPLP2; RPL27A; RPS13; TIMM10; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; CALM1; SRP14; RPL4; RPLP1; RPS17; RPS2; RAB26; RPS15A; RPL13; VAMP2; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RAB31; RPL17; ROMO1; RPS15; TIMM13; RPL36; RPS28; RPL18A; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS9; RPS5; RPL3 |
| GO:0021675 | nerve development | 3.34 | 6.59e-04 | 1.67e-02 | 9/22 | EPHB2; NRP2; CTNNB1; RPL24; CHD7; NTF3; POU4F1; SIX1; ADARB1 |
| GO:0021953 | central nervous system neuron differentiation | 3.03 | 2.36e-06 | 1.24e-04 | 20/54 | DRAXIN; EPHB2; LMO4; CNTN2; ZEB2; NRP2; MAP2; CTNNB1; ROBO1; UQCRQ; ID4; MDGA1; FZD1; MTPN; MYCBP2; POU4F1; CBLN1; MAPT; DCC; ADARB1 |
| GO:0014074 | response to purine-containing compound | 2.93 | 1.17e-04 | 4.09e-03 | 14/39 | SEPN1; JUN; RYR2; IGFBP5; SPARC; HDAC2; EZR; AKAP9; HSPA5; HSP90B1; AKAP6; NME1; BSG; SOD1 |
| GO:0050954 | sensory perception of mechanical stimulus | 2.93 | 1.17e-04 | 4.09e-03 | 14/39 | GPX1; WDR1; CASP3; FAM65B; SOBP; CHD7; NDUFB9; TIMM10; TIMM8B; SIX1; RPL38; TIMM13; RAB3A; SOD1 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 2.91 | 1.13e-08 | 9.21e-07 | 31/87 | NDUFS5; NDUFB3; NDUFAF3; COX17; NDUFB4; NDUFS6; NDUFA2; UQCC2; NDUFA5; NDUFB2; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFS8; NDUFC2; NDUFB8; NDUFB10; NDUFAB1; NDUFS7; NDUFA11; PET100; NDUFA7; NDUFB7; NDUFA13; NDUFA3; UQCR10; NDUFA6; MT-ND1; MT-ND3 |
| GO:0006401 | RNA catabolic process | 2.84 | 0.00e+00 | 0.00e+00 | 82/236 | RPL22; RPL11; YBX1; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; LSM3; RPL15; RPSA; RPL14; PCBP4; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; LSM5; LINC01420; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; EIF3E; RPL8; RPS6; RPL7A; RPLP2; RPL27A; RPS13; RNASEH2C; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SLIRP; RPL4; RPLP1; RPS17; RPS2; CARHSP1; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; CIRBP; RPS15; LSM7; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS9; RPS5; RPL3 |
| GO:2001023 | regulation of response to drug | 2.82 | 1.59e-03 | 3.65e-02 | 10/29 | PARK7; RGS2; AGT; NCOA1; SNCA; GNB2L1; TXN; SYT7; GAL; SNCG |
| GO:0006605 | protein targeting | 2.72 | 0.00e+00 | 0.00e+00 | 87/261 | RPL22; RPL11; HPCA; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; SPCS1; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; AKAP12; SEC61G; FIS1; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPL8; RPS6; SEC61B; RPL7A; RPLP2; RPL27A; RPS13; TIMM10; RPS3; TIMM8B; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; AKAP6; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; ROMO1; RPS15; TIMM13; RPL36; RPS28; UBL5; RPL18A; HOMER3; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS9; RPS5; RPL3 |
| GO:0046683 | response to organophosphorus | 2.64 | 1.70e-03 | 3.66e-02 | 11/34 | JUN; IGFBP5; SPARC; EZR; AKAP9; HSPA5; HSP90B1; AKAP6; NME1; BSG; SOD1 |
