Guided Factor Analysis on LUHMES CROP-seq Data
-- Deviance residual transformed + 4 covariates corrected
Yifan Zhou (zhouyf@uchicago.edu)
2021-11-02
1 Single cell expression data
Source:
High-throughput single-cell functional elucidation of neurodevelopmental disease-associated genes reveals convergent mechanisms altering neuronal differentiation, GEO accession: GSE142078.
Perturbations:
CRISPR knock-down of 14 autism spectrum disorder (ASD)–associated genes (3 gRNAs per gene) + 5 non-targeting gRNAs.
Cells:
Lund human mesencephalic (LUHMES) neural progenitor cell line.
Cells from 3 batches were merged together into 1 analysis. All cells have only a single type of gRNA readout. Quality control resulted in 8708 cells.
Genes:
Top 6000 genes ranked by the multinomial deviance statistics were kept.
Normalization:
Deviance residual transformation.
Batch effect, unique UMI count, library size, and mitochondria percentage were all corrected for. The corrected and scaled expression data were used as input for subsequent factor analysis.
Here, our "guide", \(G\) matrix, consists of 15 types (14 genes + NTC) of gene-level knock-down conditions across cells.
In each case, Gibbs sampling was conducted for 3000 iterations, and the posterior mean estimates were averaged over the last 1000 iterations. SVD Initialization.
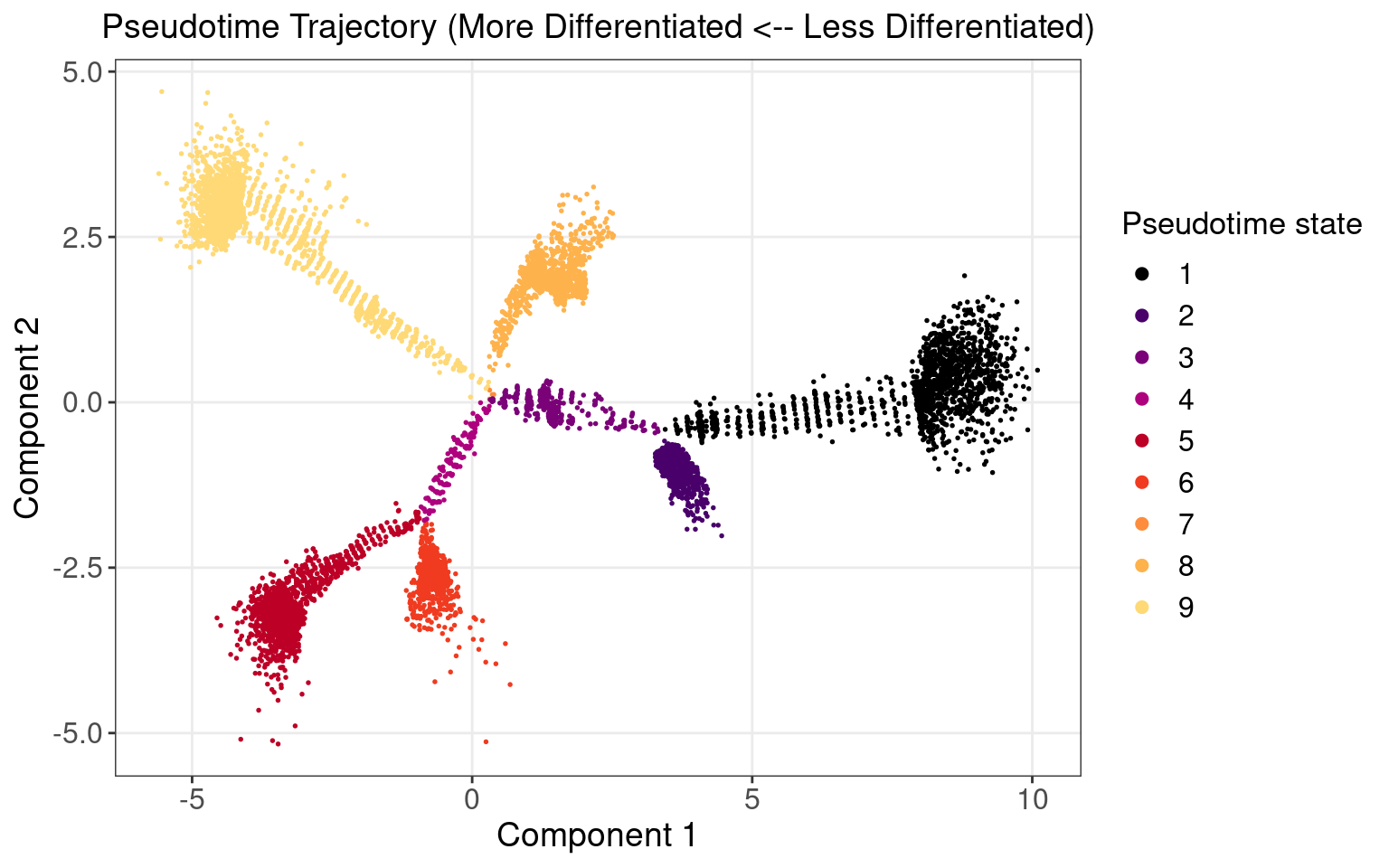
1.1 Monocle Pseudotime Trajectory
We attempted to reproduce the pseudotime trajectory of cells as reported in the original paper.
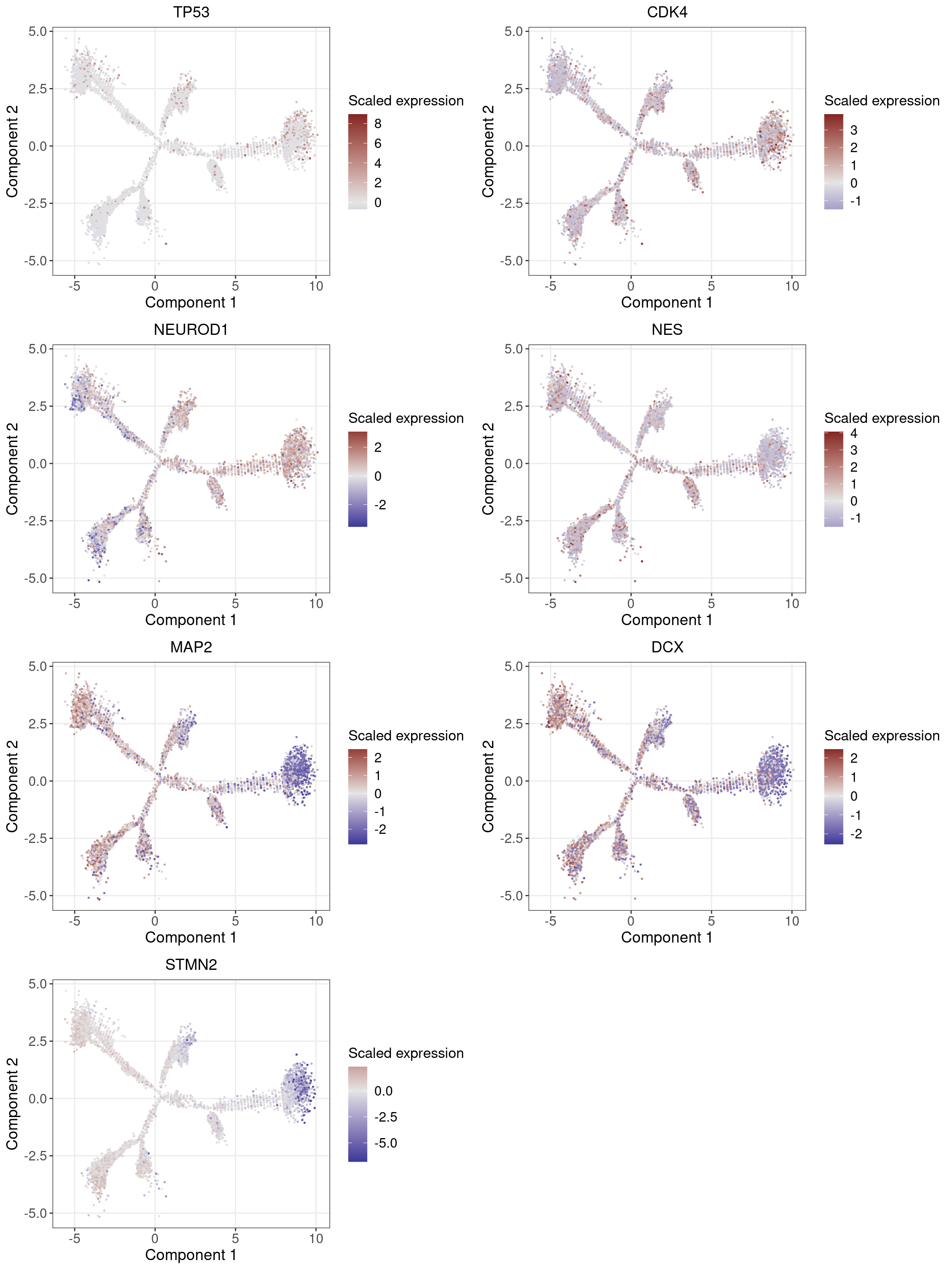
1.2 Marker Gene Expression in Pseudotime
Neuronal markers (MAP2 and DCX) increase along the pseudotime trajectory, whereas progenitor markers (TP53 and CDK4) decrease, consistent with Fig 3B of original paper.
Neuron progenitor (NPC): TP53, CDK4, NEUROD1, NES?
Post-mitotic neuron: MAP2, DCX, STMN2
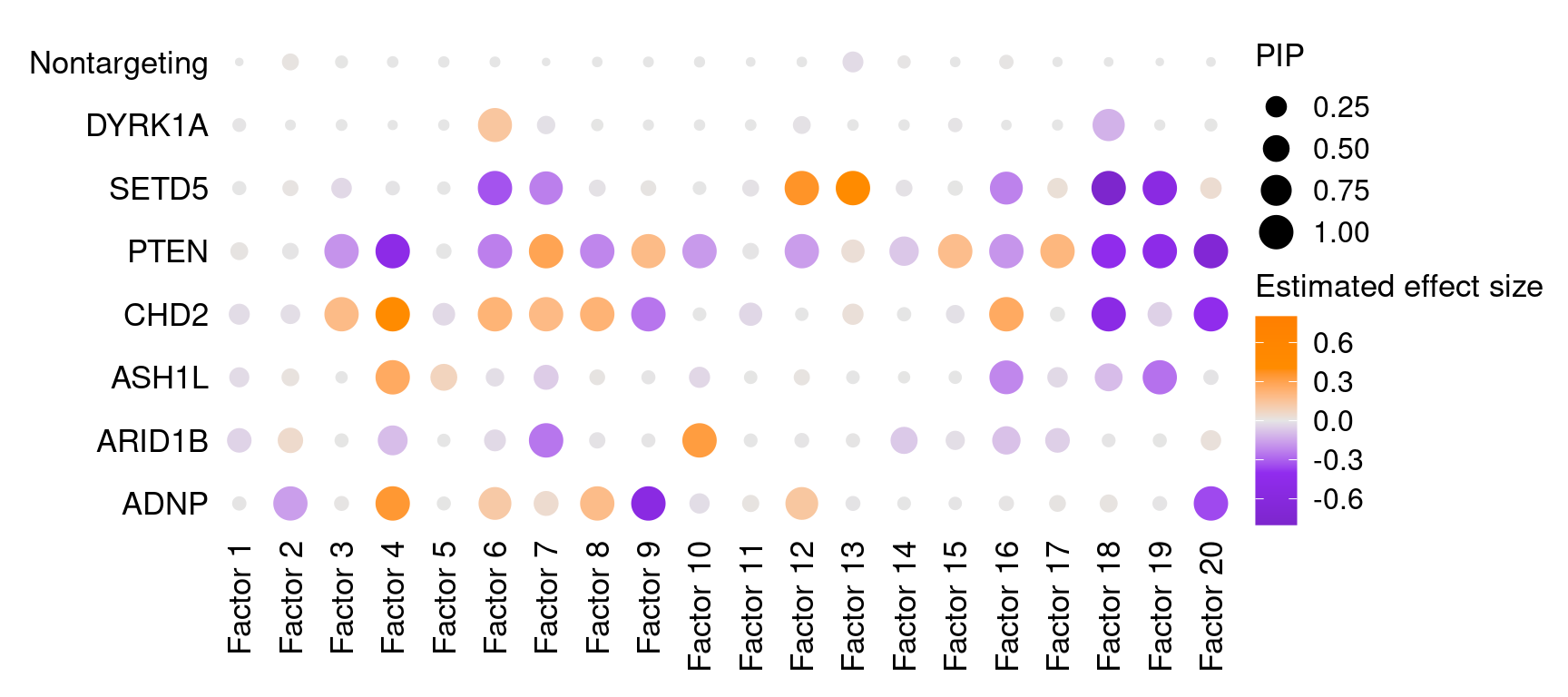
2 Factor ~ Perturbation Association
2.1 Estimate of Factor ~ Perturbation Associations (\(\beta\))
2.2 Factor ~ Perturbation Association P Values
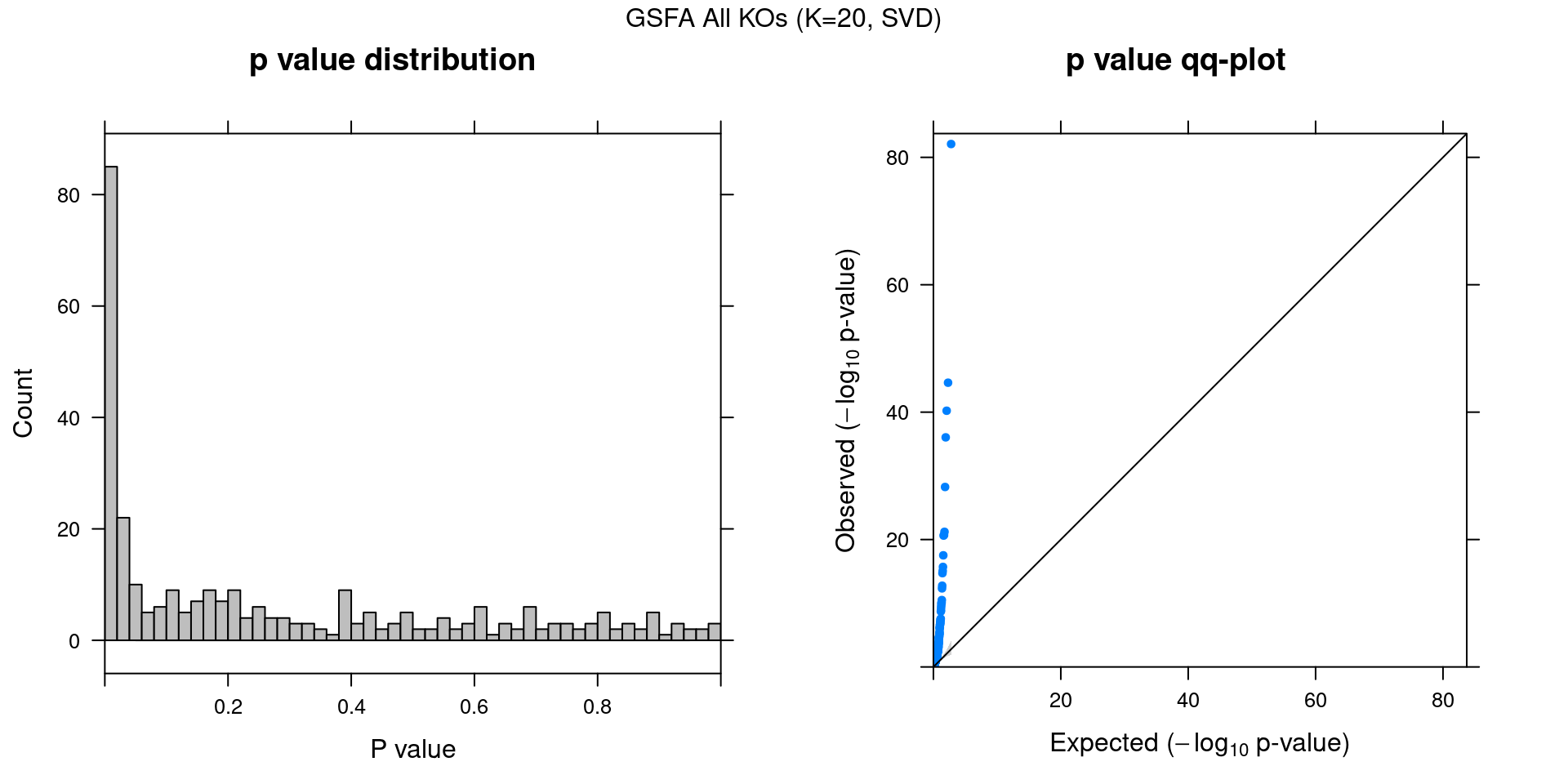
2.2.1 Beta PIP vs P-Value
2.2.2 Correlation btw Factors
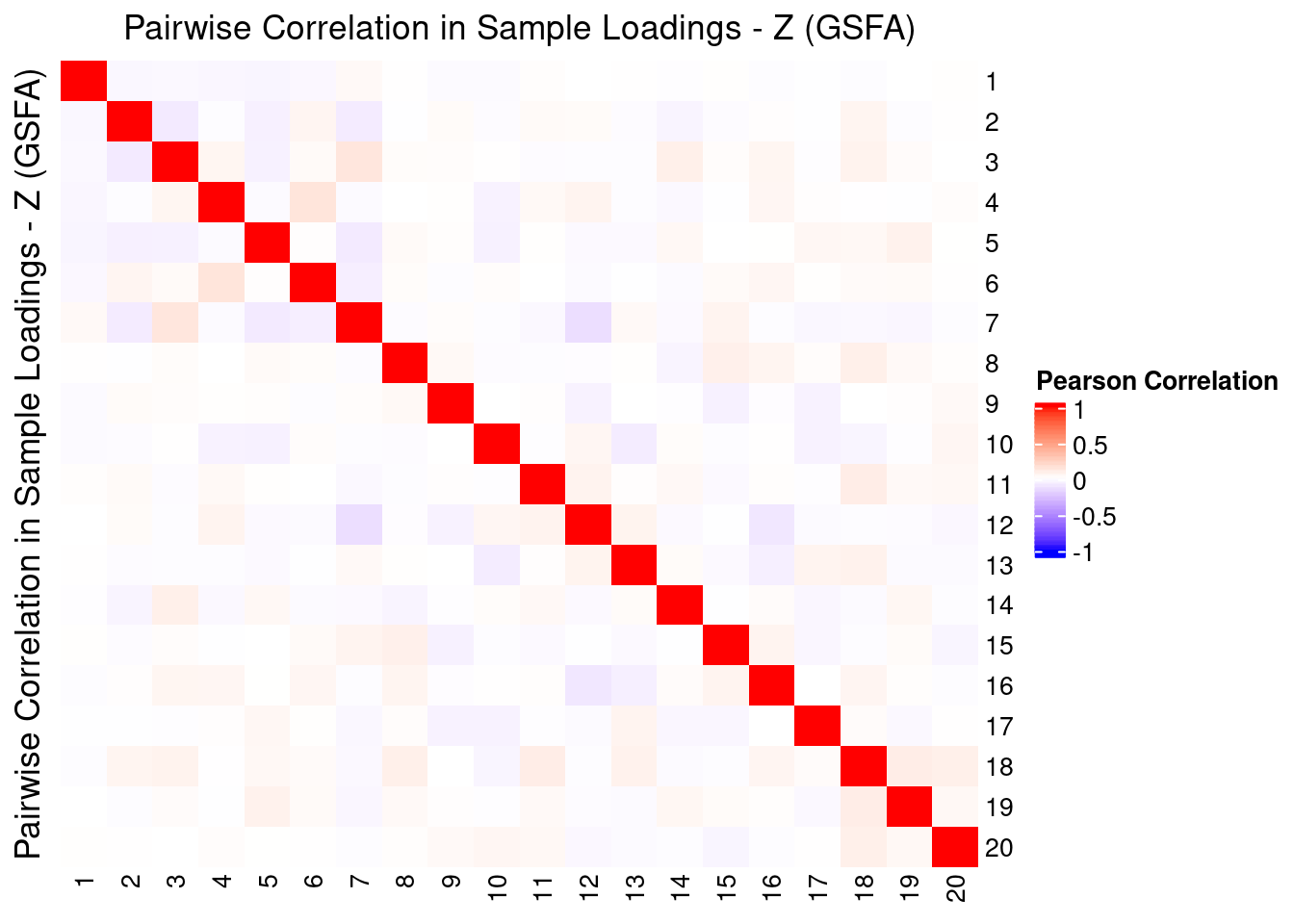
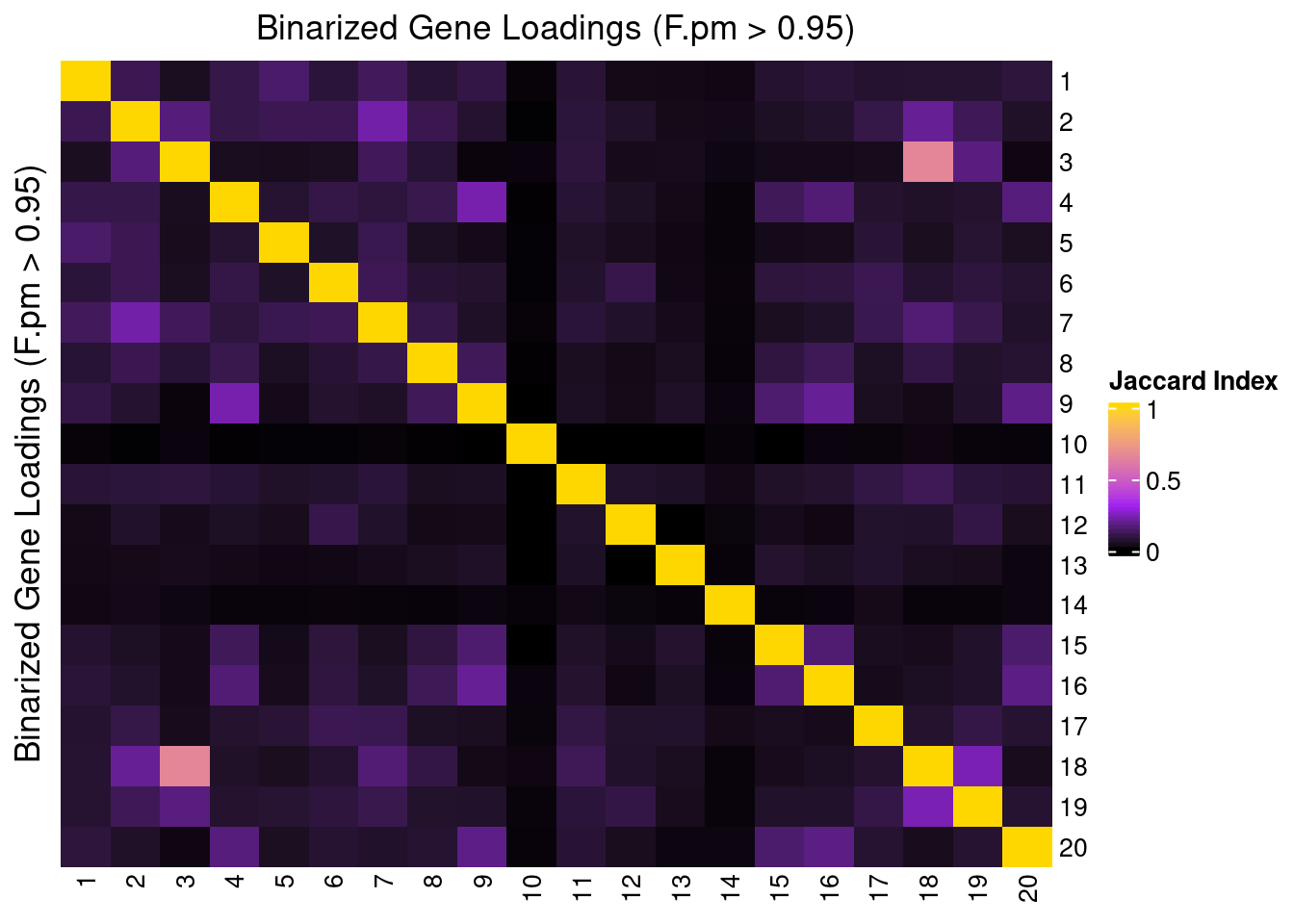
3 Factor Interpretation
3.1 Factor by gene loading
Neuronal cell markers obtained from papers and Neuronal and Glial Cell Marker Atlas:
| protein_name | gene_name | type |
|---|---|---|
| Nestin | NES | Radial Glia |
| Tenascin C | TNC | Radial Glia |
| TP53 | TP53 | NPC |
| CDK4 | CDK4 | NPC |
| NeuroD1 | NEUROD1 | Immature Neurons |
| Doublecortin | DCX | Immature, Post-mitotic |
| STMN2 | STMN2 | Post-mitotic |
| MAP2 | MAP2 | Mature, Post-mitotic |
| NeuN | RBFOX3 | Mature Neurons |
| Neurofilament-L | NEFL | Mature Neurons |
| Neurofilament-M | NEFM | Mature Neurons |
| UCHL1 | UCHL1 | Mature Neurons |
| VGLUT2 | SLC17A6 | Glutamate, Excitatory |
| GIRK2 | KCNJ6 | Dopamine, Inhibitory |
| Somatostatin | SST | GABA, Inhibitory |
| EBF1 | EBF1 | Cortical |
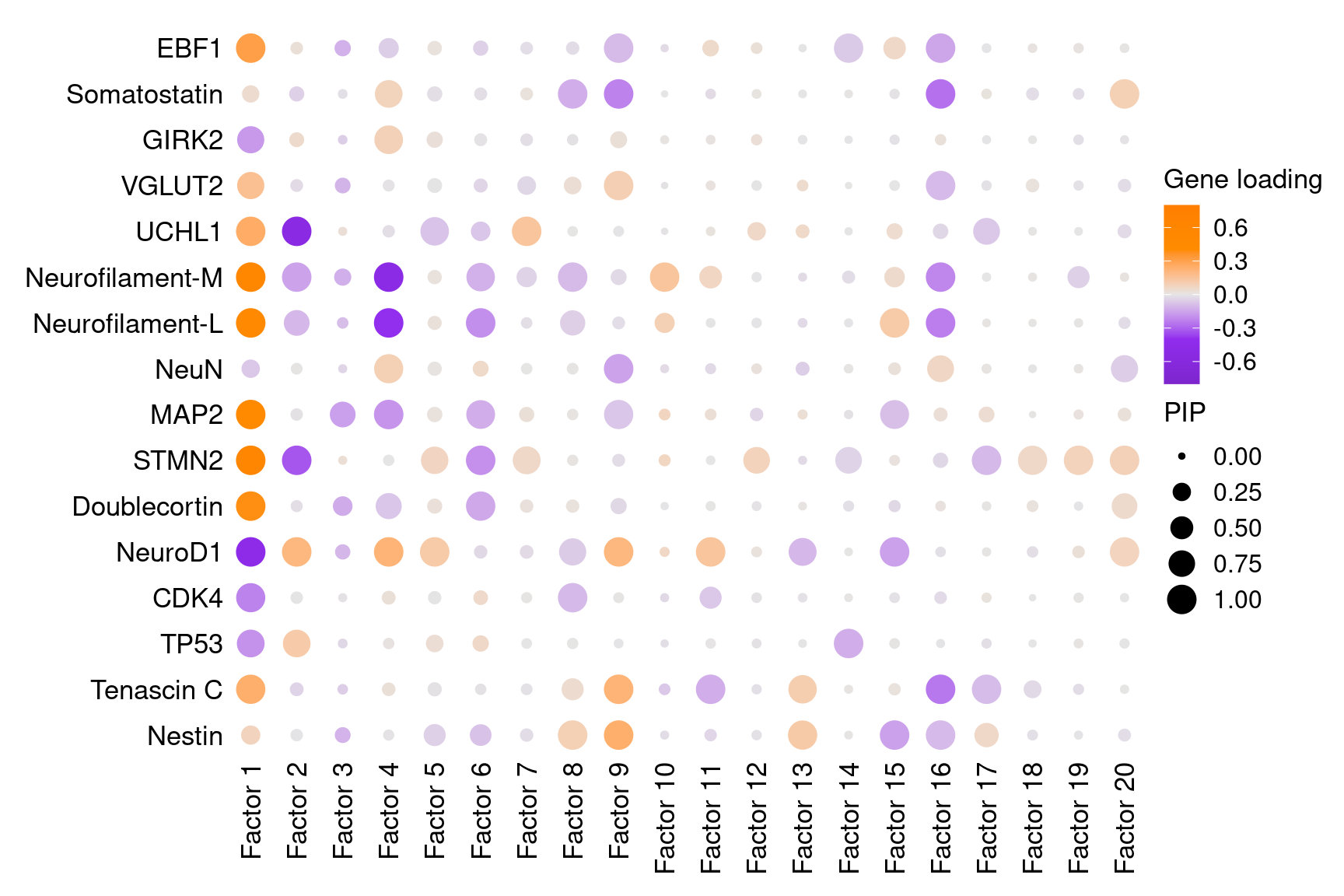
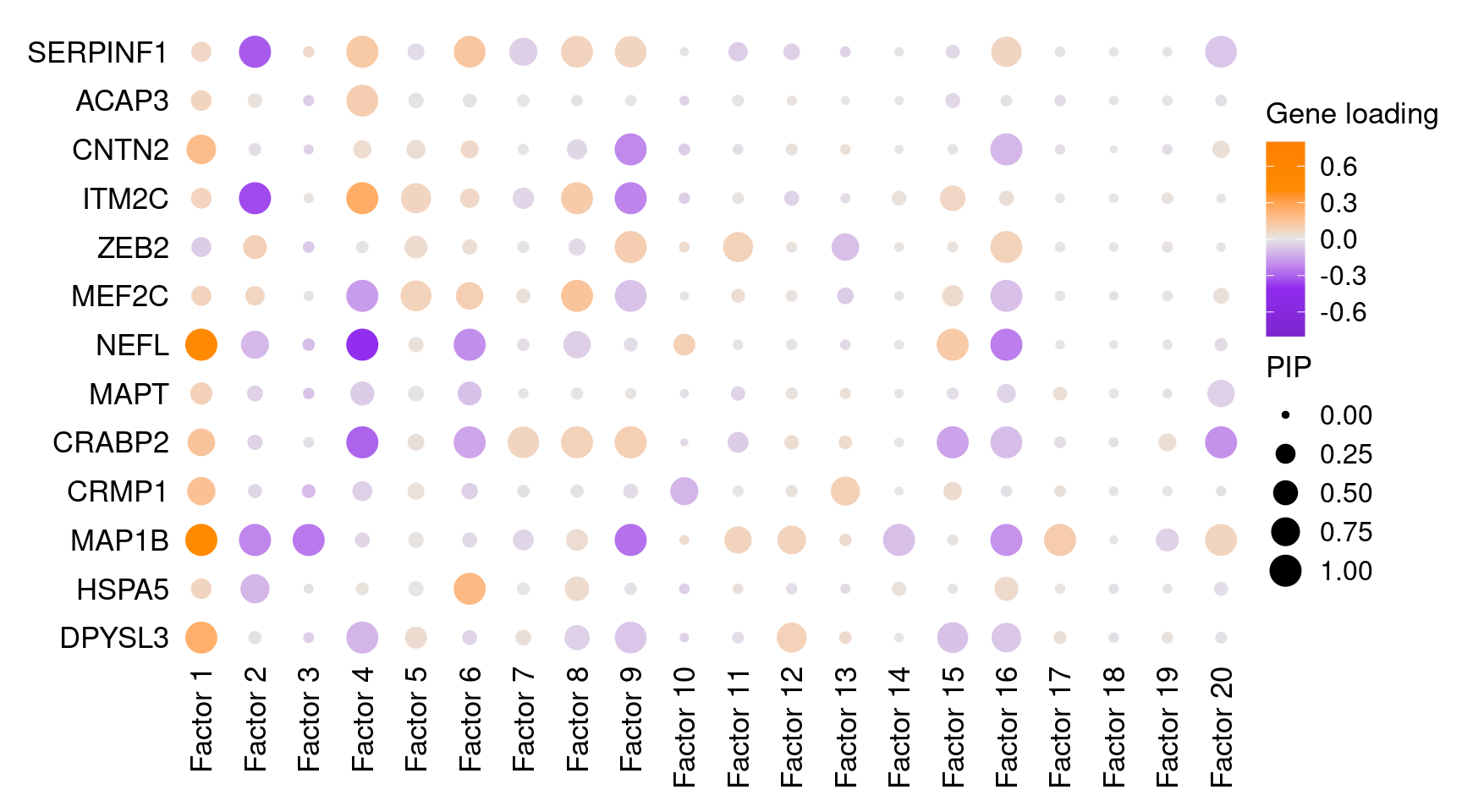
Markers in Fig 4E of original paper (Lalli et al):
SERPINF1 to ITM2C: negative regulators of neuron projection and differentiation
ZEB2 to DPYSL3: neuron maturation markers
3.2 Selected factor GO enrichment
4 DEG Interpretation
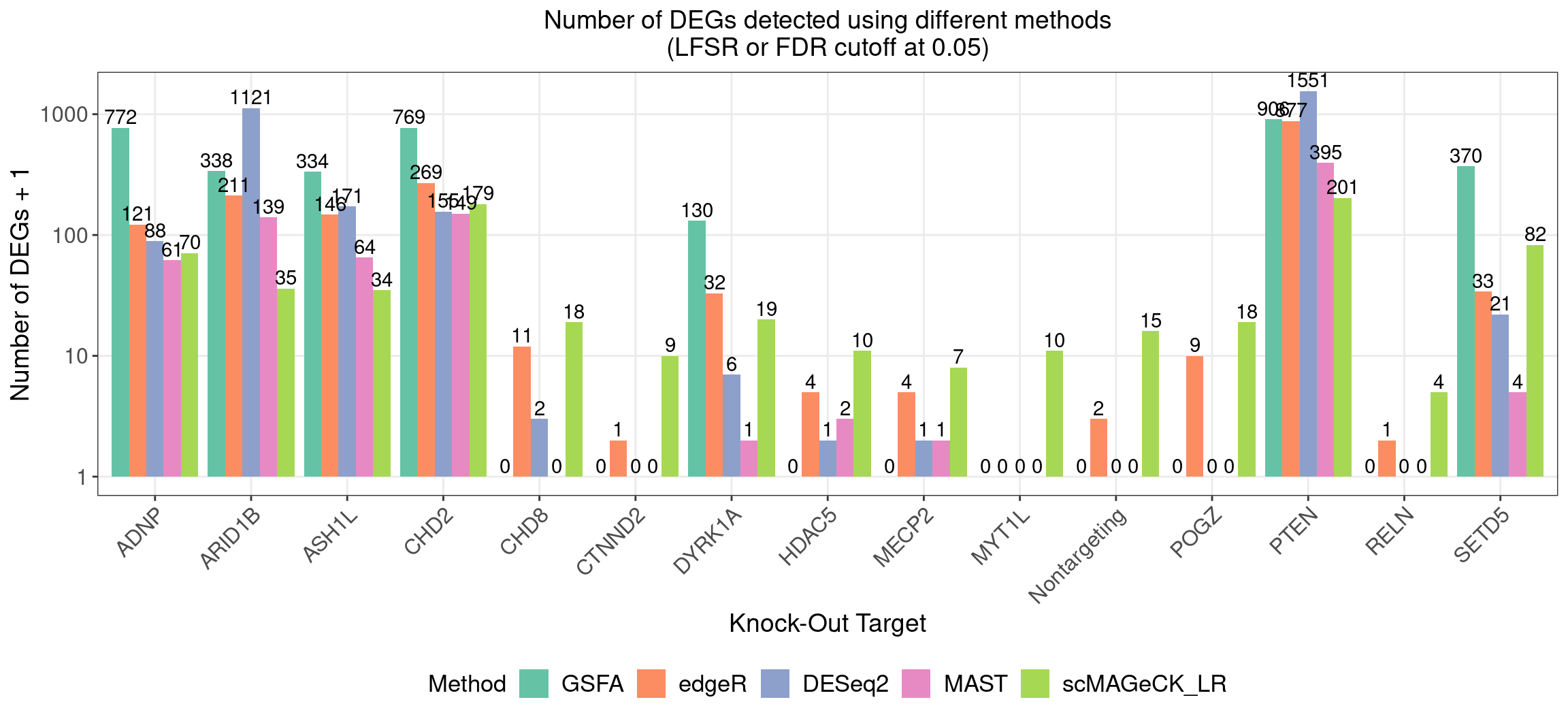
4.1 Number of DEGs detected under different methods
4.2 Perturbation effect on gene markers
Neuronal cell markers obtained from papers and Neuronal and Glial Cell Marker Atlas:
| protein_name | gene_name | type |
|---|---|---|
| Nestin | NES | Radial Glia |
| Tenascin C | TNC | Radial Glia |
| TP53 | TP53 | NPC |
| CDK4 | CDK4 | NPC |
| NeuroD1 | NEUROD1 | Immature Neurons |
| Doublecortin | DCX | Immature, Post-mitotic |
| STMN2 | STMN2 | Post-mitotic |
| MAP2 | MAP2 | Mature, Post-mitotic |
| NeuN | RBFOX3 | Mature Neurons |
| Neurofilament-L | NEFL | Mature Neurons |
| Neurofilament-M | NEFM | Mature Neurons |
| UCHL1 | UCHL1 | Mature Neurons |
| VGLUT2 | SLC17A6 | Glutamate, Excitatory |
| GIRK2 | KCNJ6 | Dopamine, Inhibitory |
| Somatostatin | SST | GABA, Inhibitory |
| EBF1 | EBF1 | Cortical |
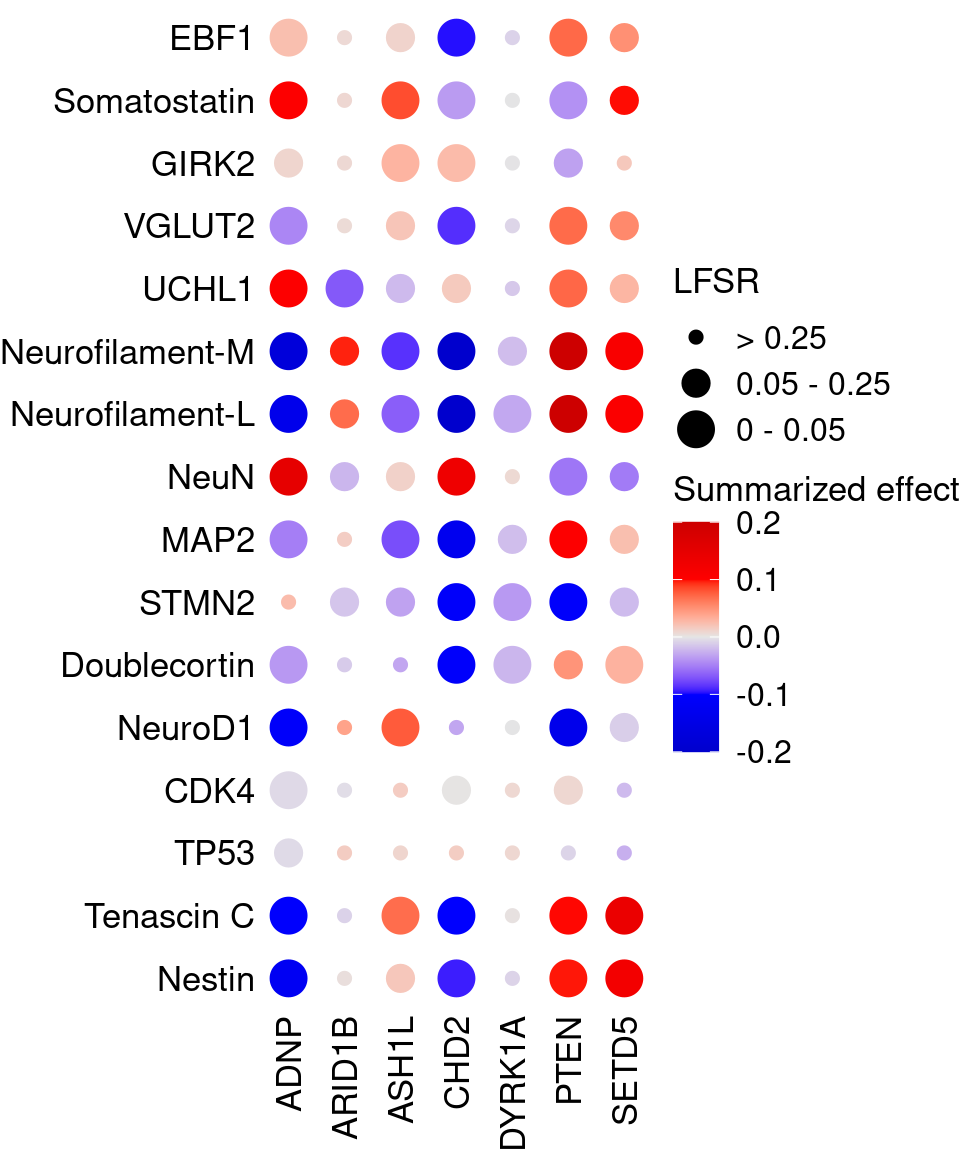
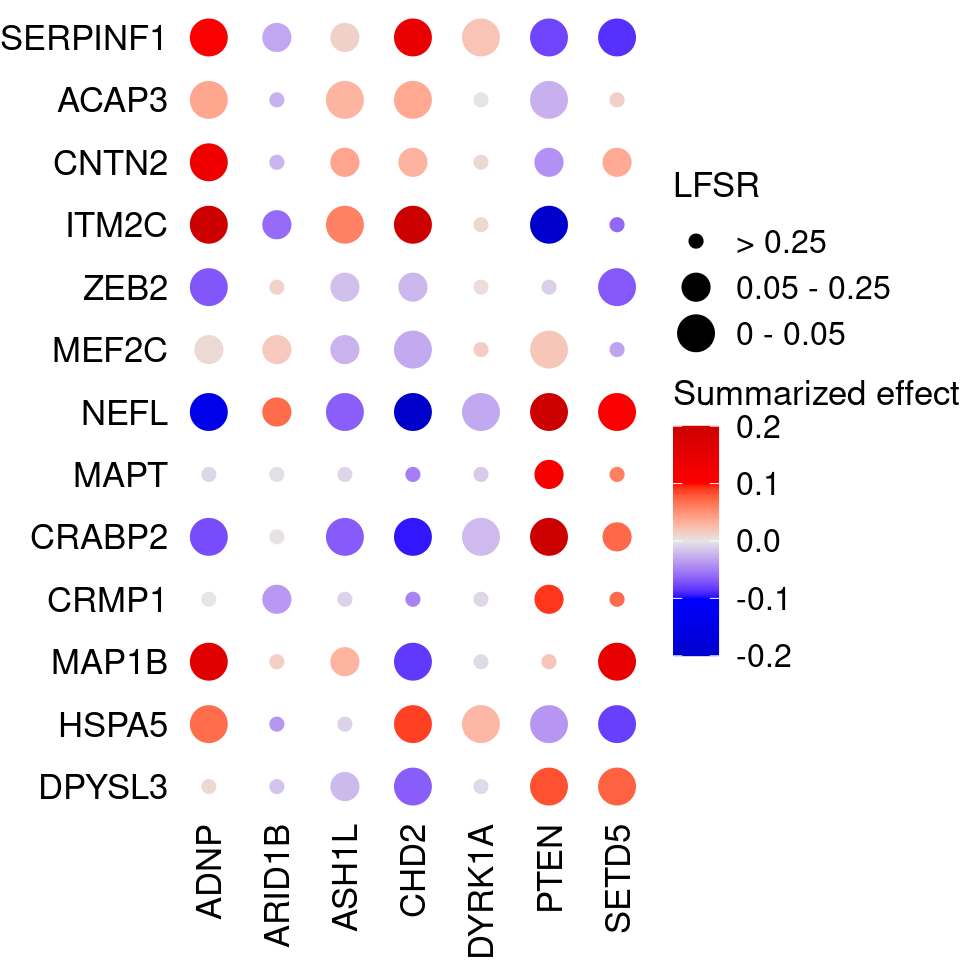
Markers in Fig 4E of original paper (Lalli et al):
SERPINF1 to ITM2C: negative regulators of neuron projection and differentiation
ZEB2 to DPYSL3: neuron maturation markers
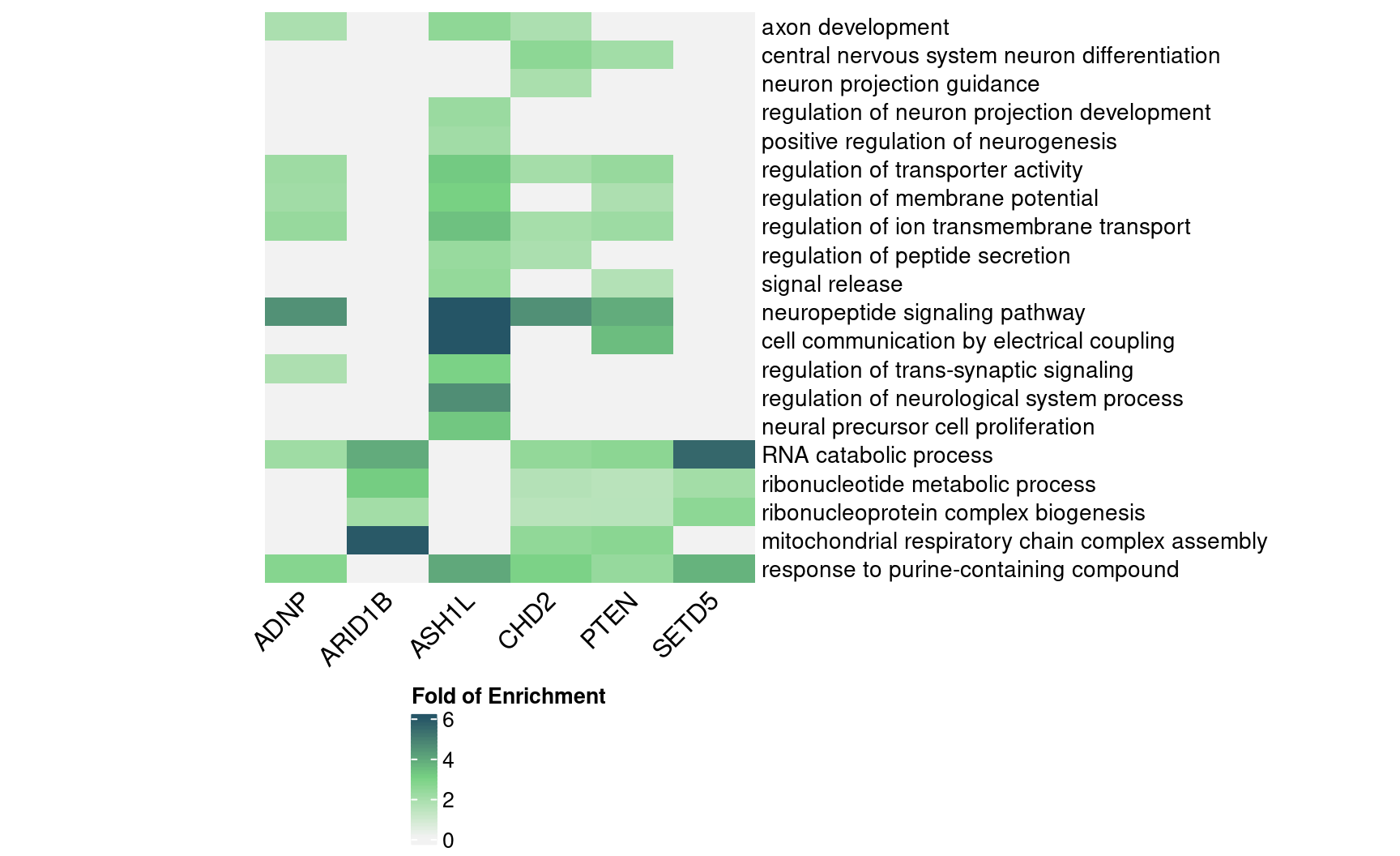
4.3 DEG GO enrichment
5 Gene Set Enrichment Analysis by Factor
Target genes: Genes w/ non-zero loadings in each factor (PIP cutoff at 0.95);
Backgroud genes: all 6000 genes used in factor analysis;
Statistical test: hypergeometric test (over-representation test);
Only GO terms/pathways that satisfy fold change \(\geq\) 2 and test FDR \(<\) 0.05 are shown below.
5.1 GO Slim Over-Representation Analysis
Gene sets: Gene ontology "Biological Process" (non-redundant).