| GO:0003012 | muscle system process | 2.55 | 6.15e-08 | 4.52e-06 | 35/112 | ENO1; TPM3; LMNA; NOS1AP; ATP1B1; RGS2; AGT; RYR2; FKBP1B; MYL1; IGFBP5; CAMK2D; SORBS2; NDUFS6; COL4A3BP; MEF2C; HDAC2; CAMK2B; SRI; CALD1; MTPN; VCL; KCNIP2; MYL6; AKAP6; CALM1; ALDOA; HSBP1; SSTR2; MYL12A; MYL12B; NEDD4L; EEF2; SOD1; PDE9A |
| GO:0032409 | regulation of transporter activity | 2.52 | 6.29e-06 | 2.57e-04 | 25/81 | PARK7; EPHB2; SEPN1; HPCA; NOS1AP; ATP1B1; RYR2; FKBP1B; COX17; SNCA; CAMK2D; MEF2C; GLRX; SRI; AKAP9; TMSB4X; GAL; FXYD6; KCNIP2; JPH4; AKAP6; CALM1; VAMP2; NEDD4L; FXYD7 |
| GO:0002576 | platelet degranulation | 2.50 | 2.83e-03 | 4.54e-02 | 11/36 | TAGLN2; MANF; WDR1; SPARC; TMSB4X; VCL; CD63; CALM1; ALDOA; LGALS3BP; SOD1 |
| GO:1902600 | proton transmembrane transport | 2.44 | 2.68e-05 | 1.03e-03 | 23/77 | PARK7; COX5B; ATP5G3; COX17; ATP5I; COX7A2; NDUFA4; ATP5J2; ATP6V1F; ATP6V0E2; TMSB4X; COX7B; COX6C; COX8A; ATP5B; COX5A; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; COX6B1; ATP5J |
| GO:0010959 | regulation of metal ion transport | 2.39 | 3.73e-06 | 1.82e-04 | 29/99 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; FKBP1B; CTNNB1; GNAI2; SNCA; CAMK2D; GLRX; CAMK2B; SRI; AKAP9; CHD7; GAL; FXYD6; KCNIP2; CD63; JPH4; AKAP6; CALM1; B2M; VAMP2; NEDD4L; NKAIN4; HOMER3; FXYD7 |
| GO:0030534 | adult behavior | 2.36 | 2.13e-03 | 4.33e-02 | 13/45 | PARK7; CNTN2; ID2; NRXN1; SNCA; HDAC2; CHD7; PBX3; SNCG; EFNB3; LGI4; SEZ6L; CSTB |
| GO:0034765 | regulation of ion transmembrane transport | 2.30 | 2.03e-06 | 1.15e-04 | 33/117 | PARK7; EPHB2; SEPN1; HPCA; NOS1AP; ATP1B1; RGS2; AGT; RYR2; FKBP1B; SCN3A; SCN9A; COX17; WDR1; SNCA; CAMK2D; MEF2C; GLRX; SRI; AKAP9; TMSB4X; CHD7; GAL; FXYD6; KCNIP2; CD63; JPH4; AKAP6; CALM1; VAMP2; NEDD4L; CACNA1A; FXYD7 |
| GO:0031099 | regeneration | 2.29 | 9.57e-04 | 2.27e-02 | 16/57 | SEPN1; PTPRF; JUN; FKBP1B; GPX1; NREP; MTPN; NEFL; TNC; GSTP1; PGF; ENO3; RPL19; PHB; RPS16; RTN4R |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.29 | 1.81e-09 | 1.66e-07 | 53/189 | PARK7; ENO1; NDUFS5; UQCRH; ATP1B1; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; SNCA; NDUFS6; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFA5; NDUFB2; TMSB4X; NDUFA1; NDUFB9; NDUFB6; AK1; NDUFS8; NDUFC2; NDUFB8; TPI1; ATP5B; RAN; COX5A; NME3; NDUFB10; NDUFAB1; ALDOA; DCTPP1; ENO3; ATP5G1; NME1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; MT-ND1; MT-ND3 |
| GO:0019932 | second-messenger-mediated signaling | 2.13 | 1.96e-04 | 6.25e-03 | 24/92 | SEPN1; HPCA; NOS1AP; ATP1B1; RGS2; AGT; RYR2; FKBP1B; GNAI2; SELK; CAMK2D; HINT1; SRI; FIS1; NRG1; GAL; JPH4; AKAP6; CALM1; MAPT; GNAS; HOMER3; APLP1; PDE9A |
| GO:0034764 | positive regulation of transmembrane transport | 2.10 | 1.91e-03 | 4.00e-02 | 17/66 | PARK7; EPHB2; NOS1AP; ATP1B1; AGT; RYR2; CAPN10; COX17; SNCA; GLRX; SRI; AKAP9; TMSB4X; GAL; KCNIP2; AKAP6; CALM1 |