Factor 1 : 12 significant GO terms| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0006414 | translational elongation | 4.14 | 1.08e-12 | 7.93e-10 | 32/109 | MRPL37; MRPL9; MRPS9; MRPS22; MRPL47; MRPS18C; MRPS30; MRPL22; MRPS18B; MRPS10; MRPL2; MRPS18A; EEF1A1; MRPL32; MRPL15; MRPL50; MRPS2; MRPL21; MRPL51; MRPL42; MRPL46; MRPS11; MRPS34; MRPL27; ICT1; MRPS7; EIF4A3; MRPL12; MRPS26; EEF2; GADD45GIP1; MRPL39 |
| GO:0140053 | mitochondrial gene expression | 3.52 | 9.44e-10 | 3.46e-07 | 29/116 | MRPL37; MRPL9; MRPS9; MRPS22; MRPL47; MRPS18C; MRPS30; MRPL22; MRPS18B; MRPS10; MRPL2; MRPS18A; MRPL32; MRPL15; MRPL50; MRPS2; MRPL21; MRPL51; MRPL42; MRPL46; MRPS11; MRPS34; MRPL27; ICT1; MRPS7; MRPL12; MRPS26; GADD45GIP1; MRPL39 |
| GO:0070972 | protein localization to endoplasmic reticulum | 3.08 | 1.43e-07 | 2.09e-05 | 26/119 | RPS7; RPS27A; RPSA; SEC62; RPL9; ANK2; RPL37; RPS10; RPS12; RPL10; RPS6; SPCS2; RPL41; RPS29; SRP14; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; ASNA1; RPS19; RPL13A; RPS5 |
| GO:0016072 | rRNA metabolic process | 2.88 | 5.49e-09 | 1.34e-06 | 35/171 | RPF1; RPS7; WDR43; MRPS9; NIFK; RPSA; EXOSC7; RRP9; ABT1; RPL7L1; DDX56; WBSCR22; DCAF13; EXOSC4; RPS6; WDR74; TRMT112; GTPBP4; DDX21; EXOSC1; NPM3; KRR1; EXOSC8; NGDN; RPS27L; RPS17; MRPS11; RPS2; RSL1D1; NOB1; DDX52; RPL27; EIF4A3; C19orf43; RPS19 |
| GO:0006413 | translational initiation | 2.60 | 1.19e-06 | 1.09e-04 | 29/157 | EIF3I; RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; HSPB1; RPL10; COPS5; RPS6; CDC123; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; EIF4A1; RPL23A; RPL19; EIF1; RPL27; EIF6; RPS19; RPL13A; RPS5; ATF4 |
| GO:0090150 | establishment of protein localization to membrane | 2.42 | 5.72e-07 | 5.99e-05 | 35/204 | ATP1B1; RPS7; RPS27A; REEP1; C2orf47; RPSA; SEC62; NSG1; RPL9; RPL37; RPS10; CD24; RPS12; RPL10; YWHAZ; RPS6; ANK3; RPL41; RPS29; SRP14; RPS27L; RPS17; RPS2; RAB26; RPL13; RPL23A; RPL19; RPL27; RAB31; ASNA1; RAB8A; RPS19; RPL13A; RPS5; TOMM22 |
| GO:0032984 | protein-containing complex disassembly | 2.40 | 4.49e-07 | 5.49e-05 | 36/211 | PEX14; STMN1; MRPL37; MRPL9; MRPS9; MRPS22; MRPL47; MRPS18C; MRPS30; MRPL22; MRPS18B; MRPS10; MRPL2; MRPS18A; MRPL32; STMN4; MRPL15; STMN2; MRPL50; SPTAN1; MRPS2; MRPL21; MRPL51; MRPL42; KLC1; MRPL46; MRPS11; MRPS34; MRPL27; ICT1; MRPS7; MRPL12; MRPS26; DSTN; GADD45GIP1; MRPL39 |
| GO:0034470 | ncRNA processing | 2.21 | 4.87e-06 | 3.97e-04 | 35/223 | RPF1; RPS7; WDR43; MRPS9; RPSA; EXOSC7; RRP9; ABT1; RPL7L1; DDX56; WBSCR22; DCAF13; EXOSC4; RPS6; WDR74; TRMT112; GTPBP4; DDX21; EXOSC1; NPM3; KRR1; EXOSC8; NGDN; RPS27L; RPS17; MRPS11; RPS2; RSL1D1; NOB1; DDX52; RPL27; EIF4A3; TP53RK; RPS19; TRMU |
| GO:0022613 | ribonucleoprotein complex biogenesis | 2.18 | 4.34e-08 | 7.96e-06 | 51/330 | EIF3I; RPF1; RPS7; WDR43; CEBPZ; MRPS9; LSM3; RPSA; EXOSC7; RRP9; BRIX1; MRPL22; ABT1; RPS10; RPL7L1; DDX56; WBSCR22; RPL10; DCAF13; EXOSC4; RPS6; MRPS2; WDR74; TRMT112; GTPBP4; DDX21; EXOSC1; NPM3; KRR1; RAN; EXOSC8; PRMT5; NGDN; RPS27L; RPS17; MRPS11; RPS2; RSL1D1; NOB1; RPL23A; DDX52; RPL27; MRPS7; EIF4A3; CELF4; SNRPB; EIF6; RPS19; RPL13A; RPS5 |
| GO:0006401 | RNA catabolic process | 2.09 | 1.76e-05 | 1.18e-03 | 35/236 | YBX1; SERBP1; RBM8A; RPS7; RPS27A; LSM3; RPSA; EXOSC7; RPL9; RPL37; RPS10; RPS12; HSPB1; RPL10; YWHAZ; EXOSC4; RPS6; EXOSC1; RPL41; EXOSC8; RPS29; SLIRP; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; EIF4A3; LSM7; C19orf43; RPS19; RPL13A; RPS5 |
| GO:0006605 | protein targeting | 2.05 | 1.17e-05 | 8.57e-04 | 38/261 | PEX14; TIMM17A; RPS7; RPS27A; RPSA; SEC62; PARL; RPL9; RPL37; RPS10; RPS12; RPL10; YWHAZ; RPS6; PMPCA; SPCS2; ANK3; RPL41; AKAP6; RPS29; SRP14; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; TIMM44; UBL5; ASNA1; TIMM50; RPS19; RPL13A; RPS5; UBE2L3; TOMM22; TSPO |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.01 | 3.13e-04 | 1.92e-02 | 27/189 | NDUFS2; ATP1B1; COX5B; NDUFB5; SNCA; NDUFS6; NDUFS4; COX7C; COX7A2; NDUFA4; TMSB4X; CYC1; NDUFS3; SDHD; NDUFA9; GAPDH; ATP5B; NDUFA12; COX6A1; RAN; NDUFB10; EIF6; ATP5E; SIRT6; UQCRFS1; TSPO; ATP5O |
Factor 2 : 44 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 8.81 | 0.00e+00 | 0.00e+00 | 84/119 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RTN4; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; BCAP31; RPL10; RPS20; RPL7; RPL30; GPAA1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; SPCS2; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; VAPA; DDRGK1; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006413 | translational initiation | 6.60 | 0.00e+00 | 0.00e+00 | 83/157 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; MCTS1; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; PPP1CA; RPS3; RPS25; CDC123; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; EIF4A1; RPL26; RPL23A; RPL23; RPL19; EIF1; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0070670 | response to interleukin-4 | 5.67 | 9.91e-04 | 2.20e-02 | 5/11 | HSP90AB1; TUBA1B; CORO1A; XBP1; RPL3 |
| GO:0090150 | establishment of protein localization to membrane | 5.45 | 0.00e+00 | 0.00e+00 | 89/204 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; YWHAQ; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; NSG1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; SDCBP; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; CALM1; HSP90AA1; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; ARL6IP1; RPL13; YWHAE; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; YWHAB; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; NDUFA13; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3; WRB |
| GO:0002181 | cytoplasmic translation | 5.30 | 5.55e-16 | 6.79e-14 | 31/73 | RPL11; RPL31; RPL32; RPL15; TMA7; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; MCTS1; RPL30; RPL8; RPL41; RPL6; RPLP0; RPS29; RPLP1; RPL26; RPL19; RPL38; RPS21; EEF2; RPL36; RPS28; RPL18A; RPL18; RPL13A |
| GO:0006401 | RNA catabolic process | 4.44 | 0.00e+00 | 0.00e+00 | 84/236 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; SSB; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; SET; RPL7A; RNH1; RPLP2; RPL27A; RPS13; RPS3; RPS25; HSPA8; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; YWHAB; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006605 | protein targeting | 4.26 | 0.00e+00 | 0.00e+00 | 89/261 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RAB7A; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; SDCBP; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; SPCS2; RPS3; RPS25; HSPA8; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; HSP90AA1; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; ARL6IP1; UQCRC2; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; YWHAB; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; NDUFA13; ECH1; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0051187 | cofactor catabolic process | 4.16 | 2.06e-03 | 3.02e-02 | 6/18 | ACOT7; PRDX1; BLVRA; PRDX4; PRDX5; PRDX2 |
| GO:0052192 | movement in environment of other organism involved in symbiotic interaction | 4.16 | 2.06e-03 | 3.02e-02 | 6/18 | RAB7A; PPIA; TSG101; SMC3; VAPA; CHMP2A |
| GO:0006081 | cellular aldehyde metabolic process | 4.16 | 1.58e-04 | 5.80e-03 | 9/27 | PARK7; AKR7A2; TKT; TALDO1; AKR1C1; PGAM1; TPI1; ESD; IDH2 |
| GO:0001906 | cell killing | 4.01 | 2.16e-04 | 6.91e-03 | 9/28 | PRDX1; TUBB; HSP90AB1; RPL30; TUBB4B; GAPDH; CORO1A; RPS19; PVRL2 |
| GO:0070671 | response to interleukin-12 | 3.87 | 2.92e-04 | 8.93e-03 | 9/29 | CDC42; HNRNPA2B1; RALA; PPIA; TALDO1; CFL1; RPLP0; PSME2; SOD1 |
| GO:0002576 | platelet degranulation | 3.81 | 7.26e-05 | 3.14e-03 | 11/36 | TAGLN2; TUBA4A; MAGED2; APLP2; PSAP; CD63; CALM1; SCG3; ALDOA; LGALS3BP; SOD1 |
| GO:1902579 | multi-organism localization | 3.74 | 3.74e-03 | 4.91e-02 | 6/20 | RPS27A; RAB7A; DYNLT1; UBC; RAN; UBA52 |
| GO:0016999 | antibiotic metabolic process | 3.66 | 1.51e-06 | 1.23e-04 | 17/58 | PARK7; AKR7A2; PRDX1; MDH1; SUCLG1; PDHB; MDH2; AKR1B1; PDHA1; PRDX4; PRDX5; AKR1C1; ALDH2; ESD; IDH2; PRDX2; SOD1 |
| GO:0048857 | neural nucleus development | 3.62 | 5.09e-04 | 1.38e-02 | 9/31 | CDC42; ATP5F1; YWHAQ; ACTB; DYNLL1; CALM1; CKB; YWHAE; ATP5J |
| GO:0006090 | pyruvate metabolic process | 3.57 | 1.78e-05 | 9.80e-04 | 14/49 | ENO1; PDHB; VDAC1; PDHA1; PGK1; LDHA; PGAM1; GAPDH; TPI1; LDHB; PKM; ALDOA; ENO3; BSG |
| GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | 3.51 | 6.59e-04 | 1.73e-02 | 9/32 | ATP6V0B; ATP5F1; ATP5G3; ATP6AP1; ATP5C1; ATP5B; ATP6V0D1; ATP5A1; ATP5O |
| GO:0072524 | pyridine-containing compound metabolic process | 3.22 | 1.87e-05 | 9.80e-04 | 16/62 | ENO1; MDH1; TKT; PDHB; MDH2; PDHA1; PGK1; TALDO1; LDHA; PGAM1; GAPDH; TPI1; PKM; IDH2; ALDOA; ENO3 |
| GO:0008037 | cell recognition | 3.12 | 1.66e-03 | 2.94e-02 | 9/36 | RTN4; CCT7; GAP43; CCT5; VDAC2; CCT2; ALDOA; CORO1A; BSG |
| GO:0030901 | midbrain development | 3.12 | 9.24e-04 | 2.19e-02 | 10/40 | CDC42; ATP5F1; YWHAQ; CTNNB1; ACTB; DYNLL1; CALM1; CKB; YWHAE; ATP5J |
| GO:0009566 | fertilization | 3.04 | 2.04e-03 | 3.02e-02 | 9/37 | PARK7; H3F3A; CCT7; CCT5; VDAC2; CCT2; REC8; ALDOA; PVRL2 |
| GO:0005996 | monosaccharide metabolic process | 2.87 | 1.14e-05 | 6.99e-04 | 20/87 | ENO1; MDH1; TKT; PDHB; MDH2; AKR1B1; PDHA1; PGK1; TALDO1; PPP1CA; PGAM1; GAPDH; TPI1; HMGB1; PKM; VIMP; ALDOA; ENO3; G6PC3; CYB5R3 |
| GO:0035821 | modification of morphology or physiology of other organism | 2.86 | 1.14e-03 | 2.38e-02 | 11/48 | RPL30; RRAGA; CFL1; HSPA8; SMC3; GAPDH; CALM1; PPIB; KPNA2; VAPA; RPS19 |
| GO:0046939 | nucleotide phosphorylation | 2.84 | 2.02e-03 | 3.02e-02 | 10/44 | ENO1; SUCLG1; PGK1; LDHA; PGAM1; GAPDH; TPI1; PKM; ALDOA; ENO3 |
| GO:0016052 | carbohydrate catabolic process | 2.75 | 1.62e-03 | 2.94e-02 | 11/50 | ENO1; PGK1; LDHA; PPP1CA; PGAM1; GAPDH; TPI1; HMGB1; PKM; ALDOA; ENO3 |
| GO:0045861 | negative regulation of proteolysis | 2.73 | 6.01e-06 | 4.01e-04 | 23/105 | PARK7; RPL11; TMEM59; RPL5; AGT; RPS7; SERPINI1; HSP90AB1; SDCBP; PRDX5; APLP2; GAPDH; PEBP1; MAP1A; ARL6IP1; HERPUD1; GABARAPL2; YWHAE; SERPINF1; RPL23; DDRGK1; CST3; CDKN2D |
| GO:0016051 | carbohydrate biosynthetic process | 2.64 | 3.88e-04 | 1.14e-02 | 15/71 | ENO1; MDH1; TKT; MDH2; AKR1B1; PGK1; TALDO1; PPP1CA; PGAM1; GAPDH; TPI1; VIMP; ALDOA; ENO3; G6PC3 |
| GO:0036230 | granulocyte activation | 2.55 | 2.48e-08 | 2.60e-06 | 39/191 | DDOST; PSMA5; ILF2; ARPC5; PSMD6; RAB7A; TUBB; CSNK2B; HSP90AB1; DYNLT1; PSMB1; PPIA; PSMC2; PRDX4; ATP6AP2; SDCBP; GGH; TUBB4B; PSMD13; GSTP1; NDUFC2; HSPA8; PSAP; PGAM1; CD63; CCT2; DYNLL1; HMGB1; ACTR10; HSP90AA1; SRP14; PKM; ALDOA; PSMD7; VAPA; CST3; EEF2; FTL; CYB5R3 |
| GO:0002446 | neutrophil mediated immunity | 2.52 | 3.35e-08 | 3.07e-06 | 39/193 | DDOST; PSMA5; ILF2; ARPC5; PSMD6; RAB7A; TUBB; CSNK2B; HSP90AB1; DYNLT1; PSMB1; PPIA; PSMC2; PRDX4; ATP6AP2; SDCBP; GGH; TUBB4B; PSMD13; GSTP1; NDUFC2; HSPA8; PSAP; PGAM1; CD63; CCT2; DYNLL1; HMGB1; ACTR10; HSP90AA1; SRP14; PKM; ALDOA; PSMD7; VAPA; CST3; EEF2; FTL; CYB5R3 |
| GO:0006909 | phagocytosis | 2.46 | 1.24e-03 | 2.53e-02 | 14/71 | CDC42; ARPC5; ARPC2; RAB7A; LMAN2; GNB2L1; HSP90AB1; ACTB; ARPC1A; ARPC3; HMGB1; HSP90AA1; CORO1A; ACTG1 |
| GO:1902600 | proton transmembrane transport | 2.43 | 9.58e-04 | 2.20e-02 | 15/77 | PARK7; ATP6V0B; ATP5F1; ATP5G3; NDUFA4; ATP6AP1; COX6C; ATP5C1; ATP5B; SLC25A3; COX5A; ATP6V0D1; ATP5A1; ATP5J; ATP5O |
| GO:0009410 | response to xenobiotic stimulus | 2.33 | 2.15e-03 | 3.10e-02 | 14/75 | AKR7A2; GSTM3; PHGDH; MGST3; EPHX1; CDO1; HSP90AB1; GSTA4; GSTP1; AKR1C1; PTGES3; CALM1; SERPINF1; SOD1 |
| GO:0019058 | viral life cycle | 2.32 | 1.96e-04 | 6.53e-03 | 21/113 | CDC42; RPS27A; RAB1A; RPSA; RAB7A; HSP90AB1; DYNLT1; PPIA; PABPC1; CHMP5; TSG101; HSPA8; SMC3; UBC; RAN; PPIB; HACD3; VAPA; UBA52; PVRL2; CHMP2A |
| GO:0052547 | regulation of peptidase activity | 2.28 | 1.30e-04 | 5.29e-03 | 23/126 | PARK7; AGT; ARL6IP5; SERPINI1; GNB2L1; CYCS; BCAP31; PRDX5; RPS3; APLP2; GAPDH; PEBP1; HMGB1; PSME2; PSMA3; ARL6IP1; HERPUD1; YWHAE; SERPINF1; DDRGK1; CST3; CDKN2D; NDUFA13 |
| GO:0070646 | protein modification by small protein removal | 2.25 | 5.75e-05 | 2.64e-03 | 26/144 | PARK7; PSMA5; PSMD4; RPS27A; COPS8; PSMD6; UCHL1; PSMB1; ACTB; COPS6; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; COPS3; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; UFD1L |
| GO:0071897 | DNA biosynthetic process | 2.23 | 2.39e-03 | 3.38e-02 | 15/84 | RPS27A; CCT7; CTNNB1; CCT5; HSP90AB1; HNRNPA2B1; PPIA; PTGES3; CCT2; UBC; HSP90AA1; TERF2IP; CDKN2D; UBA52; UFD1L |
| GO:0043687 | post-translational protein modification | 2.19 | 3.62e-05 | 1.77e-03 | 29/165 | PSMA5; PSMD4; PDIA6; RAB1A; COPS8; PSMD6; SKP1; PSMB1; COPS6; PSMC2; RAB2A; TCEB1; PSMD13; APLP2; HSP90B1; PSME2; PSMA3; SCG3; PPIB; PSMA4; PSMD7; PSMB6; COPS3; PSMB3; PSMC5; CST3; PSMA7; PSMD8; PSMC4 |
| GO:0072593 | reactive oxygen species metabolic process | 2.18 | 3.04e-03 | 4.13e-02 | 15/86 | PARK7; PRDX1; GADD45A; AGT; ARF4; DDAH2; HSP90AB1; PRDX4; PRDX5; GSTP1; HSP90AA1; VIMP; PRDX2; NDUFA13; SOD1 |
| GO:0006091 | generation of precursor metabolites and energy | 2.11 | 1.67e-06 | 1.23e-04 | 42/248 | PARK7; ENO1; AKR7A2; ATP5F1; PHGDH; MDH1; SUCLG1; UQCRC1; PDHB; SLC25A4; NDUFA4; CYCS; MDH2; AKR1B1; PDHA1; PGK1; COX6C; NDUFB6; LDHA; PPP1CA; NDUFV1; NDUFC2; ATP5C1; PGAM1; NDUFB8; GAPDH; TPI1; ATP5B; ALDH2; HMGB1; PKM; COX5A; IDH2; VIMP; UQCRC2; ALDOA; ENO3; NDUFV2; ATP5A1; ETFB; ATP5J; ATP5O |
| GO:0031098 | stress-activated protein kinase signaling cascade | 2.10 | 2.45e-03 | 3.39e-02 | 17/101 | CDC42; PRDX1; GADD45A; AGT; RPS27A; ARL6IP5; SKP1; AKR1B1; SDCBP; DNAJA1; GSTP1; RPS3; UBC; HMGB1; HACD3; UBA52; PRMT1 |
| GO:2001233 | regulation of apoptotic signaling pathway | 2.07 | 1.45e-04 | 5.59e-03 | 28/169 | PARK7; ENO1; RPL11; MLLT11; AGT; RPS7; YWHAQ; ITM2C; CTNNB1; GNB2L1; BCAP31; DNAJA1; PPP1CA; GSTP1; RPS3; VDAC2; TMBIM6; TPT1; PDIA3; VIMP; HERPUD1; YWHAE; RPL26; YWHAB; CDKN2D; NDUFA13; XBP1; SOD1 |
| GO:0046390 | ribose phosphate biosynthetic process | 2.01 | 1.80e-03 | 3.00e-02 | 20/124 | ACOT7; ENO1; ATP5F1; ATP5G3; PDHB; PDHA1; PGK1; LDHA; ATP5C1; PGAM1; GAPDH; TPI1; ATP5B; PKM; ALDOA; ENO3; ATP5A1; TECR; ATP5J; ATP5O |
| GO:1901615 | organic hydroxy compound metabolic process | 2.01 | 1.80e-03 | 3.00e-02 | 20/124 | PARK7; AKR7A2; FDPS; ACAT2; AKR1B1; EBP; NSDHL; FDFT1; LDHA; TM7SF2; DHCR7; AKR1C1; TPI1; PMEL; ALDH2; RAN; CALM1; PLTP; CYB5R3; SOD1 |
Factor 3 : 12 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 38.40 | 0.00e+00 | 0.00e+00 | 66/119 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006413 | translational initiation | 29.10 | 0.00e+00 | 0.00e+00 | 66/157 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0002181 | cytoplasmic translation | 24.70 | 0.00e+00 | 0.00e+00 | 26/73 | RPL11; RPL31; RPL32; RPL15; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; RPL30; RPL41; RPL6; RPLP0; RPLP1; RPL26; RPL19; RPL38; RPS21; RPL36; RPS28; RPL18A; RPL18; RPL13A |
| GO:0090150 | establishment of protein localization to membrane | 22.70 | 0.00e+00 | 0.00e+00 | 67/204 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006401 | RNA catabolic process | 19.40 | 0.00e+00 | 0.00e+00 | 66/236 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006605 | protein targeting | 17.80 | 0.00e+00 | 0.00e+00 | 67/261 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0016072 | rRNA metabolic process | 8.91 | 6.66e-16 | 6.11e-14 | 22/171 | RPL11; RPS8; RPL5; RPS27; RPS7; RPL14; RPL35A; RPS14; RPL10A; RPL7; RPS6; RPL35; RPL7A; RPS24; RPS17; RPL26; RPS21; RPS15; RPS28; RPS16; RPS19; RPS9 |
| GO:0034470 | ncRNA processing | 6.83 | 1.73e-13 | 1.41e-11 | 22/223 | RPL11; RPS8; RPL5; RPS27; RPS7; RPL14; RPL35A; RPS14; RPL10A; RPL7; RPS6; RPL35; RPL7A; RPS24; RPS17; RPL26; RPS21; RPS15; RPS28; RPS16; RPS19; RPS9 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 6.72 | 0.00e+00 | 0.00e+00 | 32/330 | RPL11; RPS8; RPL5; RPS27; RPS7; RPL14; RPL24; RPL35A; RPS23; RPS14; RPL10A; RPL10; RPL7; RPS6; RPL35; RPL12; RPL7A; RPS24; RPL6; RPLP0; RPS17; RPL26; RPL38; RPS21; RPS15; RPS28; RPS16; RPS19; RPL13A; RPS9; RPS5; RPL3 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 6.37 | 5.08e-10 | 3.73e-08 | 17/185 | RPL11; RPL5; RPS27; RPL24; RPS23; RPS14; RPL10; RPL12; RPL6; RPLP0; RPL38; RPS15; RPS28; RPS19; RPL13A; RPS5; RPL3 |
| GO:0034248 | regulation of cellular amide metabolic process | 3.36 | 3.50e-04 | 2.34e-02 | 11/227 | RPL5; RPS14; GNB2L1; SOX4; RPS4X; RPL10; RPS3; RPL26; RPL38; RPL13A; RPS9 |
| GO:0010608 | posttranscriptional regulation of gene expression | 2.91 | 6.88e-04 | 4.21e-02 | 12/286 | RPL5; RPS27A; RPS14; GNB2L1; SOX4; RPS4X; RPL10; RPS3; RPL26; RPL38; RPL13A; RPS9 |
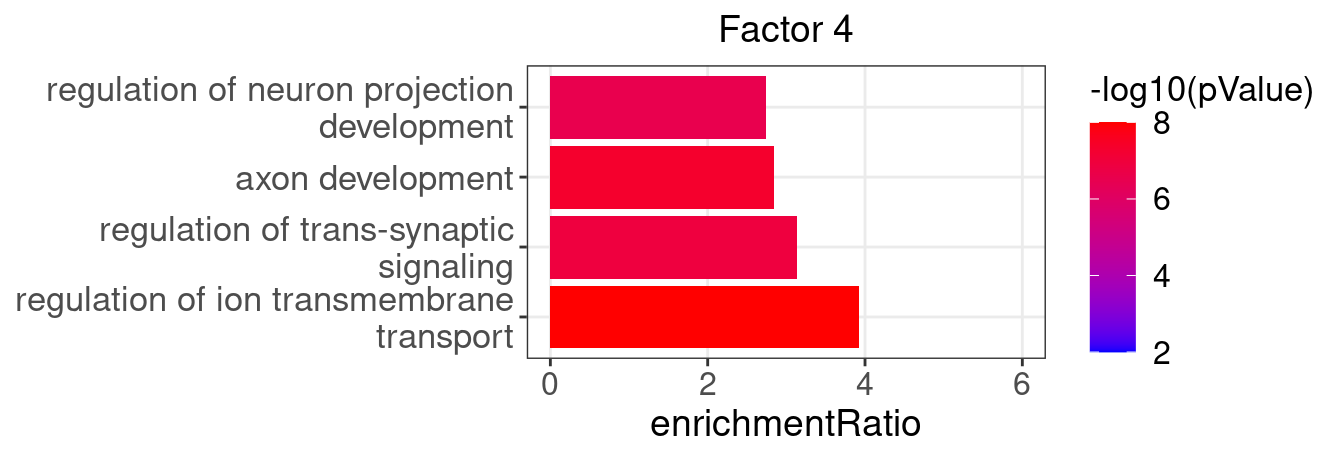
Factor 4 : 56 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0015844 | monoamine transport | 7.56 | 1.60e-05 | 1.07e-03 | 7/17 | SYT11; AGT; SNCA; ACTB; STX1A; GPM6B; SYT7 |
| GO:0007218 | neuropeptide signaling pathway | 7.34 | 1.39e-03 | 2.81e-02 | 4/10 | PCSK1N; GAL; SSTR2; PTH2 |
| GO:0010644 | cell communication by electrical coupling | 7.06 | 4.15e-04 | 1.17e-02 | 5/13 | ATP1B1; RYR2; SRI; ANK3; CALM1 |
| GO:0010737 | protein kinase A signaling | 6.67 | 2.10e-03 | 3.29e-02 | 4/11 | RAB13; AKAP12; EZR; GAL |
| GO:0034109 | homotypic cell-cell adhesion | 6.12 | 2.64e-04 | 7.74e-03 | 6/18 | CLIC1; ACTB; ILK; ANK3; ACTG1; MYL12A |
| GO:0031644 | regulation of neurological system process | 5.51 | 1.93e-05 | 1.18e-03 | 9/30 | PRKCZ; AGT; NRXN1; STX1A; AKAP9; CBLN1; EIF4A3; BAIAP2; CELF4 |
| GO:0099565 | chemical synaptic transmission, postsynaptic | 5.24 | 1.07e-05 | 7.82e-04 | 10/35 | PRKCZ; NRXN1; CHRNA1; SNCA; MEF2C; STX1A; CBLN1; EIF4A3; BAIAP2; CELF4 |
| GO:0060191 | regulation of lipase activity | 4.83 | 2.86e-03 | 3.89e-02 | 5/19 | PRKCZ; HPCA; RHOC; AGT; SNCA |
| GO:0015837 | amine transport | 4.59 | 3.65e-03 | 4.62e-02 | 5/20 | SYT11; AGT; SNCA; STX1A; SYT7 |
| GO:0035637 | multicellular organismal signaling | 4.27 | 7.54e-05 | 3.46e-03 | 10/43 | ATP1B1; AGT; RYR2; SCN3A; SCN9A; CHRNA1; SRI; AKAP9; ANK3; CALM1 |
| GO:0010959 | regulation of metal ion transport | 4.26 | 1.55e-09 | 7.02e-07 | 23/99 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; GNAI2; SNCA; TSPAN13; CAMK2B; SRI; AKAP9; G6PD; CHD7; GAL; ANK3; CD63; JPH4; CALM1; FKBP1A; NKAIN4; HOMER3; FXYD7 |
| GO:0034765 | regulation of ion transmembrane transport | 3.92 | 1.91e-09 | 7.02e-07 | 25/117 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; SCN3A; SCN9A; SNCA; MEF2C; CLIC1; TSPAN13; SRI; AKAP9; TMSB4X; G6PD; CHD7; GAL; ANK3; CD63; JPH4; CALM1; FKBP1A; CACNA1A; FXYD7 |
| GO:0048771 | tissue remodeling | 3.89 | 1.66e-03 | 3.04e-02 | 7/33 | AGT; IGFBP5; MEF2C; CHD7; CTHRC1; SYT7; CST3 |
| GO:0032409 | regulation of transporter activity | 3.85 | 1.08e-06 | 1.33e-04 | 17/81 | SEPN1; HPCA; NOS1AP; ATP1B1; RYR2; SNCA; MEF2C; ACTB; SRI; AKAP9; TMSB4X; GAL; ANK3; JPH4; CALM1; FKBP1A; FXYD7 |
| GO:0014074 | response to purine-containing compound | 3.76 | 9.82e-04 | 2.19e-02 | 8/39 | SEPN1; RYR2; IGFBP5; HDAC2; EZR; AKAP9; RAPGEF1; BSG |
| GO:0008277 | regulation of G protein-coupled receptor signaling pathway | 3.67 | 5.80e-04 | 1.42e-02 | 9/45 | STMN1; RGS16; GNG4; GNAI2; SNCA; DYNLT1; RGS10; CALM1; APLP1 |
| GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 3.67 | 1.17e-03 | 2.53e-02 | 8/40 | PTPRF; NRXN1; PTPRG; ALCAM; MDGA1; PCDH9; CBLN1; BSG |
| GO:0034764 | positive regulation of transmembrane transport | 3.61 | 4.23e-05 | 2.22e-03 | 13/66 | NOS1AP; ATP1B1; AGT; RYR2; CAPN10; SNCA; SRI; AKAP9; TMSB4X; G6PD; GAL; ANK3; CALM1 |
| GO:0003205 | cardiac chamber development | 3.51 | 8.10e-04 | 1.92e-02 | 9/47 | LMO4; RYR2; SOX11; ID2; NRP2; MEF2C; FZD1; CHD7; FKBP1A |
| GO:0033002 | muscle cell proliferation | 3.26 | 2.59e-03 | 3.73e-02 | 8/45 | SEPN1; AGT; ID2; IGFBP5; GNAI2; MEF2C; ILK; GSTP1 |
| GO:0043270 | positive regulation of ion transport | 3.24 | 2.51e-04 | 7.69e-03 | 12/68 | NOS1AP; ATP1B1; AGT; RYR2; SNCA; SRI; AKAP9; TMSB4X; G6PD; GAL; ANK3; CALM1 |
| GO:0015850 | organic hydroxy compound transport | 3.24 | 1.49e-03 | 2.81e-02 | 9/51 | SYT11; AGT; SNCA; ACTB; STX1A; GPM6B; SYT7; GAL; SELM |
| GO:0042391 | regulation of membrane potential | 3.21 | 1.41e-06 | 1.48e-04 | 21/120 | PRKCZ; NOS1AP; ATP1B1; RYR2; NRXN1; SCN3A; SCN9A; CHRNA1; SNCA; MEF2C; CLIC1; HEBP2; STX1A; SRI; AKAP9; ANK3; CBLN1; EIF4A3; BAIAP2; CELF4; CACNA1A |
| GO:0001764 | neuron migration | 3.20 | 5.00e-04 | 1.31e-02 | 11/63 | ACAP3; NAV1; NRP2; DNER; GPM6A; MEF2C; TUBB2B; MDGA1; CAMK2B; AUTS2; NEUROD4 |
| GO:0019932 | second-messenger-mediated signaling | 3.19 | 2.86e-05 | 1.62e-03 | 16/92 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; NEUROD1; GNAI2; SRI; GAL; JPH4; CALM1; FKBP1A; HOMER3; APLP1; PDE9A |
| GO:0009410 | response to xenobiotic stimulus | 3.18 | 1.68e-04 | 5.88e-03 | 13/75 | SEPN1; HPCA; EPHX1; RYR2; GPX1; SNCA; NR3C1; HSP90AB1; HDAC2; GSTP1; RGS10; CALM1; SERPINF1 |
| GO:0003007 | heart morphogenesis | 3.15 | 5.75e-04 | 1.42e-02 | 11/64 | GNG5; RYR2; SOX11; ID2; NRP2; MEF2C; FZD1; CHD7; ILK; ACTC1; FKBP1A |
| GO:0099177 | regulation of trans-synaptic signaling | 3.14 | 1.19e-07 | 2.18e-05 | 26/152 | PRKCZ; SYT11; AGT; NRXN1; GNAI2; NSG1; SNCA; MEF2C; CPLX2; TUBB2B; GRM4; AKAP12; CAMK2B; STX1A; AKAP9; SYT7; JPH4; CALM1; RAB26; CBLN1; EFNB3; EIF4A3; BAIAP2; CELF4; CACNA1A; RAB3A |
| GO:0001818 | negative regulation of cytokine production | 3.12 | 3.44e-03 | 4.43e-02 | 8/47 | SYT11; BTN2A2; HSP90AB1; EZR; TMSB4X; UBE2L6; GSTP1; HOMER3 |
| GO:0003012 | muscle system process | 3.11 | 7.35e-06 | 6.74e-04 | 19/112 | NOS1AP; ATP1B1; AGT; RYR2; CHRNA1; MYL1; IGFBP5; MEF2C; HDAC2; CAMK2B; SRI; CALD1; G6PD; MYL6; CALM1; ACTC1; SSTR2; MYL12A; PDE9A |
| GO:0072511 | divalent inorganic cation transport | 3.06 | 9.60e-06 | 7.82e-04 | 19/114 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; GNAI2; SNCA; GPM6A; TSPAN13; CAMK2B; SRI; G6PD; CHD7; JPH4; CALM1; FKBP1A; CACNA1A; HOMER3 |
| GO:0021953 | central nervous system neuron differentiation | 3.06 | 2.26e-03 | 3.32e-02 | 9/54 | DRAXIN; LMO4; NRP2; MAP2; MDGA1; HSP90AB1; FZD1; MYCBP2; CBLN1 |
| GO:0097485 | neuron projection guidance | 2.93 | 1.42e-04 | 5.20e-03 | 15/94 | DRAXIN; LHX9; NRXN1; NRP2; KLF7; ALCAM; SPON2; PDLIM7; TUBB2B; EZR; DPYSL2; FEZ1; MYCBP2; EFNB3; BSG |
| GO:0007517 | muscle organ development | 2.89 | 6.10e-05 | 2.98e-03 | 17/108 | SEPN1; CENPF; RYR2; ZBTB18; SOX11; CHRNA1; DNER; GPX1; MEF2C; SRI; FZD1; G6PD; CHD7; MYL6; ACTC1; TCF12; FKBP1A |
| GO:0061564 | axon development | 2.84 | 4.68e-08 | 1.14e-05 | 32/207 | DRAXIN; STMN1; PTPRF; S100A6; CRABP2; LHX9; NRXN1; NRP2; KLF7; MAP2; ALCAM; SPON2; PDLIM7; TUBB2B; HSP90AB1; DYNLT1; EZR; AUTS2; NEFM; NEFL; DPYSL2; ILK; FEZ1; ANK3; MYCBP2; EFNB3; RND2; NGFR; BAIAP2; BSG; RAB3A; RTN4R |
| GO:0031345 | negative regulation of cell projection organization | 2.78 | 2.74e-03 | 3.79e-02 | 10/66 | DRAXIN; MAP2; ITM2C; PTPRG; NR2F1; DPYSL3; HDAC2; EFNB3; NGFR; RTN4R |
| GO:0010975 | regulation of neuron projection development | 2.74 | 2.83e-07 | 4.16e-05 | 30/201 | ACAP3; DRAXIN; PTPRF; CRABP2; AGT; MAP2; ITM2C; PTPRG; TNIK; MEF2C; NR2F1; DPYSL3; TUBB2B; HDAC2; DYNLT1; CAMK2B; FZD1; NEFL; DPYSL2; RAPGEF1; ILK; CFL1; MYCBP2; SERPINF1; EFNB3; RND2; NGFR; BAIAP2; PREX1; RTN4R |
| GO:0060537 | muscle tissue development | 2.74 | 1.22e-04 | 4.72e-03 | 17/114 | SEPN1; CENPF; AGT; RYR2; ZBTB18; SOX11; ID2; CHRNA1; IGFBP5; DNER; GPX1; MEF2C; G6PD; CHD7; MYL6; ACTC1; FKBP1A |
| GO:0050803 | regulation of synapse structure or activity | 2.72 | 1.30e-03 | 2.72e-02 | 12/81 | NRXN1; NRP2; SNCA; GPM6A; MEF2C; TUBB; MDGA1; CAMK2B; FZD1; CFL1; CBLN1; BAIAP2 |
| GO:0060560 | developmental growth involved in morphogenesis | 2.68 | 9.41e-04 | 2.16e-02 | 13/89 | PRKCZ; DRAXIN; CRABP2; NRP2; MAP2; ALCAM; HSP90AB1; AUTS2; DPYSL2; ILK; GAL; RND2; RTN4R |
| GO:0072593 | reactive oxygen species metabolic process | 2.56 | 2.20e-03 | 3.29e-02 | 12/86 | PRDX1; GADD45A; NOS1AP; AGT; GPX1; GNAI2; SNCA; HSP90AB1; G6PD; GSTP1; SH3PXD2A; PREX1 |
| GO:0010721 | negative regulation of cell development | 2.53 | 4.82e-04 | 1.31e-02 | 16/116 | DRAXIN; PBX1; SOX11; ID2; MAP2; ITM2C; PTPRG; NR2F1; DPYSL3; HDAC2; DYNLT1; G6PD; GAL; EFNB3; NGFR; RTN4R |
| GO:0050808 | synapse organization | 2.51 | 1.06e-04 | 4.56e-03 | 20/146 | PTPRF; NOS1AP; NRXN1; CHRNA1; NRP2; DNER; SNCA; GPM6A; MEF2C; TUBB; MDGA1; ACTB; CAMK2B; FZD1; NEFL; CFL1; ANK3; CBLN1; BAIAP2; RAB3A |
| GO:0003013 | circulatory system process | 2.51 | 1.74e-03 | 3.12e-02 | 13/95 | NOS1AP; ATP1B1; AGT; RYR2; ID2; MYL1; GPX1; GNAI2; SRI; AKAP9; CHD7; CALM1; ACTC1 |
| GO:0009914 | hormone transport | 2.50 | 2.68e-03 | 3.78e-02 | 12/88 | AGT; SOX11; NEUROD1; KLF7; CAPN10; STX1A; SRI; CHD7; SYT7; GAL; CACNA1A; SELM |
| GO:0023061 | signal release | 2.42 | 1.21e-04 | 4.72e-03 | 21/159 | WLS; SYT11; AGT; SOX11; NRXN1; NEUROD1; KLF7; CAPN10; SNCA; MEF2C; CPLX2; GRM4; STX1A; SRI; CHD7; SYT7; GAL; CALM1; CACNA1A; RAB3A; SELM |
| GO:0051961 | negative regulation of nervous system development | 2.40 | 1.82e-03 | 3.15e-02 | 14/107 | DRAXIN; PBX1; SOX11; ID2; MAP2; ITM2C; PTPRG; NR2F1; DPYSL3; HDAC2; DYNLT1; EFNB3; NGFR; RTN4R |
| GO:0051235 | maintenance of location | 2.38 | 1.99e-03 | 3.24e-02 | 14/108 | SEPN1; RYR2; SNCA; MDFI; EZR; SRI; AKAP9; TMSB4X; CHD7; ANK3; JPH4; CALM1; FKBP1A; FTL |
| GO:0035690 | cellular response to drug | 2.36 | 2.17e-03 | 3.29e-02 | 14/109 | SEPN1; RYR2; GNAI2; MEF2C; NR3C1; HSP90AB1; HDAC2; ACTB; RPS3; RGS10; HNRNPA1; SERPINF1; BAIAP2; PPP1R14A |
| GO:0051098 | regulation of binding | 2.32 | 2.25e-04 | 7.50e-03 | 21/166 | STMN1; PTPRF; SOX11; ID2; NEUROD1; MAP2; HMGB2; MEF2C; MDFI; HSP90AB1; HDAC2; ACTB; SRI; TMSB4X; CTHRC1; CALM1; CCPG1; EIF4A3; FKBP1A; AES; MFNG |
| GO:0070972 | protein localization to endoplasmic reticulum | 2.31 | 1.84e-03 | 3.15e-02 | 15/119 | RYR2; RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| GO:0050769 | positive regulation of neurogenesis | 2.31 | 2.45e-04 | 7.69e-03 | 21/167 | CRABP2; AGT; SOX11; ID2; NEUROD1; MEF2C; DPYSL3; TUBB2B; HDAC2; DYNLT1; CAMK2B; FZD1; NEFL; RAPGEF1; ILK; TCF12; SERPINF1; RND2; NGFR; BAIAP2; PCP4 |
| GO:0002791 | regulation of peptide secretion | 2.27 | 3.05e-03 | 4.07e-02 | 14/113 | PRKCZ; WLS; SYT11; NEUROD1; KLF7; CAPN10; BTN2A2; EZR; STX1A; SRI; TMSB4X; CHD7; SYT7; CACNA1A |
| GO:0051346 | negative regulation of hydrolase activity | 2.23 | 1.42e-03 | 2.81e-02 | 17/140 | PRKCZ; AGT; PPP1R1C; MAP2; GPX1; GNAI2; SNCA; PTTG1; PPP1R17; PCSK1N; SERPINF1; NGFR; PPP1R27; FKBP1A; CST3; PPP1R14A; CSTB |
| GO:0032970 | regulation of actin filament-based process | 2.11 | 3.38e-03 | 4.43e-02 | 16/139 | STMN1; RHOC; NOS1AP; RYR2; CDC42EP3; MEF2C; EZR; SRI; AKAP9; TMSB4X; GPM6B; ILK; CFL1; RND2; BAIAP2; PREX1 |
| GO:0009636 | response to toxic substance | 2.06 | 1.91e-03 | 3.18e-02 | 19/169 | HPCA; PRDX1; EPHX1; SCN9A; CHRNA1; EEF1B2; GPX1; HDAC2; ACTB; G6PD; NEFL; GSTP1; RPS3; FAM213A; RGS10; HNRNPA1; CALM1; ACTC1; SNN |
Factor 5 : 13 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:1902600 | proton transmembrane transport | 3.33 | 1.51e-05 | 1.84e-03 | 16/77 | ATP6V0B; COX5B; ATP5G3; COX7C; TMSB4X; ATP5B; SLC25A3; COX6A1; ATP5A1; ATP5E; ATP5O; MT-CO1; MT-CO2; MT-CO3; MT-ND4; MT-CYB |
| GO:0010257 | NADH dehydrogenase complex assembly | 3.20 | 2.61e-04 | 1.92e-02 | 12/60 | NDUFS2; NDUFS6; NDUFB11; NDUFS8; NDUFC2; NDUFA12; NDUFB10; NDUFAB1; ATP5SL; MT-ND1; MT-ND2; MT-ND4 |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.97 | 2.82e-09 | 1.04e-06 | 35/189 | ATPIF1; NDUFS2; PARP1; COX5B; ATP5G3; SDHA; NDUFS6; COX7C; PMPCB; TMSB4X; PGK1; NDUFS8; NDUFC2; SDHD; HSPA8; PGAM1; TPI1; ATP5B; NDUFA12; COX6A1; NDUFB10; UQCRC2; NDUFAB1; COQ9; NCOR1; ATP5A1; ATP5E; ATP5O; MT-ND1; MT-ND2; MT-CO1; MT-CO2; MT-CO3; MT-ND4; MT-CYB |
| GO:0009123 | nucleoside monophosphate metabolic process | 2.88 | 6.69e-09 | 1.64e-06 | 35/195 | ATPIF1; NDUFS2; PARP1; COX5B; ATP5G3; SDHA; NDUFS6; COX7C; PMPCB; TMSB4X; PGK1; NDUFS8; NDUFC2; SDHD; HSPA8; PGAM1; TPI1; ATP5B; NDUFA12; COX6A1; NDUFB10; UQCRC2; NDUFAB1; COQ9; NCOR1; ATP5A1; ATP5E; ATP5O; MT-ND1; MT-ND2; MT-CO1; MT-CO2; MT-CO3; MT-ND4; MT-CYB |
| GO:0006091 | generation of precursor metabolites and energy | 2.84 | 8.99e-11 | 6.60e-08 | 44/248 | HMGCL; MTFR1L; ATPIF1; NDUFS2; FH; COX5B; PDHB; ADH5; SDHA; NDUFS6; COX7C; PRELID1; TBRG4; PMPCB; PGK1; PTGES2; NDUFS8; NDUFC2; SDHD; PGAM1; TPI1; ATP5B; NDUFA12; COX6A1; HMGB1; SUCLA2; NDUFB10; TRAP1; UQCRC2; NDUFAB1; CIAPIN1; COQ9; NCOR1; ATP5A1; ATP5E; GLTSCR2; ATP5O; MT-ND1; MT-ND2; MT-CO1; MT-CO2; MT-CO3; MT-ND4; MT-CYB |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 2.58 | 8.61e-04 | 4.86e-02 | 14/87 | NDUFS2; NDUFS6; NDUFB11; NDUFS8; NDUFC2; NDUFA12; NDUFB10; NDUFAB1; UQCC1; ATP5SL; SAMM50; MT-ND1; MT-ND2; MT-ND4 |
| GO:0006413 | translational initiation | 2.55 | 1.04e-05 | 1.52e-03 | 25/157 | YTHDF2; RPL5; TPR; DDX1; RPL31; EIF4A2; EIF4E; RPL34; RPS3A; HSPB1; RPS4X; RPL10; COPS5; RPL7; EIF3E; CDC123; RPS24; RPLP0; RPS29; EIF3J; EIF4A1; RPL19; EIF3D; RPL3; ATF4 |
| GO:0070646 | protein modification by small protein removal | 2.45 | 6.86e-05 | 6.30e-03 | 22/144 | USP39; PSMD6; PSMD2; COPS4; PSMB1; ACTB; PSMC2; COPS5; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; COPS3; PSMD11; PSMB3; PSMD3; PSMA7; KEAP1; PSMD8; GLTSCR2 |
| GO:0009259 | ribonucleotide metabolic process | 2.43 | 1.11e-07 | 2.03e-05 | 39/257 | HMGCL; ATPIF1; NDUFS2; PARP1; COX5B; ATP5G3; PDHB; SDHA; NDUFS6; COX7C; PMPCB; TMSB4X; PGK1; NDUFS8; NDUFC2; SDHD; HSPA8; PGAM1; TPI1; ATP5B; NDUFA12; COX6A1; SUCLA2; NDUFB10; UQCRC2; NDUFAB1; COQ9; NCOR1; ACSF2; ATP5A1; ATP5E; ATP5O; MT-ND1; MT-ND2; MT-CO1; MT-CO2; MT-CO3; MT-ND4; MT-CYB |
| GO:0043687 | post-translational protein modification | 2.43 | 2.52e-05 | 2.64e-03 | 25/165 | PSMD6; UBA3; PSMD2; COPS4; PSMB1; PSMC2; RAB2A; COPS5; DCAF13; PSMB7; PSMD13; PSMC6; KTN1; PSMC1; PSMB6; COPS3; PSMD11; PSMB3; PSMD3; PSMA7; FBXL12; KEAP1; RAB8A; PSMD8; FBXO7 |
| GO:0006457 | protein folding | 2.21 | 8.28e-04 | 4.86e-02 | 19/138 | SEP15; PFDN2; CCT7; DNAJB2; CCT5; TBCC; HSP90AB1; TCP1; HSPB1; PRDX4; VBP1; HSPA8; ERP29; PSMC1; HSP90AA1; TRAP1; EMC6; VAPA; FKBP1A |
| GO:0051098 | regulation of binding | 2.12 | 5.59e-04 | 3.73e-02 | 22/166 | PEX14; PARP1; NEUROD1; DNAJB2; CRBN; HMGB2; PPP2CA; MDFI; HSP90AB1; ACTB; TMSB4X; EIF3E; HSF1; TDG; HMGB1; CALM1; WFIKKN1; TRAF4; EIF4A3; FKBP1A; EIF3D; MFNG |
| GO:0006401 | RNA catabolic process | 2.04 | 1.21e-04 | 9.86e-03 | 30/236 | PPP1R8; YTHDF2; RPL5; RBM8A; RNASEH1; RPL31; PCBP4; RPL34; RPS3A; PPP2CA; HNRNPA0; TBRG4; HSPB1; RPS4X; RPL10; RPL7; EIF3E; ZPR1; HSPA8; RPS24; RPLP0; UBC; RPS29; SLIRP; RNPS1; RPL19; EIF4A3; HNRNPM; PPP2R1A; RPL3 |
Factor 6 : 4 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0046683 | response to organophosphorus | 6.80 | 5.51e-05 | 0.0399 | 7/34 | HSPD1; NDUFS4; EZR; HSPA5; LDHA; HSP90B1; NME1 |
| GO:0014074 | response to purine-containing compound | 5.92 | 1.39e-04 | 0.0399 | 7/39 | HSPD1; NDUFS4; EZR; HSPA5; LDHA; HSP90B1; NME1 |
| GO:0006457 | protein folding | 3.11 | 2.42e-04 | 0.0443 | 13/138 | PDIA6; FKBP1B; HSPD1; HSPE1; CANX; HSP90AB1; PRDX4; HSPA5; HSP90B1; HSP90AA1; PDIA3; PPIB; CALR |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.79 | 1.63e-04 | 0.0399 | 16/189 | ENO1; GUK1; COX5B; NDUFS4; UQCRB; AK1; LDHA; BAD; COX6A1; RAN; PKM; NME1; NDUFS7; UQCR10; ATP5O; MT-CYB |