| GO:0009123 | nucleoside monophosphate metabolic process | 2.09 | 1.30e-07 | 8.69e-06 | 50/195 | PARK7; ENO1; NDUFS5; UQCRH; ATP1B1; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; SNCA; NDUFS6; UQCRQ; NDUFA2; UQCC2; COX7A2; HDDC2; NDUFA4; CYCS; ATP5J2; NDUFA5; NDUFB2; TMSB4X; NDUFA1; NDUFB9; NDUFB6; AK1; NDUFS8; NDUFC2; NDUFB8; TPI1; ATP5B; COX5A; NDUFB10; NDUFAB1; ALDOA; ENO3; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; MT-ND1; MT-ND3 |
| GO:0097485 | neuron projection guidance | 2.09 | 2.79e-04 | 8.20e-03 | 24/94 | DRAXIN; EPHB2; SHC1; EFNA3; LHX9; CNTN2; NRXN1; KIF5C; NRP2; ROBO1; RPL24; ALCAM; SPON2; PDLIM7; TUBB2B; EZR; ETV1; DPYSL2; SPTAN1; FEZ1; MYCBP2; EFNB3; DCC; BSG |
| GO:0043270 | positive regulation of ion transport | 2.04 | 2.70e-03 | 4.54e-02 | 17/68 | PARK7; EPHB2; NOS1AP; ATP1B1; AGT; RYR2; COX17; SNCA; GLRX; SRI; AKAP9; TMSB4X; GAL; KCNIP2; AKAP6; CALM1; MIF |
PTEN : 23 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 4.79 | 0.00e+00 | 0.00e+00 | 85/119 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RYR2; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; SPCS1; RPL24; RPL35A; RPL9; RPL34; ANK2; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SEC61G; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; HSPA5; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; KDELR1; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0010644 | cell communication by electrical coupling | 4.12 | 1.52e-04 | 4.65e-03 | 8/13 | ATP1B1; RYR2; FKBP1B; DBN1; SRI; ANK3; CALM1; HRC |
| GO:0007218 | neuropeptide signaling pathway | 4.02 | 1.33e-03 | 2.78e-02 | 6/10 | PCSK1N; GAL; SCG5; NMB; SSTR2; PTH2 |
| GO:0006413 | translational initiation | 3.88 | 0.00e+00 | 0.00e+00 | 91/157 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; MCTS1; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; DENR; RPL21; RPS29; EIF3J; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; EIF2S2; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| GO:0002181 | cytoplasmic translation | 3.85 | 0.00e+00 | 0.00e+00 | 42/73 | RPL11; RPL31; RPL32; RPL15; TMA7; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; MCTS1; EIF4EBP1; RPL30; EIF3E; EIF3H; RPL8; EIF3F; EIF3M; RPS26; RPL41; RPL6; RPLP0; DENR; RPS29; EIF3J; RPLP1; RPL26; RPL19; RPL38; RPL17; RPS21; EEF2; RPL36; RPS28; RPL18A; RPL18; RPL13A; EIF3D; EIF3L |
| GO:0051339 | regulation of lyase activity | 3.35 | 2.11e-03 | 4.07e-02 | 7/14 | PARK7; HPCA; GNAI2; CAP2; AKAP9; GABBR2; PHPT1 |
| GO:0010257 | NADH dehydrogenase complex assembly | 3.24 | 7.47e-10 | 7.84e-08 | 29/60 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFC1; NDUFS6; NDUFA2; NDUFA5; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFA8; NDUFC2; NDUFB8; NDUFA12; NDUFB1; NDUFAB1; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND1; MT-ND3; MT-ND4; MT-ND6 |
| GO:0090150 | establishment of protein localization to membrane | 3.19 | 0.00e+00 | 0.00e+00 | 97/204 | RPL22; RPL11; HPCA; RPS8; RPL5; RPS27; ATP1B1; SRP9; RPS7; RPS27A; REEP1; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; CPLX1; NSG1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPS10; RPL10A; CD24; RPS12; FIS1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; TIMM10; RPS3; RPS25; ANK3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; CALM1; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; YWHAE; VAMP2; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RAB31; RPL17; ROMO1; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 2.77 | 1.50e-09 | 1.38e-07 | 36/87 | NDUFS5; COX20; NDUFB3; NDUFAF3; COX17; NDUFB4; NDUFC1; NDUFS6; NDUFA2; UQCC2; NDUFA5; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFA8; NDUFC2; NDUFB8; COX14; NDUFA12; NDUFB1; NDUFAB1; COA3; NDUFA11; PET100; NDUFA7; NDUFB7; NDUFA13; NDUFA3; UQCR10; NDUFA6; MT-ND1; MT-ND3; MT-ND4; MT-ND6 |
| GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | 2.72 | 3.63e-04 | 9.54e-03 | 13/32 | ATP6V0B; ATP2B4; ATP5G3; ATP6V1F; ATP6V0E2; ATP5L; ATP5G2; ATP5B; ATP2B1; ATP5G1; ATP5H; ATP5A1; ATP5E |
| GO:1902600 | proton transmembrane transport | 2.70 | 4.58e-08 | 3.16e-06 | 31/77 | PARK7; ATP6V0B; COX5B; ATP5G3; COX17; ATP5I; COX7C; COX7A2; NDUFA4; ATP5J2; ATP6V1F; ATP6V0E2; TMSB4X; COX7B; COX6C; ATP6V1G1; COX8A; ATP5L; ATP5G2; ATP5B; COX6A1; COX5A; ATP5G1; ATP5H; ATP5A1; ATP5E; COX6B1; ATP5J; MT-CO2; MT-CO3; MT-ND4 |
| GO:0035637 | multicellular organismal signaling | 2.65 | 6.94e-05 | 2.43e-03 | 17/43 | ATP1B1; ATP2B4; AGT; RYR2; FKBP1B; SCN3A; SCN9A; CHRNA1; ANK2; SRI; AKAP9; SCN3B; ANK3; ATP2B1; CALM1; YWHAE; HRC |
| GO:0032409 | regulation of transporter activity | 2.65 | 4.74e-08 | 3.16e-06 | 32/81 | PARK7; SEPN1; HPCA; NOS1AP; ATP1B1; RYR2; FKBP1B; COX17; SNCA; ANK2; MEF2C; GLRX; ACTB; SRI; AKAP9; TMSB4X; GAL; FXYD6; SCN3B; KIF5B; ANK3; KCNIP2; REM2; JPH4; AKAP6; CALM1; YWHAE; VAMP2; NEDD4L; FKBP1A; FXYD7; HRC |
| GO:0006401 | RNA catabolic process | 2.61 | 0.00e+00 | 0.00e+00 | 92/236 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; LSM3; RPL15; RPSA; RPL14; PCBP4; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; RPS18; RPS10; RPL10A; RPS12; LSM5; LINC01420; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; YWHAZ; EIF3E; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; MAGOHB; RPS26; RPL41; CNOT2; RPL6; RPLP0; RPL21; RPS29; SLIRP; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; NT5C3B; RPL27; RPL38; EIF4A3; RPL17; RPS21; RPS15; LSM7; RPL36; RPS28; RPL18A; LSM4; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006605 | protein targeting | 2.54 | 0.00e+00 | 0.00e+00 | 99/261 | RPL22; RPL11; ATPIF1; HPCA; RPS8; RPL5; RPS27; SRP9; TOMM20; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; SPCS1; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; AKAP12; SEC61G; FIS1; SLC25A6; TIMM17B; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; IMMP1L; TIMM10; RPS3; TIMM8B; RPS25; ANK3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; AKAP6; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; ROMO1; RPS21; RPS15; RPL36; RPS28; UBL5; RPL18A; UBA52; HOMER3; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0021953 | central nervous system neuron differentiation | 2.48 | 4.91e-05 | 1.90e-03 | 20/54 | DRAXIN; LMO4; CNTN2; NRP2; MAP2; ROBO1; UQCRQ; ID4; MDGA1; HSP90AB1; FZD1; MTPN; NFIB; C12orf57; MYCBP2; POU4F1; CBLN1; MAPT; DCC; ADARB1 |