Factor 7 : 21 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 6.64 | 0.00e+00 | 0.00e+00 | 58/119 | RPL11; RPS8; RPL5; RPS27; RTN4; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; SPCS1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SEC61G; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; SEC61B; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| GO:0010257 | NADH dehydrogenase complex assembly | 6.36 | 0.00e+00 | 0.00e+00 | 28/60 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFC1; NDUFS6; NDUFA2; NDUFB2; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFS8; NDUFC2; NDUFB8; NDUFB1; NDUFB10; NDUFAB1; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND3; MT-ND4 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 5.79 | 0.00e+00 | 0.00e+00 | 37/87 | NDUFS5; COX20; NDUFB3; NDUFAF3; COX17; NDUFB4; NDUFC1; NDUFS6; NDUFA2; UQCC2; NDUFB2; NDUFB11; NDUFA1; UQCRB; NDUFB9; TMEM261; NDUFB6; NDUFS8; NDUFC2; NDUFB8; COX14; COX16; NDUFB1; NDUFB10; NDUFAB1; COA3; NDUFS7; NDUFA11; PET100; NDUFA7; NDUFB7; NDUFA13; NDUFA3; UQCR10; NDUFA6; MT-ND3; MT-ND4 |
| GO:1902600 | proton transmembrane transport | 5.66 | 0.00e+00 | 0.00e+00 | 32/77 | PARK7; ATP6V0B; COX5B; ATP5G3; COX17; ATP5I; COX7C; COX7A2; NDUFA4; ATP5J2; ATP6V1F; TMSB4X; COX7B; COX6C; ATP6V1G1; COX8A; ATP5L; ATP5G2; SLC25A3; COX6A1; COX5A; COX4I1; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; COX6B1; ATP5J; ATP5O; MT-CO1; MT-ND4 |
| GO:0006413 | translational initiation | 5.64 | 0.00e+00 | 0.00e+00 | 65/157 | RPL11; RPS8; RPL5; RPS27; DDX1; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; EIF4A2; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; MCTS1; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL35; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; EIF3J; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; EIF1; RPL18A; EIF3K; RPS19; RPL18; RPL13A; RPS11; EIF3L; RPL3; ATF4 |
| GO:0002181 | cytoplasmic translation | 5.41 | 4.22e-15 | 2.38e-13 | 29/73 | RPL11; RPL32; RPL15; TMA7; RPL29; RPL9; RPS23; RPL10A; RPL39; MCTS1; EIF4EBP1; RPL30; EIF3E; EIF3H; EIF3F; RPS26; RPL41; RPL6; RPLP0; RPS29; EIF3J; RPL26; RPL19; EEF2; RPL18A; EIF3K; RPL18; RPL13A; EIF3L |
| GO:0090150 | establishment of protein localization to membrane | 4.54 | 0.00e+00 | 0.00e+00 | 68/204 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPL10A; CD24; RPS12; FIS1; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPS6; SEC61B; RPL35; RPL7A; RPL27A; RPS13; TIMM10; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; CALM1; HSP90AA1; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; YWHAE; VAMP2; RPL26; RPL23A; RPL23; RPL19; ROMO1; TIMM13; RPL18A; NDUFA13; PDCD5; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| GO:0017004 | cytochrome complex assembly | 4.38 | 1.10e-04 | 4.05e-03 | 9/28 | COX20; COX17; UQCC2; UQCRB; COX14; COX16; COA3; PET100; UQCR10 |
| GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | 4.26 | 5.98e-05 | 2.44e-03 | 10/32 | ATP6V0B; ATP5G3; ATP6V1F; ATP5L; ATP5G2; ATP5G1; ATP5H; ATP5A1; ATP5E; ATP5O |
| GO:0009141 | nucleoside triphosphate metabolic process | 4.11 | 0.00e+00 | 0.00e+00 | 57/189 | PARK7; ATPIF1; NDUFS5; UQCRH; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; UQCRB; NDUFB9; NDUFB6; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5G2; COX6A1; RAN; DNAJC15; NDUFB1; COX5A; NDUFB10; NDUFAB1; DCTPP1; COX4I1; ATP5G1; NME1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| GO:0001906 | cell killing | 3.89 | 6.57e-04 | 2.30e-02 | 8/28 | HMGN2; PRDX1; TUBB; RPL30; CADM1; VAMP2; ROMO1; RPS19 |
| GO:0009123 | nucleoside monophosphate metabolic process | 3.84 | 0.00e+00 | 0.00e+00 | 55/195 | PARK7; ATPIF1; NDUFS5; UQCRH; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; UQCRQ; NDUFA2; UQCC2; COX7A2; HDDC2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; UQCRB; NDUFB9; NDUFB6; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5G2; COX6A1; DNAJC15; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| GO:0006605 | protein targeting | 3.81 | 0.00e+00 | 0.00e+00 | 73/261 | RPL11; ATPIF1; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; SPCS1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; TOMM7; SEC61G; FIS1; SLC25A6; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPS6; SEC61B; RPL35; RPL7A; RPL27A; RPS13; IMMP1L; TIMM10; RPS3; TIMM8B; RPS24; RPS26; RPL41; RPL6; RPLP0; UBC; RPL21; DNAJC15; RPS29; HSP90AA1; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; ROMO1; TIMM13; UBL5; RPL18A; NDUFA13; PDCD5; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| GO:0006401 | RNA catabolic process | 3.69 | 0.00e+00 | 0.00e+00 | 64/236 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; LSM3; RPL15; RPSA; RPL29; RPL9; RPL34; LSM6; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; LSM5; LINC01420; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; EIF3E; RPS6; RPL35; RPL7A; RPL27A; RPS13; RNASEH2C; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; SLIRP; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; LSM7; RPL18A; LSM4; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| GO:0007006 | mitochondrial membrane organization | 3.49 | 1.28e-07 | 6.69e-06 | 22/86 | ATPIF1; ATP5G3; ATP5I; DYNLT1; ATP5J2; YWHAZ; TIMM10; ATP5L; ATP5G2; CALM1; HSP90AA1; YWHAE; ATP5G1; ATP5H; ATP5A1; ROMO1; ATP5E; TIMM13; NDUFA13; PDCD5; ATP5J; ATP5O |
| GO:0006091 | generation of precursor metabolites and energy | 3.41 | 0.00e+00 | 0.00e+00 | 62/248 | PARK7; SH3BGRL3; ATPIF1; NDUFS5; UQCRH; COX20; COX5B; NDUFB3; COX17; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; GLRX; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; COX7B; NDUFA1; UQCRB; COX6C; NDUFB9; NDUFB6; COX8A; NDUFS8; NDUFC2; ATP5L; CISD1; NDUFB8; TPI1; BLOC1S1; COX6A1; HMGB1; DNAJC15; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5H; ATP5A1; ATP5E; NDUFS7; UQCR11; NDUFA7; NDUFB7; COX6B1; ETFB; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| GO:0009259 | ribonucleotide metabolic process | 3.07 | 1.11e-15 | 6.79e-14 | 58/257 | PARK7; ATPIF1; NDUFS5; UQCRH; COX5B; DBI; ATP5G3; NDUFB3; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; HINT1; UQCRQ; NDUFA2; UQCC2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; UQCRB; NDUFB9; NDUFB6; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5G2; COX6A1; RAN; DNAJC15; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5G1; NME1; ATP5H; ATP5A1; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| GO:0070585 | protein localization to mitochondrion | 2.64 | 1.10e-04 | 4.05e-03 | 18/93 | ATPIF1; TOMM7; FIS1; SLC25A6; YWHAZ; IMMP1L; TIMM10; TIMM8B; DDIT3; DNAJC15; CALM1; HSP90AA1; YWHAE; ROMO1; TIMM13; UBL5; NDUFA13; PDCD5 |
| GO:0006839 | mitochondrial transport | 2.50 | 6.01e-06 | 2.60e-04 | 27/147 | ATPIF1; COX5B; ATP5I; DYNLT1; TOMM7; ATP5J2; FIS1; SLC25A6; YWHAZ; IMMP1L; TIMM10; TIMM8B; ATP5L; DNAJC15; HSP90AA1; YWHAE; ATP5H; ATP5A1; ROMO1; ATP5E; TIMM13; UBL5; NDUFA13; PDCD5; SMDT1; ATP5J; ATP5O |
| GO:0071826 | ribonucleoprotein complex subunit organization | 2.36 | 3.14e-06 | 1.44e-04 | 32/185 | MRPL20; RPL11; RPL5; RPS27; SNRPE; DDX1; SNRPG; LSM3; RPSA; RPS23; RPS14; LINC01420; MCTS1; RPL10; EIF3E; EIF3H; EIF3F; SNRPF; RPL6; RPLP0; HSP90AA1; EIF3J; RPL23A; SNRPD1; LSM4; EIF3K; RPS19; SNRPD2; RPL13A; EIF3L; RPL3; SNU13 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 2.02 | 8.46e-07 | 4.14e-05 | 49/330 | MRPL20; RPL11; RPS8; RPL5; RPS27; SNRPE; DDX1; SNRPG; LSM3; RPSA; LSM6; RPS23; RPS14; NHP2; RPL10A; GTF2H5; LINC01420; MCTS1; RPL10; RPL7; EIF3E; EIF3H; RPS6; RPL35; RPL7A; EIF3F; TRMT112; RPS24; PA2G4; SNRPF; RPL6; RPLP0; RAN; HSP90AA1; NOP10; EIF3J; RPS17; RPS2; RPL26; RPL23A; SNRPD1; LSM4; EIF3K; RPS19; SNRPD2; RPL13A; EIF3L; RPL3; SNU13 |
Factor 8 : 7 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0050954 | sensory perception of mechanical stimulus | 4.42 | 3.33e-04 | 3.49e-02 | 8/39 | CASP3; FAM65B; POU3F4; NAV2; COL2A1; CACNB3; SRRM4; SIX1 |
| GO:0070972 | protein localization to endoplasmic reticulum | 4.17 | 2.92e-09 | 2.14e-06 | 23/119 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; OS9; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| GO:0002181 | cytoplasmic translation | 3.55 | 1.10e-04 | 1.61e-02 | 12/73 | RPL32; RPL24; RPL36A; RPL8; ZNF385A; RPS26; RPL41; RPL6; RPLP0; DENR; RPL26; RPL36 |
| GO:0006413 | translational initiation | 3.16 | 6.32e-07 | 1.55e-04 | 23/157 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; DENR; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| GO:0090150 | establishment of protein localization to membrane | 2.85 | 5.20e-07 | 1.55e-04 | 27/204 | RPL32; RPL24; NSG1; RPL34; GNB2L1; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; CACNB3; RPS26; RPL41; RPL6; RPLP0; CALM1; RPS17; RPL13; VAMP2; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| GO:0006401 | RNA catabolic process | 2.47 | 9.11e-06 | 1.67e-03 | 27/236 | RPL32; RPL24; RPL34; RPS18; LIN28B; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RNASEH2C; RPS3; MAGOHB; PYM1; RPS26; RPL41; CNOT2; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| GO:0006605 | protein targeting | 2.15 | 1.48e-04 | 1.81e-02 | 26/261 | RPL32; RPL24; RPL34; RPS18; TOMM7; SLC25A6; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; CACNB3; RPS26; RPL41; OS9; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
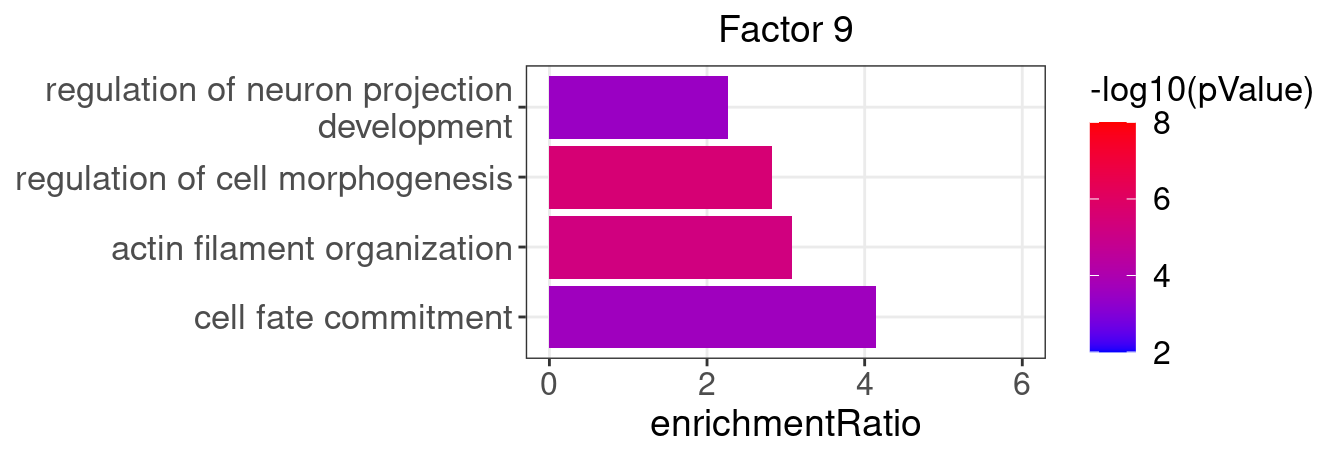
Factor 9 : 72 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0048645 | animal organ formation | 9.85 | 7.37e-05 | 0.00492 | 5/11 | GNG5; CTNNB1; ROBO1; MEF2C; SIX1 |
| GO:0007218 | neuropeptide signaling pathway | 8.67 | 7.45e-04 | 0.01709 | 4/10 | GAL; NMB; SSTR2; PTH2 |
| GO:0034109 | homotypic cell-cell adhesion | 7.22 | 1.05e-04 | 0.00552 | 6/18 | CLIC1; ACTB; ILK; ANK3; VCL; ACTG1 |
| GO:0007586 | digestion | 6.67 | 2.27e-03 | 0.02979 | 4/13 | NEUROD1; SST; NEUROG1; LIMA1 |
| GO:0048483 | autonomic nervous system development | 6.67 | 2.27e-03 | 0.02979 | 4/13 | CTNNB1; SIX1; INSM1; ADARB1 |
| GO:0021675 | nerve development | 4.92 | 2.77e-03 | 0.03444 | 5/22 | CTNNB1; NEUROG1; ILK; SIX1; ADARB1 |
| GO:0030048 | actin filament-based movement | 4.64 | 9.52e-05 | 0.00552 | 9/42 | ANK2; CAMK2D; CACNA2D1; TPM2; MYL6; ACTC1; EMP2; TNNC2; TPM4 |
| GO:0010927 | cellular component assembly involved in morphogenesis | 4.60 | 6.21e-04 | 0.01519 | 7/33 | WDR1; ANK2; MEF2C; ILK; ACTC1; ACTG1; EPB41L3 |
| GO:0035249 | synaptic transmission, glutamatergic | 4.51 | 4.14e-03 | 0.04473 | 5/24 | NRXN1; GRIA2; MEF2C; SLC17A6; SYT1 |
| GO:0001763 | morphogenesis of a branching structure | 4.33 | 9.03e-04 | 0.01842 | 7/35 | PBX1; CTNNB1; TNC; ILK; SIX1; PGF; PKD1 |
| GO:0045165 | cell fate commitment | 4.15 | 2.38e-04 | 0.00864 | 9/47 | ID2; NEUROD1; CTNNB1; CASP3; MEF2C; NEUROG1; EBF2; SIX1; ONECUT2 |
| GO:0003007 | heart morphogenesis | 4.06 | 2.72e-05 | 0.00249 | 12/64 | JUN; GNG5; ID2; CTNNB1; ROBO1; IFT57; COL4A3BP; MEF2C; OLFM1; ILK; SIX1; ACTC1 |
| GO:0035270 | endocrine system development | 4.06 | 2.99e-03 | 0.03565 | 6/32 | PBX1; NEUROD1; CDKN1C; SIX1; ONECUT2; INSM1 |
| GO:0031032 | actomyosin structure organization | 4.04 | 6.25e-05 | 0.00492 | 11/59 | STMN1; EPB41; RHOC; CD47; WDR1; MEF2C; ZYX; ACTC1; PFN1; ACTG1; EPB41L3 |
| GO:0060560 | developmental growth involved in morphogenesis | 3.89 | 2.18e-06 | 0.00069 | 16/89 | DRAXIN; CRABP2; ZEB2; MAP2; CTNNB1; ALCAM; MAP1B; TNC; OLFM1; ILK; GAL; VCL; SYT1; SIX1; SYT4; DCC |
| GO:0007272 | ensheathment of neurons | 3.89 | 1.77e-03 | 0.02699 | 7/39 | POU3F1; CNTN2; CTNNB1; ANK2; ILK; EPB41L3; LGI4 |
| GO:0030534 | adult behavior | 3.85 | 8.92e-04 | 0.01842 | 8/45 | CNTN2; ID2; NRXN1; HDAC2; PBX3; PCDH17; LGI4; SEZ6L |
| GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 3.79 | 2.06e-03 | 0.02838 | 7/40 | CNTN2; NRXN1; ROBO1; ALCAM; CADM1; PCDH17; PKD1 |
| GO:1904029 | regulation of cyclin-dependent protein kinase activity | 3.79 | 2.06e-03 | 0.02838 | 7/40 | GADD45A; CCNI; CASP3; ACTB; CDKN1C; CCNY; PKD1 |
| GO:0003012 | muscle system process | 3.68 | 5.99e-07 | 0.00044 | 19/112 | CHRNA1; ANK2; CAMK2D; SORBS2; COL4A3BP; MEF2C; NEUROG1; HDAC2; CACNA2D1; TPM2; VCL; MYL6; AKAP6; CALM1; ACTC1; SSTR2; TNNC2; TPM4; FXYD1 |
| GO:0008544 | epidermis development | 3.61 | 1.80e-04 | 0.00817 | 11/66 | POU3F1; CRABP2; PKP4; SATB1; CTNNB1; CASP3; FAM65B; HDAC2; AGPAT2; GAL; TNFRSF19 |
| GO:0021953 | central nervous system neuron differentiation | 3.61 | 7.00e-04 | 0.01658 | 9/54 | DRAXIN; CNTN2; ZEB2; MAP2; CTNNB1; ROBO1; POU3F4; DCC; ADARB1 |
| GO:0090130 | tissue migration | 3.57 | 1.38e-05 | 0.00145 | 15/91 | JUN; ZEB2; SCG2; IQSEC1; ROBO1; MEF2C; SPARC; MEOX2; TMSB4X; CORO1C; ACTC1; EMP2; SERPINF1; PFN1; MAPRE2 |
| GO:0001655 | urogenital system development | 3.56 | 1.05e-04 | 0.00552 | 12/73 | PBX1; ID2; CTNNB1; MEF2C; TNC; CDKN1C; ILK; EMX2; SIX1; PGF; PKD1; SERPINF1 |
| GO:0035637 | multicellular organismal signaling | 3.53 | 3.16e-03 | 0.03683 | 7/43 | CHRNA1; ANK2; CAMK2D; CACNA2D1; ANK3; ATP2B1; CALM1 |
| GO:0048568 | embryonic organ development | 3.51 | 4.68e-06 | 0.00081 | 17/105 | MFAP2; NES; PBX1; VASH2; ID2; NCOA1; NEUROD1; CTNNB1; IFT57; MEF2C; NEUROG1; FAM65B; POU3F4; CTHRC1; CDKN1C; SIX1; PKD1 |
| GO:0032409 | regulation of transporter activity | 3.48 | 7.01e-05 | 0.00492 | 13/81 | ANK2; CAMK2D; MEF2C; GLRX; ACTB; CACNA2D1; TMSB4X; GAL; FXYD6; ANK3; AKAP6; CALM1; FXYD1 |
| GO:0043588 | skin development | 3.42 | 1.05e-03 | 0.02028 | 9/57 | STMN1; POU3F1; PKP4; CTNNB1; CASP3; HDAC2; GAL; TNFRSF19; PKD1 |
| GO:0009612 | response to mechanical stimulus | 3.40 | 2.08e-03 | 0.02838 | 8/51 | JUN; GADD45A; NRXN1; NEUROG1; FAM65B; ETV1; TNC; PKD1 |
| GO:0015850 | organic hydroxy compound transport | 3.40 | 2.08e-03 | 0.02838 | 8/51 | NCOA1; ACTB; SYT7; GAL; LIMA1; SYT1; SYT4; SELM |
| GO:0070661 | leukocyte proliferation | 3.33 | 2.36e-03 | 0.03035 | 8/52 | SATB1; CTNNB1; CASP3; MEF2C; BTN2A2; GAL; RPS3; CADM1 |
| GO:0061351 | neural precursor cell proliferation | 3.27 | 2.67e-03 | 0.03376 | 8/53 | GNG5; NES; ID2; ZEB2; CTNNB1; ILK; EMX2; INSM1 |
| GO:0043270 | positive regulation of ion transport | 3.19 | 9.90e-04 | 0.01964 | 10/68 | ANK2; GLRX; CACNA2D1; TMSB4X; GAL; ANK3; AKAP6; CALM1; SYT4; FXYD1 |
| GO:0006814 | sodium ion transport | 3.15 | 3.39e-03 | 0.03883 | 8/55 | CAMK2D; GLRX; SLC17A6; FXYD6; ANK3; ASIC1; NKAIN4; FXYD1 |
| GO:0097305 | response to alcohol | 3.10 | 1.25e-03 | 0.02217 | 10/70 | EEF1B2; CTNNB1; SPARC; HDAC2; LANCL2; TNC; ATP2B1; CALM1; ACTC1; MT-ND4 |
| GO:0007015 | actin filament organization | 3.07 | 5.52e-06 | 0.00081 | 20/141 | STMN1; RHOC; ARPC5; CDC42EP3; RND3; CD47; WDR1; SORBS2; DPYSL3; CAP2; ZYX; TMSB4X; TPM2; CFL1; LIMA1; ACTC1; EMP2; PFN1; PREX1; TPM4 |
| GO:0060485 | mesenchyme development | 3.05 | 1.39e-03 | 0.02271 | 10/71 | ZEB2; CTNNB1; ROBO1; MEF2C; HDAC2; OLFM1; CFL1; CORO1C; SIX1; ACTC1 |
| GO:0050673 | epithelial cell proliferation | 3.00 | 3.29e-04 | 0.01051 | 13/94 | JUN; VASH2; ID2; SCG2; CTNNB1; ROBO1; IFT57; MEF2C; SPARC; CDKN1C; SIX1; PGF; SERPINF1 |
| GO:0090596 | sensory organ morphogenesis | 2.99 | 4.73e-03 | 0.04825 | 8/58 | MFAP2; CTNNB1; NEUROG1; FAM65B; HDAC2; POU3F4; CTHRC1; SIX1 |
| GO:0099504 | synaptic vesicle cycle | 2.99 | 5.72e-04 | 0.01493 | 12/87 | NRXN1; CTNNB1; DNM1; SLC17A6; SYT7; SYT1; PCDH17; TRIM9; CALM1; DOC2B; ACTG1; SYT4 |
| GO:0034330 | cell junction organization | 2.97 | 2.11e-04 | 0.00817 | 14/102 | RHOC; PKP4; IQSEC1; CTNNB1; WDR1; ANK2; ACTB; ILK; CADM1; VCL; CORO1C; ACTG1; EPB41L3; MAPRE2 |
| GO:0034764 | positive regulation of transmembrane transport | 2.95 | 3.01e-03 | 0.03565 | 9/66 | ANK2; GLRX; CACNA2D1; TMSB4X; GAL; ANK3; AKAP6; CALM1; FXYD1 |
| GO:0071241 | cellular response to inorganic substance | 2.95 | 3.01e-03 | 0.03565 | 9/66 | JUN; ID2; CAMK2D; MEF2C; SYT7; ANK3; SYT1; SERPINF1; SYT4 |
| GO:0031589 | cell-substrate adhesion | 2.87 | 1.35e-03 | 0.02271 | 11/83 | CTNNB1; ZYX; NINJ1; ILK; RSU1; VCL; CORO1C; PKD1; EMP2; ONECUT2; PREX1 |