| GO:0010959 | regulation of metal ion transport | 2.37 | 2.90e-07 | 1.77e-05 | 35/99 | SEPN1; NKAIN1; HPCA; NOS1AP; ATP1B1; ATP2B4; AGT; RYR2; FKBP1B; GNAI2; SNCA; ANK2; GLRX; SRI; AKAP9; CHD7; GAL; FXYD6; SCN3B; KIF5B; ANK3; KCNIP2; CD63; REM2; JPH4; AKAP6; CALM1; YWHAE; VAMP2; NEDD4L; FKBP1A; NKAIN4; HOMER3; FXYD7; HRC |
| GO:0034765 | regulation of ion transmembrane transport | 2.23 | 3.60e-07 | 1.89e-05 | 39/117 | PARK7; SEPN1; HPCA; NOS1AP; ATP1B1; RGS2; ATP2B4; AGT; RYR2; FKBP1B; SCN3A; SCN9A; COX17; SNCA; ANK2; MEF2C; GLRX; SRI; AKAP9; TMSB4X; CHD7; GAL; FXYD6; SCN3B; KIF5B; ANK3; KCNIP2; CD63; REM2; JPH4; AKAP6; CALM1; YWHAE; VAMP2; NEDD4L; FKBP1A; CACNA1A; FXYD7; HRC |
| GO:0072507 | divalent inorganic cation homeostasis | 2.21 | 2.63e-06 | 1.21e-04 | 34/103 | SEPN1; SV2A; ATP1B1; ATP2B4; AGT; RYR2; FKBP1B; SELK; SNCA; ANK2; CD24; SRI; FIS1; CHD7; GNG3; ANK3; ATP2B1; HSP90B1; HMGB1; TPT1; JPH4; AKAP6; CALM1; NMB; MT1F; MT1X; YWHAE; EIF5A; FKBP1A; CALR; CACNA1A; HRC; KCTD17; SMDT1 |
| GO:0019932 | second-messenger-mediated signaling | 2.11 | 3.84e-05 | 1.56e-03 | 29/92 | SEPN1; HPCA; NOS1AP; ATP1B1; RGS2; ATP2B4; AGT; RYR2; FKBP1B; NEUROD1; GNAI2; SELK; ANK2; HINT1; DDAH2; SRI; FIS1; NRG1; NCALD; GAL; PEBP1; JPH4; AKAP6; CALM1; MAPT; FKBP1A; HOMER3; APLP1; HRC |
| GO:0009914 | hormone transport | 2.06 | 1.19e-04 | 3.79e-03 | 27/88 | PARK7; SLC16A1; MPC2; AGT; FKBP1B; NEUROD1; CAPN10; CPLX1; UQCC2; HMGN3; SRI; SLC25A6; SLC25A5; CHD7; ENY2; PHPT1; SYT7; GAL; KIF5B; DYNLL1; SCG5; NMB; DOC2B; VAMP2; MIDN; CACNA1A; SELM |
| GO:0034764 | positive regulation of transmembrane transport | 2.03 | 1.06e-03 | 2.50e-02 | 20/66 | PARK7; NOS1AP; ATP1B1; AGT; RYR2; CAPN10; COX17; SNCA; ANK2; GLRX; SRI; AKAP9; TMSB4X; GAL; KIF5B; ANK3; KCNIP2; OSBPL8; AKAP6; CALM1 |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.02 | 4.11e-08 | 3.16e-06 | 57/189 | PARK7; ATPIF1; NDUFS5; ATP1B1; COX5B; ATP5G3; NDUFB3; IMPDH2; NDUFB4; ATP5I; SNCA; NDUFC1; NDUFS6; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFA5; TMSB4X; NDUFA1; NDUFB9; NDUFB6; NDUFA8; NDUFC2; ATP5L; PGAM1; NDUFB8; ENO2; ATP5G2; ATP5B; NDUFA12; COX6A1; NDUFB1; COX5A; NME3; NDUFAB1; DCTPP1; ENO3; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; MT-ND1; MT-CO2; MT-CO3; MT-ND3; MT-ND4 |
SETD5 : 25 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 6.62 | 0.00e+00 | 0.00e+00 | 83/119 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RTN4; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; SPCS1; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; RPL35; HSPA5; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; KDELR1; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006413 | translational initiation | 5.56 | 0.00e+00 | 0.00e+00 | 92/157 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; DDX1; RPS27A; EIF5B; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; EIF3A; RPL41; RPL6; RPLP0; RPL21; RPS29; EIF3J; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; EIF2S2; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3; ATF4 |
| GO:0002181 | cytoplasmic translation | 4.94 | 0.00e+00 | 0.00e+00 | 38/73 | RPL11; RPL31; RPL32; RPL15; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; EIF4EBP1; RPL30; EIF3E; EIF3H; RPL8; EIF3M; EIF3A; RPL41; RPL6; RPLP0; RPS29; EIF3J; RPLP1; RPL26; RPL19; RPL38; RPL17; RPS21; EEF2; RPL36; RPS28; RPL18A; RPL18; RPL13A; EIF3D; EIF3L |
| GO:0090150 | establishment of protein localization to membrane | 4.14 | 0.00e+00 | 0.00e+00 | 89/204 | RPL22; RPL11; RPS8; RPL5; RPS27; ATP1B1; CNST; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPS10; RPL10A; RPS12; FIS1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; TIMM10; BAD; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; ROMO1; RPS21; RPS15; TIMM13; RPL36; RPS28; RPL18A; UBA52; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0010257 | NADH dehydrogenase complex assembly | 4.11 | 4.75e-11 | 3.44e-09 | 26/60 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFA2; NDUFB2; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFA8; NDUFS8; NDUFC2; NDUFB1; NDUFB10; NDUFAB1; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND2; MT-ND3 |
| GO:0006401 | RNA catabolic process | 3.90 | 0.00e+00 | 0.00e+00 | 97/236 | RPL22; RPL11; YBX1; RPS8; SERBP1; RPL5; RPS27; RPS7; RPS27A; RPL31; SSB; RPL37A; RPL32; LSM3; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; HNRNPD; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; RPS18; RPS10; RPL10A; RPS12; LSM5; RPS4X; RPL36A; RPL39; RPL10; DKC1; RPS20; RPL7; RPL30; PABPC1; EIF3E; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RNASEH2C; RPS3; RPS25; HSPA8; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; HNRNPC; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; DDX5; RPL38; RPL17; RPS21; CIRBP; RPS15; LSM7; RPL36; RPS28; HNRNPM; C19orf43; RPL18A; LSM4; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006959 | humoral immune response | 3.56 | 4.55e-04 | 1.45e-02 | 9/24 | HMGN2; SPON2; RPL39; RPL30; SERPING1; C1QBP; PHB; ROMO1; RPS19 |