| GO:0001667 | ameboidal-type cell migration | 2.86 | 1.17e-04 | 0.00574 | 16/121 | JUN; ZEB2; SCG2; IQSEC1; ROBO1; MEF2C; SPARC; MEOX2; TMSB4X; ILK; CFL1; CORO1C; EMP2; SERPINF1; PFN1; MAPRE2 |
| GO:0010959 | regulation of metal ion transport | 2.84 | 5.51e-04 | 0.01493 | 13/99 | CTNNB1; ANK2; CAMK2D; GLRX; CACNA2D1; GAL; FXYD6; ANK3; AKAP6; CALM1; NKAIN4; HOMER3; FXYD1 |
| GO:0022604 | regulation of cell morphogenesis | 2.83 | 2.82e-06 | 0.00069 | 24/184 | DRAXIN; EPB41; RHOC; S100A13; CRABP2; CNTN2; CDC42EP3; ZEB2; RND3; MAP2; ROBO1; WDR1; MAP1B; SPARC; OLFM1; ILK; CFL1; SYT1; CORO1C; LINGO1; EPB41L3; SYT4; DCC; PREX1 |
| GO:0010721 | negative regulation of cell development | 2.80 | 2.47e-04 | 0.00864 | 15/116 | DRAXIN; PBX1; CNTN2; ID2; MAP2; ITM2C; CTNNB1; NR2F1; DPYSL3; HDAC2; GAL; CORO1C; LINGO1; SYT4; DCC |
| GO:0001525 | angiogenesis | 2.76 | 4.69e-04 | 0.01378 | 14/110 | JUN; SHC1; VASH2; SCG2; CTNNB1; ROBO1; SPARC; THSD7A; MEOX2; SAT1; VAV2; PGF; EMP2; SERPINF1 |
| GO:0048638 | regulation of developmental growth | 2.71 | 8.88e-04 | 0.01842 | 13/104 | DRAXIN; CRABP2; MAP2; MAP1B; MEF2C; OLFM1; ILK; GAL; SYT1; AKAP6; SIX1; SYT4; DCC |
| GO:0048880 | sensory system development | 2.65 | 1.67e-03 | 0.02610 | 12/98 | MFAP2; JUN; NES; ZEB2; NEUROD1; CTNNB1; HDAC2; NINJ1; CDKN1C; ATP2B1; MAB21L1; SERPINF1 |
| GO:0051961 | negative regulation of nervous system development | 2.63 | 1.16e-03 | 0.02133 | 13/107 | DRAXIN; PBX1; CNTN2; ID2; MAP2; ITM2C; CTNNB1; NR2F1; DPYSL3; HDAC2; LINGO1; SYT4; DCC |
| GO:0061564 | axon development | 2.62 | 7.11e-06 | 0.00087 | 25/207 | DRAXIN; STMN1; JUN; SHC1; CRABP2; LHX9; CNTN2; NRXN1; ZEB2; MAP2; ROBO1; ALCAM; CASP3; MAP1B; PDLIM7; ETV1; TNC; OLFM1; ILK; NCAM1; ANK3; VCL; LINGO1; DCC; ADARB1 |
| GO:0007517 | muscle organ development | 2.61 | 1.27e-03 | 0.02217 | 13/108 | CHRNA1; CTNNB1; MEF2C; NEUROG1; FAM65B; ETV1; MEOX2; MYL6; AKAP6; SIX1; ACTC1; TCF12; ADARB1 |
| GO:0007163 | establishment or maintenance of cell polarity | 2.60 | 1.99e-03 | 0.02838 | 12/100 | RHOC; ARPC5; RND3; MAP2; FRMD4B; WDR1; MAP1B; CAP2; FAM65B; ILK; CFL1; PKD1 |
| GO:0034765 | regulation of ion transmembrane transport | 2.59 | 8.81e-04 | 0.01842 | 14/117 | WDR1; ANK2; CAMK2D; MEF2C; GLRX; CLIC1; CACNA2D1; TMSB4X; GAL; FXYD6; ANK3; AKAP6; CALM1; FXYD1 |
| GO:0002009 | morphogenesis of an epithelium | 2.55 | 4.61e-04 | 0.01378 | 16/136 | RHOC; PBX1; ZEB2; CTNNB1; IFT57; WDR1; MEF2C; CTHRC1; TNC; ILK; CFL1; VCL; SIX1; PGF; PKD1; PFN1 |
| GO:0097485 | neuron projection guidance | 2.54 | 3.71e-03 | 0.04125 | 11/94 | DRAXIN; SHC1; LHX9; CNTN2; NRXN1; ROBO1; ALCAM; PDLIM7; ETV1; NCAM1; DCC |
| GO:0042391 | regulation of membrane potential | 2.53 | 1.13e-03 | 0.02133 | 14/120 | JUN; NRXN1; CHRNA1; WDR1; ANK2; CAMK2D; MEF2C; GLRX; CLIC1; CACNA2D1; ANK3; ASIC1; AKAP6; FXYD1 |
| GO:0001503 | ossification | 2.51 | 4.03e-03 | 0.04413 | 11/95 | PBX1; ID2; CTNNB1; MEF2C; SPARC; PDLIM7; CLIC1; CTHRC1; TNC; ILK; TPM4 |
| GO:0032970 | regulation of actin filament-based process | 2.49 | 5.90e-04 | 0.01493 | 16/139 | STMN1; RHOC; ARPC5; CDC42EP3; RND3; CD47; WDR1; ANK2; CAMK2D; MEF2C; TMSB4X; ILK; CFL1; LIMA1; PFN1; PREX1 |
| GO:0051271 | negative regulation of cellular component movement | 2.48 | 4.37e-03 | 0.04648 | 11/96 | SRGAP3; ROBO1; MEF2C; DPYSL3; FAM65B; MEOX2; ILK; VCL; CORO1C; SERPINF1; ADARB1 |
| GO:0051098 | regulation of binding | 2.48 | 1.95e-04 | 0.00817 | 19/166 | STMN1; EPB41; JUN; NES; ID2; NEUROD1; MAP2; CTNNB1; MEF2C; FAM65B; HDAC2; ACTB; TMSB4X; EBF2; CTHRC1; CALM1; CCPG1; WFIKKN1; PKD1 |
| GO:0010038 | response to metal ion | 2.47 | 2.09e-03 | 0.02838 | 13/114 | JUN; S100A13; ID2; CAMK2D; CASP3; MEF2C; SPARC; SYT7; ANK3; SYT1; CALM1; SERPINF1; SYT4 |
| GO:0050769 | positive regulation of neurogenesis | 2.46 | 2.11e-04 | 0.00817 | 19/167 | CRABP2; ID2; NCOA1; ZEB2; NEUROD1; CTNNB1; ROBO1; MAP1B; MEF2C; NEUROG1; DPYSL3; HDAC2; ILK; SYT1; TCF12; SERPINF1; SYT4; TCF4; PCP4 |
| GO:0007389 | pattern specification process | 2.36 | 4.44e-03 | 0.04655 | 12/110 | PBX1; ZEB2; NEUROD1; CTNNB1; ROBO1; IFT57; MEF2C; NEUROG1; MEOX2; PBX3; EMX2; SIX1 |
| GO:0010975 | regulation of neuron projection development | 2.26 | 3.26e-04 | 0.01051 | 21/201 | DRAXIN; CRABP2; CNTN2; ZEB2; MAP2; ITM2C; ROBO1; MAP1B; MEF2C; NR2F1; DPYSL3; HDAC2; OLFM1; ILK; CFL1; SYT1; LINGO1; SERPINF1; SYT4; DCC; PREX1 |
| GO:0030900 | forebrain development | 2.25 | 3.49e-03 | 0.03940 | 14/135 | DRAXIN; POU3F1; ARPC5; CNTN2; ID2; ZEB2; NEUROD1; CTNNB1; ROBO1; CASP3; POU3F4; EMX2; TACC2; SSTR2 |
| GO:0090066 | regulation of anatomical structure size | 2.21 | 1.62e-03 | 0.02591 | 17/167 | DRAXIN; CRABP2; ARPC5; CNTN2; CDC42EP3; MAP2; WDR1; MAP1B; TMSB4X; VAV2; OLFM1; ILK; CFL1; LIMA1; PFN1; DCC; PREX1 |
| GO:0016049 | cell growth | 2.18 | 1.37e-03 | 0.02271 | 18/179 | DRAXIN; CRABP2; ZEB2; MAP2; CTNNB1; ALCAM; CAMK2D; SORBS2; MAP1B; OLFM1; ILK; GAL; VCL; SYT1; AKAP6; EPB41L3; SYT4; DCC |
| GO:0051493 | regulation of cytoskeleton organization | 2.13 | 5.62e-04 | 0.01493 | 22/224 | STMN1; RHOC; NES; ARPC5; CDC42EP3; RND3; MAP2; CTNNB1; CD47; WDR1; MEF2C; TMSB4X; STMN4; ILK; CFL1; RPS3; BICD1; LIMA1; PKD1; PFN1; MAPRE2; PREX1 |
| GO:0009636 | response to toxic substance | 2.05 | 4.60e-03 | 0.04758 | 16/169 | JUN; EPHX1; CHRNA1; EEF1B2; CASP3; SPARC; HDAC2; ACTB; PON2; TNC; RPS3; ASIC1; CALM1; ACTC1; NXN; MT-ND4 |
Factor 11 : 8 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0006959 | humoral immune response | 13.50 | 2.61e-05 | 3.83e-03 | 5/24 | HMGN2; RPL39; CD59; GAPDH; RPS19 |
| GO:0001906 | cell killing | 11.60 | 5.75e-05 | 7.03e-03 | 5/28 | HMGN2; PRDX1; TUBB; GAPDH; RPS19 |
| GO:0006413 | translational initiation | 7.42 | 1.14e-11 | 8.37e-09 | 18/157 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; EIF4EBP1; RPL12; RPS24; RPL21; EIF5; RPS2; EIF1; RPL27; EIF2S2; RPS19; RPS11; ATF4 |
| GO:0070972 | protein localization to endoplasmic reticulum | 7.07 | 2.13e-08 | 7.82e-06 | 13/119 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| GO:0048871 | multicellular organismal homeostasis | 4.62 | 2.93e-04 | 2.69e-02 | 8/112 | STMN1; PRDX1; NEUROD1; ACTB; GADD45G; DDIT3; DYNC1H1; ATF4 |
| GO:0090150 | establishment of protein localization to membrane | 4.44 | 2.02e-06 | 4.94e-04 | 14/204 | RPL37A; RPL35A; RPL37; GNB2L1; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| GO:0006401 | RNA catabolic process | 3.84 | 1.12e-05 | 2.05e-03 | 14/236 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; SLIRP; RPS2; RPL27; RPS19; RPS11 |
| GO:0006605 | protein targeting | 3.22 | 1.48e-04 | 1.55e-02 | 13/261 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
Factor 13 : 2 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0015844 | monoamine transport | 45.7 | 3.21e-05 | 0.0141 | 3/17 | ACTB; SNCG; SYT1 |
| GO:0034109 | homotypic cell-cell adhesion | 43.1 | 3.85e-05 | 0.0141 | 3/18 | CLIC1; ACTB; ACTG1 |
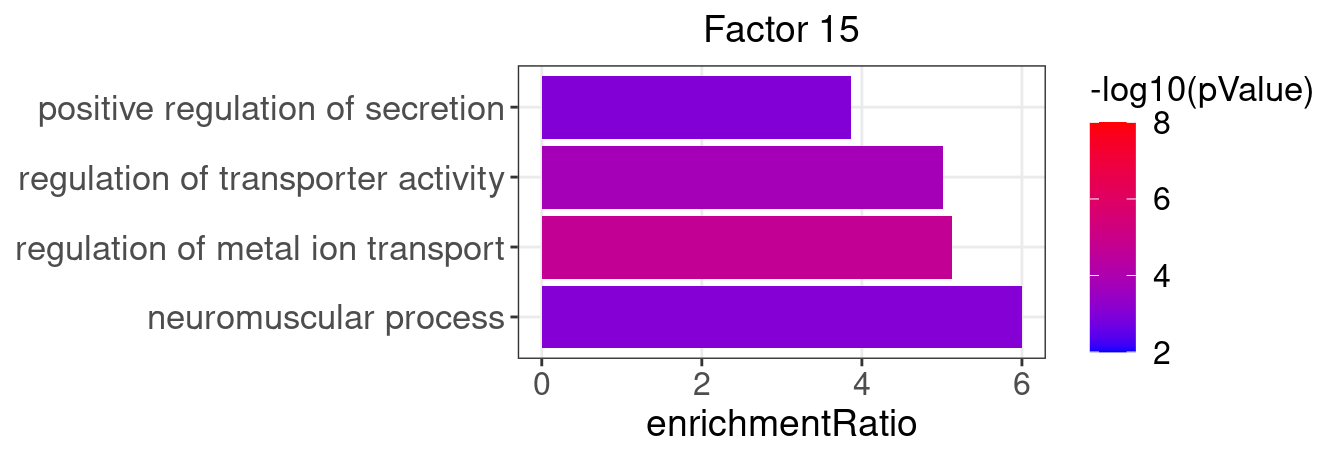
Factor 15 : 20 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0015844 | monoamine transport | 14.90 | 1.38e-05 | 0.00748 | 5/17 | SNCA; ACTB; GPM6B; SYT1; SYT4 |
| GO:0034109 | homotypic cell-cell adhesion | 11.30 | 3.52e-04 | 0.02169 | 4/18 | ACTB; HSPB1; ACTG1; MYL12A |
| GO:0060191 | regulation of lipase activity | 10.70 | 4.39e-04 | 0.02169 | 4/19 | HPCA; RHOC; SNCA; NTF3 |
| GO:0015696 | ammonium transport | 8.82 | 9.44e-04 | 0.03829 | 4/23 | SNCA; GPM6B; SYT1; SYT4 |
| GO:2001023 | regulation of response to drug | 8.74 | 2.20e-04 | 0.02169 | 5/29 | NFE2L2; SNCA; GAL; SYT1; SYT4 |
| GO:0050905 | neuromuscular process | 6.50 | 9.11e-04 | 0.03829 | 5/39 | CHRNA1; NEUROG1; NEFL; POU4F1; IGDCC3 |
| GO:0043583 | ear development | 6.21 | 3.55e-04 | 0.02169 | 6/49 | HPCA; NEUROD1; NEUROG1; SPARC; FAM65B; CTHRC1 |
| GO:0015893 | drug transport | 6.04 | 1.28e-03 | 0.04714 | 5/42 | SNCA; SLC25A6; GAL; SYT1; SYT4 |
| GO:0015850 | organic hydroxy compound transport | 5.96 | 4.43e-04 | 0.02169 | 6/51 | SNCA; ACTB; GPM6B; GAL; SYT1; SYT4 |
| GO:0010959 | regulation of metal ion transport | 5.12 | 2.04e-05 | 0.00748 | 10/99 | HPCA; ATP2B4; SNCA; SRI; GAL; REM2; B2M; NKAIN4; FXYD7; HRC |
| GO:0032409 | regulation of transporter activity | 5.01 | 1.69e-04 | 0.02062 | 8/81 | HPCA; SNCA; ACTB; SRI; GAL; REM2; FXYD7; HRC |
| GO:0007517 | muscle organ development | 4.69 | 4.37e-05 | 0.01042 | 10/108 | CHRNA1; BASP1; NEUROG1; FAM65B; UQCC2; SRI; CNTFR; MYL6; POU4F1; EFNB2 |
| GO:0048880 | sensory system development | 4.66 | 1.15e-04 | 0.01686 | 9/98 | HPCA; NES; ATP2B4; SIX3; NEUROD1; MAB21L2; NEUROD4; POU4F1; CELF4 |
| GO:0009914 | hormone transport | 4.61 | 3.01e-04 | 0.02169 | 8/88 | NEUROD1; FOXD1; SOX4; UQCC2; SRI; SLC25A6; GAL; DYNLL1 |
| GO:0050878 | regulation of body fluid levels | 4.02 | 7.66e-04 | 0.03516 | 8/101 | NFE2L2; CDO1; NEUROG1; ACTB; HSPB1; ZNF385A; ACTG1; MYL12A |
| GO:0034765 | regulation of ion transmembrane transport | 3.90 | 4.43e-04 | 0.02169 | 9/117 | HPCA; ATP2B4; SCN9A; SNCA; SRI; GAL; REM2; FXYD7; HRC |
| GO:0051047 | positive regulation of secretion | 3.86 | 9.91e-04 | 0.03829 | 8/105 | SNCA; SOX4; SRI; GAL; CADM1; SYT1; DYNLL1; SYT4 |
| GO:0023061 | signal release | 3.83 | 5.68e-05 | 0.01042 | 12/159 | NEUROD1; SNCA; FOXD1; SOX4; UQCC2; SRI; SLC25A6; GAL; ASIC1; SYT1; DYNLL1; SYT4 |
| GO:0051098 | regulation of binding | 3.36 | 3.73e-04 | 0.02169 | 11/166 | EPB41; NES; NEUROD1; FAM65B; ACTB; SRI; CTHRC1; POU4F1; B2M; CCPG1; WFIKKN1 |
| GO:0009636 | response to toxic substance | 3.30 | 4.36e-04 | 0.02169 | 11/169 | HPCA; PRDX1; SCN9A; CHRNA1; NFE2L2; CDO1; SPARC; ACTB; NEFL; RPS3; ASIC1 |
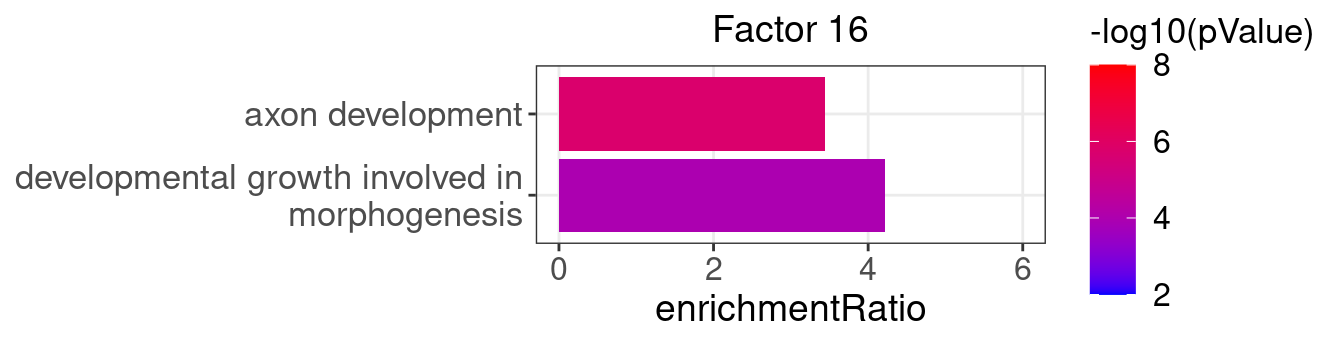
Factor 16 : 2 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0060560 | developmental growth involved in morphogenesis | 4.22 | 1.08e-04 | 0.03980 | 10/89 | DRAXIN; CRABP2; ZEB2; NRP2; ALCAM; MAP1B; TNC; GAL; SYT1; SYT4 |
| GO:0061564 | axon development | 3.45 | 1.73e-06 | 0.00127 | 19/207 | DRAXIN; EPHB2; JUN; S100A6; CRABP2; CNTN2; ZEB2; NRP2; ALCAM; GAP43; SPON2; CASP3; MAP1B; ETV1; NEFM; NEFL; TNC; FEZ1; MAP1A |
Factor 17 : 2 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0009141 | nucleoside triphosphate metabolic process | 4.22 | 1.98e-05 | 0.00997 | 12/189 | ENO1; ATP5I; TMSB4X; UQCRB; LDHA; NDUFC2; HSPA8; ATP5B; PKM; COX4I1; ATP5E; UQCR10 |
| GO:0009123 | nucleoside monophosphate metabolic process | 4.09 | 2.72e-05 | 0.00997 | 12/195 | ENO1; ATP5I; TMSB4X; UQCRB; LDHA; NDUFC2; HSPA8; ATP5B; PKM; COX4I1; ATP5E; UQCR10 |
Factor 18 : 12 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 30.60 | 0.00e+00 | 0.00e+00 | 77/119 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006413 | translational initiation | 23.20 | 0.00e+00 | 0.00e+00 | 77/157 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0002181 | cytoplasmic translation | 19.40 | 0.00e+00 | 0.00e+00 | 30/73 | RPL11; RPL31; RPL32; RPL15; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; RPL30; RPL8; RPL41; RPL6; RPLP0; RPS29; RPLP1; RPL26; RPL19; RPL38; RPL17; RPS21; EEF2; RPL36; RPS28; RPL18A; RPL18; RPL13A |
| GO:0090150 | establishment of protein localization to membrane | 18.10 | 0.00e+00 | 0.00e+00 | 78/204 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006401 | RNA catabolic process | 15.80 | 0.00e+00 | 0.00e+00 | 79/236 | RPL22; RPL11; YBX1; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; DDX5; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006605 | protein targeting | 14.10 | 0.00e+00 | 0.00e+00 | 78/261 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0016072 | rRNA metabolic process | 6.91 | 4.22e-15 | 3.87e-13 | 25/171 | RPL11; RPS8; RPL5; RPS27; RPS7; RPSA; RPL14; RPL35A; RPS14; RPL10A; RPL7; RPS6; RPL35; RPL7A; RPS24; RPS17; RPS2; RPL26; RPL27; RPS21; RPS15; RPS28; RPS16; RPS19; RPS9 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 5.30 | 0.00e+00 | 0.00e+00 | 37/330 | RPL11; RPS8; RPL5; RPS27; RPS7; RPSA; RPL14; RPL24; RPL35A; RPS23; RPS14; RPS10; RPL10A; RPL10; RPL7; RPS6; RPL35; RPL12; RPL7A; RPS24; RPL6; RPLP0; RPS17; RPS2; RPL26; RPL23A; RPL27; RPL38; RPS21; RPS15; RPS28; RPS16; RPS19; RPL13A; RPS9; RPS5; RPL3 |
| GO:0034470 | ncRNA processing | 5.30 | 2.16e-12 | 1.76e-10 | 25/223 | RPL11; RPS8; RPL5; RPS27; RPS7; RPSA; RPL14; RPL35A; RPS14; RPL10A; RPL7; RPS6; RPL35; RPL7A; RPS24; RPS17; RPS2; RPL26; RPL27; RPS21; RPS15; RPS28; RPS16; RPS19; RPS9 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 5.11 | 9.40e-10 | 6.90e-08 | 20/185 | RPL11; RPL5; RPS27; RPSA; RPL24; RPS23; RPS14; RPS10; RPL10; RPL12; RPL6; RPLP0; RPL23A; RPL38; RPS15; RPS28; RPS19; RPL13A; RPS5; RPL3 |
| GO:0034248 | regulation of cellular amide metabolic process | 2.92 | 2.60e-04 | 1.59e-02 | 14/227 | RPL5; RPS14; GNB2L1; SOX4; EEF1A1; RPS4X; RPL10; RPS3; GAPDH; RPL26; RPL38; EEF2; RPL13A; RPS9 |
| GO:0010608 | posttranscriptional regulation of gene expression | 2.81 | 8.45e-05 | 5.64e-03 | 17/286 | YBX1; RPL5; RPS27A; RPS14; GNB2L1; SOX4; RPS4X; RPL10; RPS3; GAPDH; RPL26; DDX5; RPL38; EEF2; UBA52; RPL13A; RPS9 |
Factor 19 : 11 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 12.30 | 0.00e+00 | 0.00e+00 | 28/119 | RPL5; SRP9; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; KDELR1; RPL13A; RPS9; RPL3 |
| GO:0006413 | translational initiation | 11.30 | 0.00e+00 | 0.00e+00 | 34/157 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; EIF4A2; RPS3A; NPM1; RPL10A; RPS4X; RPL10; RPS20; RPL7; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| GO:0002181 | cytoplasmic translation | 10.80 | 3.13e-12 | 3.82e-10 | 15/73 | RPL15; RPL10A; EIF3E; EIF3H; EIF3F; EIF3M; RPL41; RPL6; RPS29; EEF2; RPL36; RPS28; RPL13A; EIF3D; EIF3L |
| GO:0090150 | establishment of protein localization to membrane | 7.44 | 0.00e+00 | 0.00e+00 | 29/204 | RPL5; SRP9; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; GNB2L1; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; HSP90AA1; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| GO:0006401 | RNA catabolic process | 6.65 | 0.00e+00 | 0.00e+00 | 30/236 | YBX1; RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; NPM1; RPL10A; RPS4X; RPL10; RPS20; RPL7; EIF3E; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; CIRBP; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| GO:0006605 | protein targeting | 5.81 | 1.55e-15 | 2.28e-13 | 29/261 | RPL5; SRP9; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; SLC25A6; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; HSP90AA1; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 5.09 | 7.36e-09 | 6.75e-07 | 18/185 | RPL5; RPSA; NPM1; RBMX; RPL10; EIF3E; EIF3H; EIF3F; EIF3M; RPL6; HSP90AA1; CIRBP; RPS28; GLTSCR2; RPL13A; EIF3D; EIF3L; RPL3 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 4.12 | 1.96e-10 | 2.05e-08 | 26/330 | RPL5; RPS7; RPSA; RPL14; NPM1; RPL10A; RBMX; RPL10; RPL7; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPL6; HSP90AA1; RPS2; CIRBP; RPS28; GLTSCR2; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| GO:0034248 | regulation of cellular amide metabolic process | 3.92 | 9.48e-07 | 7.73e-05 | 17/227 | RPL5; SRP9; EIF4A2; NPM1; GNB2L1; EEF1A1; RBM3; RPS4X; RPL10; EIF3E; EIF3H; RPS3; CIRBP; EEF2; RPL13A; RPS9; EIF3D |
| GO:0016072 | rRNA metabolic process | 3.67 | 8.45e-05 | 5.64e-03 | 12/171 | RPL5; RPS7; RPSA; RPL14; RPL10A; RPL7; RPL35; RPL7A; RPS2; RPS28; GLTSCR2; RPS9 |
| GO:0010608 | posttranscriptional regulation of gene expression | 3.29 | 5.45e-06 | 4.00e-04 | 18/286 | YBX1; RPL5; SRP9; H3F3A; EIF4A2; NPM1; GNB2L1; RBM3; RPS4X; RPL10; EIF3E; EIF3H; RPS3; CIRBP; EEF2; RPL13A; RPS9; EIF3D |
| Factor | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 |
| Signif_GO_terms | 12 | 44 | 12 | 56 | 13 | 4 | 21 | 7 | 72 | 0 | 8 | 0 | 2 | 0 | 20 | 2 | 2 | 12 | 11 |
5.1.1 Factors of interest
4, 9, 15, 16
5.2 Reactome Pathway Over-Representation Analysis
Gene sets: The Reactome pathway database.
Factor 1 : 165 significant Reactome pathways| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-8876725 | Protein methylation | 5.34 | 1.40e-03 | 8.12e-03 | 5/12 | CALM2; EEF1A1; RPS2; EEF2; CALM3 |
| R-HSA-450604 | KSRP (KHSRP) binds and destabilizes mRNA | 5.34 | 1.40e-03 | 8.12e-03 | 5/12 | EXOSC7; YWHAZ; EXOSC4; EXOSC1; EXOSC8 |
| R-HSA-392851 | Prostacyclin signalling through prostacyclin receptor | 5.13 | 5.21e-03 | 2.68e-02 | 4/10 | GNG3; GNG2; GNAS; GNG8 |
| R-HSA-420092 | Glucagon-type ligand receptors | 5.13 | 5.21e-03 | 2.68e-02 | 4/10 | GNG3; GNG2; GNAS; GNG8 |
| R-HSA-445095 | Interaction between L1 and Ankyrins | 5.13 | 5.84e-04 | 3.70e-03 | 6/15 | NFASC; SCN3A; ANK2; SPTAN1; ANK3; ACTG1 |
| R-HSA-373080 | Class B/2 (Secretin family receptors) | 4.93 | 2.12e-03 | 1.18e-02 | 5/13 | GNG3; GNG2; GNAS; GNG8; PTH2 |
| R-HSA-3000178 | ECM proteoglycans | 4.93 | 2.12e-03 | 1.18e-02 | 5/13 | VCAN; TNC; NCAM1; APP; COL6A1 |
| R-HSA-5419276 | Mitochondrial translation termination | 4.59 | 3.70e-13 | 1.53e-10 | 29/81 | MRPL37; MRPL9; MRPS9; MRPS22; MRPL47; MRPS18C; MRPS30; MRPL22; MRPS18B; MRPS10; MRPL2; MRPS18A; MRPL32; MRPL15; MRPL50; MRPS2; MRPL21; MRPL51; MRPL42; MRPL46; MRPS11; MRPS34; MRPL27; ICT1; MRPS7; MRPL12; MRPS26; GADD45GIP1; MRPL39 |
| R-HSA-5368286 | Mitochondrial translation initiation | 4.53 | 5.33e-13 | 1.53e-10 | 29/82 | MRPL37; MRPL9; MRPS9; MRPS22; MRPL47; MRPS18C; MRPS30; MRPL22; MRPS18B; MRPS10; MRPL2; MRPS18A; MRPL32; MRPL15; MRPL50; MRPS2; MRPL21; MRPL51; MRPL42; MRPL46; MRPS11; MRPS34; MRPL27; ICT1; MRPS7; MRPL12; MRPS26; GADD45GIP1; MRPL39 |
| R-HSA-380994 | ATF4 activates genes | 4.53 | 1.26e-03 | 7.60e-03 | 6/17 | EXOSC7; EXOSC4; EXOSC1; DDIT3; EXOSC8; ATF4 |
| R-HSA-5389840 | Mitochondrial translation elongation | 4.48 | 7.63e-13 | 1.64e-10 | 29/83 | MRPL37; MRPL9; MRPS9; MRPS22; MRPL47; MRPS18C; MRPS30; MRPL22; MRPS18B; MRPS10; MRPL2; MRPS18A; MRPL32; MRPL15; MRPL50; MRPS2; MRPL21; MRPL51; MRPL42; MRPL46; MRPS11; MRPS34; MRPL27; ICT1; MRPS7; MRPL12; MRPS26; GADD45GIP1; MRPL39 |
| R-HSA-5368287 | Mitochondrial translation | 4.32 | 2.15e-12 | 3.71e-10 | 29/86 | MRPL37; MRPL9; MRPS9; MRPS22; MRPL47; MRPS18C; MRPS30; MRPL22; MRPS18B; MRPS10; MRPL2; MRPS18A; MRPL32; MRPL15; MRPL50; MRPS2; MRPL21; MRPL51; MRPL42; MRPL46; MRPS11; MRPS34; MRPL27; ICT1; MRPS7; MRPL12; MRPS26; GADD45GIP1; MRPL39 |
| R-HSA-69229 | Ubiquitin-dependent degradation of Cyclin D1 | 4.08 | 3.01e-06 | 8.95e-05 | 14/44 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; CDK4; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-75815 | Ubiquitin-dependent degradation of Cyclin D | 4.08 | 3.01e-06 | 8.95e-05 | 14/44 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; CDK4; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-389957 | Prefoldin mediated transfer of substrate to CCT/TriC | 4.08 | 9.98e-04 | 6.09e-03 | 7/22 | PFDN2; CCT5; PFDN1; TUBB2B; CCT6A; TUBA1A; PFDN4 |
| R-HSA-381042 | PERK regulates gene expression | 4.05 | 2.42e-03 | 1.30e-02 | 6/19 | EXOSC7; EXOSC4; EXOSC1; DDIT3; EXOSC8; ATF4 |
| R-HSA-451326 | Activation of kainate receptors upon glutamate binding | 4.01 | 5.93e-03 | 3.02e-02 | 5/16 | CALM2; GNG3; GNG2; CALM3; GNG8 |
| R-HSA-1236978 | Cross-presentation of soluble exogenous antigens (endosomes) | 3.95 | 2.30e-05 | 3.42e-04 | 12/39 | PSMD4; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5578775 | Ion homeostasis | 3.90 | 1.34e-03 | 7.91e-03 | 7/23 | ATP1B1; CALM2; SRI; FXYD6; ATP2B1; FXYD7; CALM3 |
| R-HSA-389958 | Cooperation of Prefoldin and TriC/CCT in actin and tubulin folding | 3.90 | 1.34e-03 | 7.91e-03 | 7/23 | PFDN2; CCT5; PFDN1; TUBB2B; CCT6A; TUBA1A; PFDN4 |
| R-HSA-180534 | Vpu mediated degradation of CD4 | 3.88 | 1.29e-05 | 2.37e-04 | 13/43 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-211733 | Regulation of activated PAK-2p34 by proteasome mediated degradation | 3.88 | 1.29e-05 | 2.37e-04 | 13/43 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69601 | Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | 3.88 | 1.29e-05 | 2.37e-04 | 13/43 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69610 | p53-Independent DNA Damage Response | 3.88 | 1.29e-05 | 2.37e-04 | 13/43 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69613 | p53-Independent G1/S DNA damage checkpoint | 3.88 | 1.29e-05 | 2.37e-04 | 13/43 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-187577 | SCF(Skp2)-mediated degradation of p27/p21 | 3.82 | 7.23e-06 | 1.78e-04 | 14/47 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; CDK4; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-450408 | AUF1 (hnRNP D0) binds and destabilizes mRNA | 3.82 | 7.23e-06 | 1.78e-04 | 14/47 | PSMD4; RPS27A; PSMB1; PSMA2; HSPB1; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-9604323 | Negative regulation of NOTCH4 signaling | 3.82 | 7.23e-06 | 1.78e-04 | 14/47 | PSMD4; RPS27A; PSMB1; PSMA2; YWHAZ; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-500792 | GPCR ligand binding | 3.80 | 7.40e-04 | 4.62e-03 | 8/27 | GNG3; PSAP; GNG2; SSTR2; GNAS; GNG8; PTH2; APP |
| R-HSA-169911 | Regulation of Apoptosis | 3.79 | 1.71e-05 | 2.94e-04 | 13/44 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-174113 | SCF-beta-TrCP mediated degradation of Emi1 | 3.79 | 1.71e-05 | 2.94e-04 | 13/44 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 3.74 | 9.50e-06 | 2.23e-04 | 14/48 | EIF3I; RPS7; RPS27A; RPSA; RPS10; RPS12; RPS6; RPS29; RPS27L; RPS17; RPS2; RPS19; RPS5 |
| R-HSA-936837 | Ion transport by P-type ATPases | 3.74 | 1.77e-03 | 1.00e-02 | 7/24 | ATP1B1; CALM2; SRI; FXYD6; ATP2B1; FXYD7; CALM3 |
| R-HSA-349425 | Autodegradation of the E3 ubiquitin ligase COP1 | 3.70 | 2.24e-05 | 3.42e-04 | 13/45 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-4641258 | Degradation of DVL | 3.70 | 2.24e-05 | 3.42e-04 | 13/45 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-68827 | CDT1 association with the CDC6:ORC:origin complex | 3.70 | 2.24e-05 | 3.42e-04 | 13/45 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-8854050 | FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | 3.70 | 2.24e-05 | 3.42e-04 | 13/45 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-8941858 | Regulation of RUNX3 expression and activity | 3.70 | 2.24e-05 | 3.42e-04 | 13/45 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-156902 | Peptide chain elongation | 3.66 | 8.65e-09 | 7.45e-07 | 24/84 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; EEF1A1; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; EEF2; RPS19; RPL13A; RPS5 |
| R-HSA-180585 | Vif-mediated degradation of APOBEC3G | 3.62 | 2.90e-05 | 4.10e-04 | 13/46 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-4641257 | Degradation of AXIN | 3.62 | 2.90e-05 | 4.10e-04 | 13/46 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-450531 | Regulation of mRNA stability by proteins that bind AU-rich elements | 3.58 | 4.87e-07 | 2.00e-05 | 19/68 | PSMD4; RPS27A; EXOSC7; PSMB1; PSMA2; HSPB1; YWHAZ; EXOSC4; PSMB7; EXOSC1; EXOSC8; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 3.58 | 1.45e-08 | 1.03e-06 | 24/86 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; EEF1A1; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; EEF2; RPS19; RPL13A; RPS5 |
| R-HSA-350562 | Regulation of ornithine decarboxylase (ODC) | 3.58 | 6.80e-05 | 6.97e-04 | 12/43 | PSMD4; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5362768 | Hh mutants that don't undergo autocatalytic processing are degraded by ERAD | 3.55 | 3.74e-05 | 4.73e-04 | 13/47 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5387390 | Hh mutants abrogate ligand secretion | 3.55 | 3.74e-05 | 4.73e-04 | 13/47 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5607761 | Dectin-1 mediated noncanonical NF-kB signaling | 3.55 | 3.74e-05 | 4.73e-04 | 13/47 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5676590 | NIK-->noncanonical NF-kB signaling | 3.55 | 3.74e-05 | 4.73e-04 | 13/47 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-68867 | Assembly of the pre-replicative complex | 3.55 | 3.74e-05 | 4.73e-04 | 13/47 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-1168372 | Downstream signaling events of B Cell Receptor (BCR) | 3.54 | 4.84e-06 | 1.30e-04 | 16/58 | PSMD4; CALM2; RPS27A; PSMB1; PSMA2; PPIA; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4; CALM3 |
| R-HSA-8852276 | The role of GTSE1 in G2/M progression after G2 checkpoint | 3.50 | 1.13e-05 | 2.37e-04 | 15/55 | PSMD4; RPS27A; TUBB2B; PSMB1; PSMA2; PSMB7; TUBA1A; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-72649 | Translation initiation complex formation | 3.50 | 1.13e-05 | 2.37e-04 | 15/55 | EIF3I; RPS7; RPS27A; RPSA; RPS10; RPS12; RPS6; RPS29; RPS27L; RPS17; RPS2; EIF4A1; RPS19; RPS5 |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 3.50 | 1.13e-05 | 2.37e-04 | 15/55 | EIF3I; RPS7; RPS27A; RPSA; RPS10; RPS12; RPS6; RPS29; RPS27L; RPS17; RPS2; EIF4A1; RPS19; RPS5 |
| R-HSA-68949 | Orc1 removal from chromatin | 3.47 | 4.77e-05 | 5.34e-04 | 13/48 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-1169091 | Activation of NF-kappaB in B cells | 3.47 | 4.77e-05 | 5.34e-04 | 13/48 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-4608870 | Asymmetric localization of PCP proteins | 3.47 | 4.77e-05 | 5.34e-04 | 13/48 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5610780 | Degradation of GLI1 by the proteasome | 3.47 | 4.77e-05 | 5.34e-04 | 13/48 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5668541 | TNFR2 non-canonical NF-kB pathway | 3.47 | 4.77e-05 | 5.34e-04 | 13/48 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69541 | Stabilization of p53 | 3.47 | 4.77e-05 | 5.34e-04 | 13/48 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 3.43 | 1.43e-05 | 2.57e-04 | 15/56 | EIF3I; RPS7; RPS27A; RPSA; RPS10; RPS12; RPS6; RPS29; RPS27L; RPS17; RPS2; EIF4A1; RPS19; RPS5 |
| R-HSA-72764 | Eukaryotic Translation Termination | 3.43 | 7.08e-08 | 4.36e-06 | 23/86 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; TRMT112; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-418346 | Platelet homeostasis | 3.42 | 1.59e-03 | 9.11e-03 | 8/30 | CALM2; SRI; GNG3; ATP2B1; GNG2; GNAS; CALM3; GNG8 |
| R-HSA-5610783 | Degradation of GLI2 by the proteasome | 3.40 | 6.05e-05 | 6.28e-04 | 13/49 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5610785 | GLI3 is processed to GLI3R by the proteasome | 3.40 | 6.05e-05 | 6.28e-04 | 13/49 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5678895 | Defective CFTR causes cystic fibrosis | 3.40 | 6.05e-05 | 6.28e-04 | 13/49 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-192823 | Viral mRNA Translation | 3.40 | 1.65e-07 | 8.86e-06 | 22/83 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-1236974 | ER-Phagosome pathway | 3.33 | 7.61e-05 | 7.53e-04 | 13/50 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5358346 | Hedgehog ligand biogenesis | 3.33 | 7.61e-05 | 7.53e-04 | 13/50 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-174154 | APC/C:Cdc20 mediated degradation of Securin | 3.32 | 4.16e-05 | 5.12e-04 | 14/54 | PSMD4; RPS27A; PTTG1; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-174178 | APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | 3.32 | 4.16e-05 | 5.12e-04 | 14/54 | PSMD4; RPS27A; PTTG1; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-2408557 | Selenocysteine synthesis | 3.32 | 2.61e-07 | 1.32e-05 | 22/85 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-9013694 | Signaling by NOTCH4 | 3.32 | 2.27e-05 | 3.42e-04 | 15/58 | PSMD4; RPS27A; PSMB1; PSMA2; YWHAZ; PSMB7; PSMB5; PSMA3; SNW1; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5619084 | ABC transporter disorders | 3.27 | 9.52e-05 | 8.53e-04 | 13/51 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-1236975 | Antigen processing-Cross presentation | 3.27 | 9.52e-05 | 8.53e-04 | 13/51 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-8939902 | Regulation of RUNX2 expression and activity | 3.27 | 9.52e-05 | 8.53e-04 | 13/51 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-176409 | APC/C:Cdc20 mediated degradation of mitotic proteins | 3.26 | 5.20e-05 | 5.66e-04 | 14/55 | PSMD4; RPS27A; PTTG1; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-176814 | Activation of APC/C and APC/C:Cdc20 mediated degradation of mitotic proteins | 3.26 | 5.20e-05 | 5.66e-04 | 14/55 | PSMD4; RPS27A; PTTG1; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5658442 | Regulation of RAS by GAPs | 3.21 | 1.18e-04 | 9.69e-04 | 13/52 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69002 | DNA Replication Pre-Initiation | 3.21 | 1.18e-04 | 9.69e-04 | 13/52 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69563 | p53-Dependent G1 DNA Damage Response | 3.21 | 1.18e-04 | 9.69e-04 | 13/52 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69580 | p53-Dependent G1/S DNA damage checkpoint | 3.21 | 1.18e-04 | 9.69e-04 | 13/52 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69615 | G1/S DNA Damage Checkpoints | 3.21 | 1.18e-04 | 9.69e-04 | 13/52 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-1234176 | Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | 3.21 | 1.18e-04 | 9.69e-04 | 13/52 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-174084 | Autodegradation of Cdh1 by Cdh1:APC/C | 3.21 | 1.18e-04 | 9.69e-04 | 13/52 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5632684 | Hedgehog 'on' state | 3.21 | 1.18e-04 | 9.69e-04 | 13/52 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 3.21 | 5.04e-07 | 2.00e-05 | 22/88 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-418555 | G alpha (s) signalling events | 3.21 | 8.63e-03 | 4.32e-02 | 6/24 | REEP1; GNG3; GNG2; GNAS; GNG8; PTH2 |
| R-HSA-5607764 | CLEC7A (Dectin-1) signaling | 3.21 | 3.52e-05 | 4.73e-04 | 15/60 | PSMD4; CALM2; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4; CALM3 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 3.18 | 5.61e-11 | 8.04e-09 | 37/149 | RBM8A; PSMD4; RPS7; RPS27A; RPSA; ROBO2; RPL9; RPL37; RPS10; RPS12; PSMB1; PSMA2; RPL10; RPS6; PSMB7; RPL41; PSMB5; RPS29; PSMA3; PSMC1; RPS27L; PSMA4; RPS17; RPS2; RPL13; PSMB6; RPL23A; RPL19; RPL27; EIF4A3; PSMA7; PSMD8; PSMC4; RPS19; RPL13A; RPS5 |
| R-HSA-983705 | Signaling by the B Cell Receptor (BCR) | 3.16 | 2.37e-05 | 3.47e-04 | 16/65 | PSMD4; CALM2; RPS27A; PSMB1; PSMA2; PPIA; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4; CALM3 |
| R-HSA-69202 | Cyclin E associated events during G1/S transition | 3.15 | 7.96e-05 | 7.53e-04 | 14/57 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; CDK4; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69656 | Cyclin A:Cdk2-associated events at S phase entry | 3.15 | 7.96e-05 | 7.53e-04 | 14/57 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; CDK4; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-174143 | APC/C-mediated degradation of cell cycle proteins | 3.15 | 7.96e-05 | 7.53e-04 | 14/57 | PSMD4; RPS27A; PTTG1; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-453276 | Regulation of mitotic cell cycle | 3.15 | 7.96e-05 | 7.53e-04 | 14/57 | PSMD4; RPS27A; PTTG1; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-2871837 | FCERI mediated NF-kB activation | 3.15 | 1.46e-04 | 1.14e-03 | 13/53 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69017 | CDK-mediated phosphorylation and removal of Cdc6 | 3.15 | 1.46e-04 | 1.14e-03 | 13/53 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-202424 | Downstream TCR signaling | 3.15 | 1.46e-04 | 1.14e-03 | 13/53 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-2408522 | Selenoamino acid metabolism | 3.11 | 2.78e-07 | 1.33e-05 | 24/99 | RPS7; RPS27A; RPSA; RPL9; RPL37; EEF1E1; RPS10; RPS12; AIMP2; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 3.10 | 5.11e-07 | 2.00e-05 | 23/95 | EIF3I; RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-72766 | Translation | 3.09 | 0.00e+00 | 0.00e+00 | 62/257 | EIF3I; MRPL37; MRPL9; RPS7; RPS27A; MRPS9; RPSA; MRPS22; MRPL47; RPL9; MRPS18C; PPA2; RPL37; MRPS30; MRPL22; EEF1E1; MRPS18B; RPS10; MRPS10; MRPL2; MRPS18A; EEF1A1; RPS12; AIMP2; MRPL32; RPL10; MRPL15; RPS6; MRPL50; MRPS2; TRMT112; MRPL21; SPCS2; MRPL51; RPL41; MRPL42; RPS29; SRP14; RPS27L; RPS17; MRPL46; MRPS11; MRPS34; RPS2; RPL13; EIF4A1; RPL23A; RPL19; RPL27; MRPL27; ICT1; MRPS7; MRPL12; MRPS26; EEF2; FARSA; GADD45GIP1; RPS19; RPL13A; RPS5; MRPL39 |
| R-HSA-174184 | Cdc20:Phospho-APC/C mediated degradation of Cyclin A | 3.09 | 1.79e-04 | 1.35e-03 | 13/54 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-179419 | APC:Cdc20 mediated degradation of cell cycle proteins prior to satisfation of the cell cycle checkpoint | 3.09 | 1.79e-04 | 1.35e-03 | 13/54 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-8878159 | Transcriptional regulation by RUNX3 | 3.04 | 1.19e-04 | 9.69e-04 | 14/59 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; SNW1; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-9020702 | Interleukin-1 signaling | 3.04 | 1.19e-04 | 9.69e-04 | 14/59 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4; APP |
| R-HSA-8948751 | Regulation of PTEN stability and activity | 3.03 | 2.18e-04 | 1.59e-03 | 13/55 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5619115 | Disorders of transmembrane transporters | 3.03 | 2.18e-04 | 1.59e-03 | 13/55 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-8939236 | RUNX1 regulates transcription of genes involved in differentiation of HSCs | 2.99 | 1.45e-04 | 1.14e-03 | 14/60 | PSMD4; H3F3A; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-6790901 | rRNA modification in the nucleus and cytosol | 2.98 | 1.34e-03 | 7.91e-03 | 10/43 | RPS7; WDR43; RRP9; WBSCR22; DCAF13; RPS6; TRMT112; KRR1; RPS2; DDX52 |
| R-HSA-382556 | ABC-family proteins mediated transport | 2.98 | 2.65e-04 | 1.87e-03 | 13/56 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-351202 | Metabolism of polyamines | 2.98 | 2.65e-04 | 1.87e-03 | 13/56 | PSMD4; PSMB1; PSMA2; SAT1; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-176408 | Regulation of APC/C activators between G1/S and early anaphase | 2.98 | 2.65e-04 | 1.87e-03 | 13/56 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5610787 | Hedgehog 'off' state | 2.97 | 5.26e-05 | 5.66e-04 | 16/69 | PSMD4; RPS27A; TUBB2B; PSMB1; PSMA2; PSMB7; TUBA1A; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; GNAS; PSMA7; PSMD8; PSMC4 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 2.96 | 7.41e-07 | 2.77e-05 | 24/104 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; SPCS2; RPL41; RPS29; SRP14; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 2.93 | 8.94e-07 | 2.96e-05 | 24/105 | EIF3I; RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; EIF4A1; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 2.93 | 8.94e-07 | 2.96e-05 | 24/105 | RBM8A; RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; EIF4A3; RPS19; RPL13A; RPS5 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 2.93 | 8.94e-07 | 2.96e-05 | 24/105 | RBM8A; RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; EIF4A3; RPS19; RPL13A; RPS5 |
| R-HSA-5576891 | Cardiac conduction | 2.93 | 4.53e-03 | 2.38e-02 | 8/35 | ATP1B1; CALM2; SCN3A; SRI; FXYD6; ATP2B1; FXYD7; CALM3 |
| R-HSA-1234174 | Regulation of Hypoxia-inducible Factor (HIF) by oxygen | 2.92 | 3.19e-04 | 2.16e-03 | 13/57 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-2262749 | Cellular response to hypoxia | 2.92 | 3.19e-04 | 2.16e-03 | 13/57 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 2.90 | 1.08e-06 | 3.43e-05 | 24/106 | EIF3I; RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; EIF4A1; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-4086400 | PCP/CE pathway | 2.85 | 2.52e-04 | 1.82e-03 | 14/63 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; DAAM1; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-3858494 | Beta-catenin independent WNT signaling | 2.83 | 2.17e-05 | 3.42e-04 | 19/86 | PSMD4; CALM2; RPS27A; PSMB1; PSMA2; PSMB7; GNG3; PSMB5; GNG2; PSMA3; DAAM1; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4; CALM3; GNG8 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 2.83 | 1.02e-08 | 7.95e-07 | 34/154 | RPS7; WDR43; RPS27A; RPSA; EXOSC7; RRP9; RPL9; RPL37; RPS10; RPS12; WBSCR22; RPL10; DCAF13; EXOSC4; RPS6; DDX21; EXOSC1; RPL41; KRR1; EXOSC8; RPS29; RPS27L; RPS17; RPS2; NOB1; RPL13; RPL23A; DDX52; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-69052 | Switching of origins to a post-replicative state | 2.83 | 4.58e-04 | 2.99e-03 | 13/59 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-376176 | Signaling by ROBO receptors | 2.81 | 9.28e-10 | 1.14e-07 | 39/178 | RBM8A; PSMD4; RPS7; RPS27A; SRGAP3; RPSA; ROBO2; RPL9; RPL37; RPS10; RPS12; PSMB1; PSMA2; RPL10; RPS6; PSMB7; RPL41; PSMB5; RPS29; PSMA3; PSMC1; RPS27L; PSMA4; RPS17; RPS2; RPL13; PSMB6; RPL23A; RPL19; RPL27; EIF4A3; DCC; PSMA7; PSMD8; PSMC4; RPS19; RPL13A; RPS5 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 2.80 | 7.80e-09 | 7.45e-07 | 35/160 | RPS7; WDR43; RPS27A; RPSA; EXOSC7; RRP9; RPL9; RPL37; RPS10; RPS12; WBSCR22; RPL10; DCAF13; EXOSC4; RPS6; TRMT112; DDX21; EXOSC1; RPL41; KRR1; EXOSC8; RPS29; RPS27L; RPS17; RPS2; NOB1; RPL13; RPL23A; DDX52; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-5689603 | UCH proteinases | 2.79 | 1.97e-04 | 1.46e-03 | 15/69 | PSMD4; RPS27A; UCHL1; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; ADRM1; PSMD8; PSMC4 |