| GO:0006605 | protein targeting | 3.53 | 0.00e+00 | 0.00e+00 | 97/261 | RPL22; RPL11; ATPIF1; RPS8; RPL5; RPS27; TOMM20; RPS7; RPS27A; RPL31; HSPD1; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; SPCS1; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; LARS; RPS14; RPS18; RPS10; RPL10A; RPS12; FIS1; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; IMMP1L; TIMM10; RPS3; TIMM8B; RPS25; HSPA8; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; AKAP6; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; ROMO1; RPS21; RPS15; TIMM13; RPL36; RPS28; UBL5; RPL18A; UBA52; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 3.49 | 5.15e-11 | 3.44e-09 | 32/87 | NDUFS5; COX20; NDUFB3; NDUFAF3; COX17; NDUFB4; NDUFA2; UQCC2; NDUFB2; NDUFB11; NDUFA1; UQCRB; NDUFB9; TMEM261; NDUFB6; NDUFA8; NDUFS8; NDUFC2; NDUFB1; NDUFB10; NDUFAB1; COA3; NDUFS7; NDUFA11; PET100; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND2; MT-ND3 |
| GO:0046683 | response to organophosphorus | 3.35 | 1.02e-04 | 4.00e-03 | 12/34 | JUN; HSPD1; EZR; HSPA5; RAPGEF1; LDHA; HSP90B1; AKAP6; NME1; BSG; JUND; SOD1 |
| GO:0014074 | response to purine-containing compound | 3.16 | 1.04e-04 | 4.00e-03 | 13/39 | SEPN1; JUN; HSPD1; EZR; HSPA5; RAPGEF1; LDHA; HSP90B1; AKAP6; NME1; BSG; JUND; SOD1 |
| GO:1902600 | proton transmembrane transport | 3.08 | 1.25e-07 | 5.74e-06 | 25/77 | PARK7; COX7A2L; COX5B; ATP5G3; COX17; COX7C; COX7A2; NDUFA4; ATP5J2; ATP6V1F; COX7B; COX6C; COX8A; ATP5L; COX5A; COX4I1; ATP5G1; ATP5H; CYB5A; ATP5E; ATP5D; NDUFS7; COX6B1; ATP5J; MT-CYB |
| GO:0001906 | cell killing | 3.05 | 1.64e-03 | 4.29e-02 | 9/28 | HMGN2; PRDX1; HSP90AB1; RPL30; BAD; CADM1; EMP2; ROMO1; RPS19 |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.81 | 1.04e-13 | 1.09e-11 | 56/189 | PARK7; ENO1; ATPIF1; NDUFS5; UQCRH; ATP1B1; COX7A2L; COX5B; ATP5G3; NDUFB3; IMPDH2; NDUFB4; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; NDUFA1; UQCRB; NDUFB9; NDUFB6; NDUFA8; LDHA; BAD; NDUFS8; NDUFC2; ATP5L; HSPA8; DDIT4; RAN; NDUFB1; COX5A; NME4; NDUFB10; NDUFAB1; DCTPP1; COX4I1; ENO3; ATP5G1; NME1; ATP5H; ATP5E; ATP5D; NDUFS7; NDUFA7; NDUFB7; NDUFA3; NDUFA6; ATP5J; MT-ND2; MT-ND3; MT-CYB |
| GO:0009123 | nucleoside monophosphate metabolic process | 2.63 | 6.01e-12 | 5.52e-10 | 54/195 | PARK7; ENO1; ATPIF1; NDUFS5; UQCRH; ATP1B1; COX7A2L; COX5B; ATP5G3; NDUFB3; IMPDH2; NDUFB4; PAICS; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; HDDC2; NDUFA4; CYCS; ATP5J2; NDUFB2; NDUFA1; UQCRB; NDUFB9; NDUFB6; NDUFA8; LDHA; BAD; NDUFS8; NDUFC2; ATP5L; HSPA8; DDIT4; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ENO3; ATP5G1; ATP5H; ATP5E; ATP5D; NDUFS7; NDUFA7; NDUFB7; NDUFA3; NDUFA6; ATP5J; MT-ND2; MT-ND3; MT-CYB |
| GO:0032103 | positive regulation of response to external stimulus | 2.47 | 1.52e-03 | 4.14e-02 | 13/50 | PARK7; AGT; FKBP1B; HSPD1; TUBB2B; FAM65B; NTF3; HMGB1; C1QBP; CALR; RPS19; MIF; DDT |
| GO:0016072 | rRNA metabolic process | 2.33 | 7.38e-08 | 3.61e-06 | 42/171 | RPL11; RPS8; RPL5; RPS27; RPS7; WDR43; NOP58; NCL; RPSA; RPL14; RPL35A; LYAR; RPS14; NHP2; RPL10A; GTF2H5; DKC1; RPL7; RPS6; RPL35; RPL7A; TRMT112; DDX21; RPS24; NPM3; PA2G4; RPS17; RPS2; RPL26; RPL27; MAPT; RPS21; RPS15; RPS28; C19orf43; RPS16; FBL; RPS19; CD3EAP; GLTSCR2; RPS9; SNU13 |
| GO:0031099 | regeneration | 2.33 | 1.84e-03 | 4.51e-02 | 14/57 | SEPN1; PTPRF; JUN; FKBP1B; GPX1; GAP43; NEFL; TNC; GSTP1; ENO3; RPL19; PHB; RPS16; MT-CYB |
| GO:0072593 | reactive oxygen species metabolic process | 2.32 | 1.60e-04 | 5.60e-03 | 21/86 | PARK7; ATPIF1; PRDX1; PRDX6; AGT; HSPD1; GPX1; DDAH2; HSP90AB1; PRDX4; PRDX5; GSTP1; DDIT4; SH3PXD2A; EIF5A; MAPT; ROMO1; PRDX2; NDUFA13; SOD1; MT-ND2 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 2.31 | 3.37e-08 | 1.77e-06 | 45/185 | MRPL20; RPL11; RPL5; RPS27; SNRPE; DDX1; SNRPG; LSM3; RPSA; RPL24; RPS23; RPS14; NPM1; RPS10; HSP90AB1; RBMX; RPL10; EIF3E; EIF3H; RPL12; EIF3M; EIF3A; SNRPF; RPL6; RPLP0; EIF3J; C1QBP; RPL23A; MAPT; RPL38; SNRPD1; SNRPB; CIRBP; RPS15; RPS28; LSM4; RPS19; SNRPD2; GLTSCR2; RPL13A; RPS5; EIF3D; EIF3L; RPL3; SNU13 |
| GO:0009259 | ribonucleotide metabolic process | 2.22 | 8.25e-10 | 5.05e-08 | 60/257 | PARK7; ENO1; ATPIF1; NDUFS5; UQCRH; ATP1B1; COX7A2L; COX5B; DBI; ATP5G3; NDUFB3; IMPDH2; NDUFB4; PAICS; COX7C; HINT1; UQCRQ; NDUFA2; UQCC2; COX7A2; ELOVL4; NDUFA4; CYCS; ATP5J2; NDUFB2; NDUFA1; UQCRB; NDUFB9; NDUFB6; NDUFA8; LDHA; BAD; NDUFS8; NDUFC2; ATP5L; HSPA8; DDIT4; SCD; RAN; NDUFB1; COX5A; NME4; NDUFB10; NDUFAB1; COX4I1; ENO3; ATP5G1; NME1; ATP5H; ATP5E; ATP5D; NDUFS7; NDUFA7; NDUFB7; NDUFA3; NDUFA6; ATP5J; MT-ND2; MT-ND3; MT-CYB |
| GO:0022613 | ribonucleoprotein complex biogenesis | 2.19 | 6.82e-12 | 5.57e-10 | 76/330 | MRPL20; RPL11; RPS8; RPL5; RPS27; SNRPE; RPS7; DDX1; WDR43; SNRPG; NOP58; LSM3; RPSA; RPL14; RPL24; RPL35A; LYAR; RPS23; RPS14; NPM1; NOP16; NHP2; PAK1IP1; RPS10; RPL10A; HSP90AB1; GTF2H5; RBMX; RPL10; DKC1; RPL7; EIF3E; EIF3H; RPS6; RPL35; RPL12; RPL7A; EIF3M; TRMT112; DDX21; RPS24; NPM3; EIF3A; PA2G4; SNRPF; RPL6; RPLP0; RAN; EIF3J; RPS17; RPS2; C1QBP; RPL26; RPL23A; RPL27; MAPT; RPL38; SNRPD1; SNRPB; RPS21; CIRBP; RPS15; RPS28; LSM4; RPS16; FBL; RPS19; SNRPD2; GLTSCR2; RPL13A; RPS9; RPS5; EIF3D; EIF3L; RPL3; SNU13 |
| GO:0001101 | response to acid chemical | 2.18 | 2.08e-04 | 6.94e-03 | 23/100 | FABP3; LAMTOR5; LAMTOR2; TOMM20; SST; HNRNPD; CASP3; LARS; ASNS; LAMTOR4; FIS1; TNC; BAD; CFL1; GSTP1; SCD; SERPINF1; NME1; BAIAP2; EEF2; NDUFA13; TEAD2; XBP1 |
| GO:0006091 | generation of precursor metabolites and energy | 2.18 | 4.25e-09 | 2.40e-07 | 57/248 | PARK7; ENO1; ATPIF1; NDUFS5; UQCRH; COX20; COX7A2L; COX5B; NDUFB3; COX17; NDUFB4; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; COX7B; NDUFA1; UQCRB; COX6C; NDUFB9; NDUFB6; NDUFA8; LDHA; COX8A; NDUFS8; NDUFC2; ATP5L; CISD1; DDIT4; HMGB1; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ENO3; ATP5H; CYB5A; ATP5E; ATP5D; NDUFS7; UQCR11; NDUFA7; NDUFB7; COX6B1; GLTSCR2; NDUFA3; NDUFA6; ATP5J; MT-ND2; MT-ND3; MT-CYB |
| GO:0001819 | positive regulation of cytokine production | 2.09 | 9.88e-04 | 2.90e-02 | 20/91 | PARK7; AGT; DDX1; HSPD1; SELK; SPON2; HMGB2; AGPAT2; RPS3; CADM1; DDX21; DDIT3; HMGB1; C1QBP; GLTSCR2; MIF; DDT; XBP1; ATF4; SOD1 |
| Marker | ADNP | CHD2 | PTEN | SETD5 |
| Signif_GO_terms | 69 | 32 | 23 | 25 |