| R-HSA-5687128 | MAPK6/MAPK4 signaling | 2.76 | 3.57e-04 | 2.40e-03 | 14/65 | PSMD4; RPS27A; PSMB1; PSMA2; HSPB1; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-72312 | rRNA processing | 2.74 | 1.55e-08 | 1.03e-06 | 35/164 | RPS7; WDR43; RPS27A; RPSA; EXOSC7; RRP9; RPL9; RPL37; RPS10; RPS12; WBSCR22; RPL10; DCAF13; EXOSC4; RPS6; TRMT112; DDX21; EXOSC1; RPL41; KRR1; EXOSC8; RPS29; RPS27L; RPS17; RPS2; NOB1; RPL13; RPL23A; DDX52; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-202403 | TCR signaling | 2.73 | 6.44e-04 | 4.05e-03 | 13/61 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 2.72 | 3.64e-06 | 1.01e-04 | 24/113 | EIF3I; RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; EIF4A1; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 2.72 | 3.64e-06 | 1.01e-04 | 24/113 | EIF3I; RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; EIF4A1; RPL23A; RPL19; RPL27; RPS19; RPL13A; RPS5 |
| R-HSA-8878166 | Transcriptional regulation by RUNX2 | 2.72 | 4.21e-04 | 2.79e-03 | 14/66 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; CDK4; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-446652 | Interleukin-1 family signaling | 2.72 | 4.21e-04 | 2.79e-03 | 14/66 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4; APP |
| R-HSA-5621481 | C-type lectin receptors (CLRs) | 2.71 | 2.76e-04 | 1.93e-03 | 15/71 | PSMD4; CALM2; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4; CALM3 |
| R-HSA-195253 | Degradation of beta-catenin by the destruction complex | 2.69 | 7.59e-04 | 4.70e-03 | 13/62 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-373760 | L1CAM interactions | 2.64 | 5.82e-04 | 3.70e-03 | 14/68 | NFASC; SCN3A; ALCAM; GAP43; ANK2; TUBB2B; CD24; DCX; DPYSL2; SPTAN1; NCAM1; ANK3; TUBA1A; ACTG1 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 2.61 | 1.24e-05 | 2.37e-04 | 23/113 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RAE1; RPS19; RPL13A; RPS5 |
| R-HSA-2454202 | Fc epsilon receptor (FCERI) signaling | 2.60 | 2.90e-04 | 2.01e-03 | 16/79 | PSMD4; CALM2; RPS27A; MAPK10; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4; CALM3 |
| R-HSA-611105 | Respiratory electron transport | 2.56 | 9.36e-05 | 8.53e-04 | 19/95 | SDHB; NDUFS2; COX5B; NDUFB5; NDUFS6; NDUFS4; COX7C; NDUFA4; COX6C; CYC1; NDUFS3; COX8A; TMEM126B; SDHD; NDUFA9; NDUFA12; COX6A1; NDUFB10; UQCRFS1 |
| R-HSA-168255 | Influenza Life Cycle | 2.54 | 1.27e-05 | 2.37e-04 | 24/121 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RAN; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RAE1; RPS19; RPL13A; RPS5 |
| R-HSA-69206 | G1/S Transition | 2.53 | 9.18e-04 | 5.64e-03 | 14/71 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; CDK4; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-69239 | Synthesis of DNA | 2.53 | 1.41e-03 | 8.13e-03 | 13/66 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5358351 | Signaling by Hedgehog | 2.50 | 4.52e-04 | 2.97e-03 | 16/82 | PSMD4; RPS27A; TUBB2B; PSMB1; PSMA2; PSMB7; TUBA1A; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; GNAS; PSMA7; PSMD8; PSMC4 |
| R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 2.47 | 4.64e-05 | 5.34e-04 | 22/114 | SDHB; NDUFS2; COX5B; NDUFB5; NDUFS6; NDUFS4; COX7C; NDUFA4; COX6C; CYC1; NDUFS3; COX8A; TMEM126B; SDHD; NDUFA9; ATP5B; NDUFA12; COX6A1; NDUFB10; ATP5E; UQCRFS1; ATP5O |
| R-HSA-5673001 | RAF/MAP kinase cascade | 2.47 | 7.12e-05 | 7.21e-04 | 21/109 | PSMD4; CALM2; RPS27A; PSMB1; PSMA2; NEFL; NRG1; PSMB7; SPTAN1; NCAM1; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PHB; ACTG1; PSMA7; PSMD8; PSMC4; CALM3 |
| R-HSA-109581 | Apoptosis | 2.46 | 1.67e-04 | 1.28e-03 | 19/99 | PSMD4; RPS27A; SATB1; CASP3; PSMB1; PSMA2; FNTA; YWHAZ; PSMB7; SPTAN1; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; DCC; PSMA7; PSMD8; PSMC4 |
| R-HSA-5357801 | Programmed Cell Death | 2.46 | 1.67e-04 | 1.28e-03 | 19/99 | PSMD4; RPS27A; SATB1; CASP3; PSMB1; PSMA2; FNTA; YWHAZ; PSMB7; SPTAN1; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; DCC; PSMA7; PSMD8; PSMC4 |
| R-HSA-5684996 | MAPK1/MAPK3 signaling | 2.45 | 8.18e-05 | 7.66e-04 | 21/110 | PSMD4; CALM2; RPS27A; PSMB1; PSMA2; NEFL; NRG1; PSMB7; SPTAN1; NCAM1; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PHB; ACTG1; PSMA7; PSMD8; PSMC4; CALM3 |
| R-HSA-391251 | Protein folding | 2.43 | 4.41e-03 | 2.33e-02 | 11/58 | PFDN2; CCT5; PFDN1; TUBB2B; TBCC; CCT6A; GNG3; TUBA1A; GNG2; PFDN4; GNG8 |
| R-HSA-390466 | Chaperonin-mediated protein folding | 2.42 | 6.79e-03 | 3.42e-02 | 10/53 | PFDN2; CCT5; PFDN1; TUBB2B; CCT6A; GNG3; TUBA1A; GNG2; PFDN4; GNG8 |
| R-HSA-69306 | DNA Replication | 2.42 | 2.15e-03 | 1.19e-02 | 13/69 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-168254 | Influenza Infection | 2.40 | 3.42e-05 | 4.73e-04 | 24/128 | RPS7; RPS27A; RPSA; RPL9; RPL37; RPS10; RPS12; RPL10; RPS6; RPL41; RAN; RPS29; RPS27L; RPS17; RPS2; RPL13; RPL23A; RPL19; RPL27; RAE1; RPS19; RPL13A; RPS5 |
| R-HSA-157118 | Signaling by NOTCH | 2.37 | 1.88e-04 | 1.41e-03 | 20/108 | YBX1; PSMD4; PBX1; H3F3A; RPS27A; PSMB1; PSMA2; YWHAZ; PSMB7; PSMB5; PSMA3; SNW1; PSMC1; PSMA4; PSMB6; PSMA7; SIRT6; PSMD8; PSMC4; MFNG |
| R-HSA-453279 | Mitotic G1-G1/S phases | 2.36 | 1.84e-03 | 1.03e-02 | 14/76 | PSMD4; RPS27A; PSMB1; PSMA2; PSMB7; CDK4; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 2.33 | 3.34e-07 | 1.52e-05 | 38/209 | PSMD4; RPS7; RPS27A; RPSA; RPL9; RPL37; EEF1E1; RPS10; SERINC1; RPS12; PSMB1; AIMP2; PSMA2; SAT1; RPL10; RPS6; PSMB7; RPL41; PSMB5; RPS29; PSMA3; PSMC1; RPS27L; PSMA4; RPS17; RPS2; RPL13; PSMB6; RPL23A; RPL19; RPL27; PSMA7; PSMD8; PSMC4; RPS19; RPL13A; RPS5 |
| R-HSA-381119 | Unfolded Protein Response (UPR) | 2.31 | 6.57e-03 | 3.33e-02 | 11/61 | DCTN1; EXOSC7; SEC62; EXOSC4; YIF1A; EXOSC1; DDIT3; EXOSC8; MYDGF; ASNA1; ATF4 |
| R-HSA-69481 | G2/M Checkpoints | 2.30 | 2.37e-03 | 1.29e-02 | 14/78 | PSMD4; RPS27A; PSMB1; PSMA2; YWHAZ; PSMB7; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-162909 | Host Interactions of HIV factors | 2.23 | 1.67e-03 | 9.52e-03 | 16/92 | PSMD4; RPS27A; PSMB1; PSMA2; PPIA; PSMB7; RAN; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; RAE1; PSMA7; PSMD8; PSMC4 |
| R-HSA-422475 | Axon guidance | 2.23 | 1.26e-09 | 1.36e-07 | 57/328 | RBM8A; PSMD4; NFASC; RPS7; RPS27A; SCN3A; SRGAP3; RPSA; ROBO2; ALCAM; GAP43; RPL9; ANK2; RPL37; RGMB; DPYSL3; TUBB2B; RPS10; CD24; RPS12; PSMB1; PSMA2; DCX; RPL10; DPYSL2; RPS6; PSMB7; SPTAN1; NCAM1; ANK3; TUBA1A; RPL41; MYL6; PSMB5; RPS29; PSMA3; PSMC1; RPS27L; PSMA4; RPS17; RPS2; RPL13; PSMB6; RPL23A; RPL19; RPL27; EIF4A3; ACTG1; DCC; PSMA7; PSMD8; PSMC4; RPS19; RPL13A; RPS5; COL6A1 |
| R-HSA-2467813 | Separation of Sister Chromatids | 2.14 | 1.07e-03 | 6.51e-03 | 19/114 | NUDC; PSMD4; PMF1; RPS27A; PTTG1; TUBB2B; PSMB1; PSMA2; PSMB7; TUBA1A; PSMB5; PSMA3; PSMC1; RPS27L; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-5683057 | MAPK family signaling cascades | 2.12 | 4.86e-04 | 3.15e-03 | 22/133 | PSMD4; CALM2; RPS27A; PSMB1; PSMA2; HSPB1; NEFL; NRG1; PSMB7; SPTAN1; NCAM1; PSMB5; PSMA3; PSMC1; PSMA4; PSMB6; PHB; ACTG1; PSMA7; PSMD8; PSMC4; CALM3 |
| R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | 2.06 | 5.49e-04 | 3.53e-03 | 23/143 | SDHB; NDUFS2; COX5B; NDUFB5; NDUFS6; NDUFS4; COX7C; NDUFA4; COX6C; CYC1; NDUFS3; COX8A; TMEM126B; SDHD; NDUFA9; ATP5B; NDUFA12; COX6A1; NDUFB10; IDH3B; ATP5E; UQCRFS1; ATP5O |
| R-HSA-2555396 | Mitotic Metaphase and Anaphase | 2.01 | 2.22e-03 | 1.21e-02 | 19/121 | NUDC; PSMD4; PMF1; RPS27A; PTTG1; TUBB2B; PSMB1; PSMA2; PSMB7; TUBA1A; PSMB5; PSMA3; PSMC1; RPS27L; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
| R-HSA-68882 | Mitotic Anaphase | 2.01 | 2.22e-03 | 1.21e-02 | 19/121 | NUDC; PSMD4; PMF1; RPS27A; PTTG1; TUBB2B; PSMB1; PSMA2; PSMB7; TUBA1A; PSMB5; PSMA3; PSMC1; RPS27L; PSMA4; PSMB6; PSMA7; PSMD8; PSMC4 |
Factor 2 : 217 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-156902 | Peptide chain elongation | 9.54 | 0.00e+00 | 0.00e+00 | 77/84 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-192823 | Viral mRNA Translation | 9.53 | 0.00e+00 | 0.00e+00 | 76/83 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 9.32 | 0.00e+00 | 0.00e+00 | 77/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-2408557 | Selenocysteine synthesis | 9.31 | 0.00e+00 | 0.00e+00 | 76/85 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72764 | Eukaryotic Translation Termination | 9.20 | 0.00e+00 | 0.00e+00 | 76/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 9.11 | 0.00e+00 | 0.00e+00 | 77/88 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 8.33 | 0.00e+00 | 0.00e+00 | 76/95 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 8.31 | 0.00e+00 | 0.00e+00 | 83/104 | RPL22; DDOST; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; SSR4; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; SPCS2; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; SEC11C; RPN2; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 8.10 | 9.52e-12 | 2.34e-10 | 14/18 | CDC42; CALM2; TUBA4A; CTNNB1; TUBB2A; TUBB2B; ACTB; TUBB4B; TUBA1B; TUBA1A; CALM1; TUBB3; ACTG1; CALM3 |
| R-HSA-2408522 | Selenoamino acid metabolism | 7.99 | 0.00e+00 | 0.00e+00 | 76/99 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 7.83 | 0.00e+00 | 0.00e+00 | 79/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; EIF4A1; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 7.73 | 0.00e+00 | 0.00e+00 | 78/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 7.73 | 0.00e+00 | 0.00e+00 | 78/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 7.66 | 0.00e+00 | 0.00e+00 | 78/106 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; EIF4A1; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 7.28 | 0.00e+00 | 0.00e+00 | 79/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; EIF4A1; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 7.28 | 0.00e+00 | 0.00e+00 | 79/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; EIF4A1; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 7.09 | 0.00e+00 | 0.00e+00 | 77/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; HSP90AA1; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-168255 | Influenza Life Cycle | 6.80 | 0.00e+00 | 0.00e+00 | 79/121 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; CLTA; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RAN; RPL21; RPS29; HSP90AA1; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 6.78 | 0.00e+00 | 0.00e+00 | 97/149 | RPL22; RPL11; RPS8; RPL5; PSMA5; PSMD4; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; PSMD6; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; PSMB1; PSMC2; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; PSMD13; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; MSI1; UBC; RPL21; PSME2; RPS29; PSMA3; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; PSMB6; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; PSMC5; RPL38; EIF4A3; PSMA7; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; PSMD8; RPS16; PSMC4; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 6.51 | 0.00e+00 | 0.00e+00 | 30/48 | RPS8; RPS27; RPS7; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-168254 | Influenza Infection | 6.51 | 0.00e+00 | 0.00e+00 | 80/128 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; CLTA; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RAN; RPL21; RPS29; HSP90AA1; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; KPNA2; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-389960 | Formation of tubulin folding intermediates by CCT/TriC | 6.51 | 2.79e-07 | 1.95e-06 | 10/16 | CCT7; TUBA4A; CCT5; TUBB2A; TUBB2B; TUBB4B; TUBA1B; TUBA1A; CCT2; TUBB3 |
| R-HSA-389977 | Post-chaperonin tubulin folding pathway | 6.41 | 5.61e-06 | 3.22e-05 | 8/13 | TUBA4A; TUBB2A; TUBB2B; TUBB4B; TUBA1B; TUBA1A; TUBB3; TBCB |
| R-HSA-190828 | Gap junction trafficking | 6.36 | 9.46e-08 | 8.76e-07 | 11/18 | TUBA4A; AP2M1; TUBB2A; TUBB2B; ACTB; CLTA; TUBB4B; TUBA1B; TUBA1A; TUBB3; ACTG1 |
| R-HSA-72649 | Translation initiation complex formation | 6.25 | 0.00e+00 | 0.00e+00 | 33/55 | RPS8; RPS27; RPS7; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; PABPC1; RPS6; RPS13; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; EIF4A1; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 6.13 | 0.00e+00 | 0.00e+00 | 33/56 | RPS8; RPS27; RPS7; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; PABPC1; RPS6; RPS13; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; EIF4A1; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-8876725 | Protein methylation | 6.07 | 3.68e-05 | 1.98e-04 | 7/12 | CALM2; HSPA8; CALM1; RPS2; EEF2; CALM3; ETFB |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 6.06 | 0.00e+00 | 0.00e+00 | 32/55 | RPS8; RPS27; RPS7; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; EIF4A1; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-157858 | Gap junction trafficking and regulation | 6.03 | 2.05e-07 | 1.57e-06 | 11/19 | TUBA4A; AP2M1; TUBB2A; TUBB2B; ACTB; CLTA; TUBB4B; TUBA1B; TUBA1A; TUBB3; ACTG1 |
| R-HSA-8955332 | Carboxyterminal post-translational modifications of tubulin | 5.95 | 1.20e-05 | 6.66e-05 | 8/14 | TUBA4A; TUBB2A; TUBB2B; TUBB4B; TUBA1B; TUBA1A; TUBB3; TPGS2 |
| R-HSA-376176 | Signaling by ROBO receptors | 5.73 | 0.00e+00 | 0.00e+00 | 98/178 | RPL22; CDC42; RPL11; RPS8; RPL5; PSMA5; PSMD4; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; PSMD6; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; PSMB1; PSMC2; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; PSMD13; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; MSI1; UBC; RPL21; PSME2; RPS29; PSMA3; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; PSMB6; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; PSMC5; RPL38; EIF4A3; PSMA7; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; PSMD8; RPS16; PSMC4; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-70263 | Gluconeogenesis | 5.48 | 2.47e-06 | 1.49e-05 | 10/19 | ENO1; MDH1; MDH2; PGK1; PGAM1; GAPDH; TPI1; ALDOA; ENO3; G6PC3 |
| R-HSA-8852276 | The role of GTSE1 in G2/M progression after G2 checkpoint | 5.30 | 3.89e-15 | 9.84e-14 | 28/55 | PSMA5; PSMD4; RPS27A; TUBA4A; PSMD6; TUBB2A; TUBB2B; HSP90AB1; PSMB1; PSMC2; TUBB4B; PSMD13; TUBA1B; TUBA1A; UBC; PSME2; PSMA3; HSP90AA1; PSMA4; PSMD7; TUBB3; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-175474 | Assembly Of The HIV Virion | 5.20 | 1.33e-03 | 6.01e-03 | 5/10 | RPS27A; PPIA; TSG101; UBC; UBA52 |
| R-HSA-5663084 | Diseases of carbohydrate metabolism | 5.20 | 1.33e-03 | 6.01e-03 | 5/10 | RPS27A; TALDO1; UBC; G6PC3; UBA52 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 5.14 | 0.00e+00 | 0.00e+00 | 76/154 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-389958 | Cooperation of Prefoldin and TriC/CCT in actin and tubulin folding | 4.98 | 2.58e-06 | 1.54e-05 | 11/23 | CCT7; TUBA4A; CCT5; TUBB2A; TUBB2B; ACTB; TUBB4B; TUBA1B; TUBA1A; CCT2; TUBB3 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 4.94 | 0.00e+00 | 0.00e+00 | 76/160 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 4.93 | 0.00e+00 | 0.00e+00 | 99/209 | RPL22; RPL11; RPS8; RPL5; PSMA5; PHGDH; PSMD4; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; PDHB; PSMD6; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; CDO1; RPS14; RPS18; RPS10; RPL10A; RPS12; PSMB1; ASNS; PSMC2; PDHA1; HSD17B10; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; PSMD13; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; PSME2; RPS29; PSMA3; CKB; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; PSMB6; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; PSMC5; RPL38; PSMA7; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; PSMD8; RPS16; PSMC4; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72312 | rRNA processing | 4.89 | 0.00e+00 | 0.00e+00 | 77/164 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; HSD17B10; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-180534 | Vpu mediated degradation of CD4 | 4.84 | 3.20e-10 | 7.06e-09 | 20/43 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-3371497 | HSP90 chaperone cycle for steroid hormone receptors (SHR) | 4.78 | 9.10e-09 | 1.10e-07 | 17/37 | CAPZB; TUBA4A; TUBB2A; TUBB2B; HSP90AB1; DNAJA1; DCTN3; TUBB4B; HSPA8; TUBA1B; TUBA1A; PTGES3; DCTN2; DYNLL1; ACTR10; HSP90AA1; TUBB3 |
| R-HSA-174113 | SCF-beta-TrCP mediated degradation of Emi1 | 4.73 | 5.36e-10 | 1.15e-08 | 20/44 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-389957 | Prefoldin mediated transfer of substrate to CCT/TriC | 4.73 | 1.33e-05 | 7.27e-05 | 10/22 | CCT7; TUBA4A; CCT5; TUBB2A; TUBB2B; ACTB; TUBB4B; TUBA1A; CCT2; TUBB3 |
| R-HSA-450408 | AUF1 (hnRNP D0) binds and destabilizes mRNA | 4.65 | 2.90e-10 | 6.57e-09 | 21/47 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PABPC1; PSMD13; HSPA8; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-8854050 | FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | 4.63 | 8.81e-10 | 1.85e-08 | 20/45 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-211733 | Regulation of activated PAK-2p34 by proteasome mediated degradation | 4.60 | 2.67e-09 | 4.26e-08 | 19/43 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69601 | Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | 4.60 | 2.67e-09 | 4.26e-08 | 19/43 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69610 | p53-Independent DNA Damage Response | 4.60 | 2.67e-09 | 4.26e-08 | 19/43 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69613 | p53-Independent G1/S DNA damage checkpoint | 4.60 | 2.67e-09 | 4.26e-08 | 19/43 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-180585 | Vif-mediated degradation of APOBEC3G | 4.53 | 1.42e-09 | 2.85e-08 | 20/46 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; TCEB1; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69229 | Ubiquitin-dependent degradation of Cyclin D1 | 4.49 | 4.29e-09 | 5.87e-08 | 19/44 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-75815 | Ubiquitin-dependent degradation of Cyclin D | 4.49 | 4.29e-09 | 5.87e-08 | 19/44 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-169911 | Regulation of Apoptosis | 4.49 | 4.29e-09 | 5.87e-08 | 19/44 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-8982491 | Glycogen metabolism | 4.46 | 1.16e-03 | 5.28e-03 | 6/14 | CALM2; RPS27A; UBC; CALM1; UBA52; CALM3 |
| R-HSA-187577 | SCF(Skp2)-mediated degradation of p27/p21 | 4.43 | 2.26e-09 | 3.90e-08 | 20/47 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5607761 | Dectin-1 mediated noncanonical NF-kB signaling | 4.43 | 2.26e-09 | 3.90e-08 | 20/47 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5676590 | NIK-->noncanonical NF-kB signaling | 4.43 | 2.26e-09 | 3.90e-08 | 20/47 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-9604323 | Negative regulation of NOTCH4 signaling | 4.43 | 2.26e-09 | 3.90e-08 | 20/47 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-349425 | Autodegradation of the E3 ubiquitin ligase COP1 | 4.39 | 6.79e-09 | 8.35e-08 | 19/45 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-4641258 | Degradation of DVL | 4.39 | 6.79e-09 | 8.35e-08 | 19/45 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-68827 | CDT1 association with the CDC6:ORC:origin complex | 4.39 | 6.79e-09 | 8.35e-08 | 19/45 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-8941858 | Regulation of RUNX3 expression and activity | 4.39 | 6.79e-09 | 8.35e-08 | 19/45 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-1236974 | ER-Phagosome pathway | 4.37 | 1.18e-09 | 2.43e-08 | 21/50 | PSMA5; PSMD4; RPS27A; PSMD6; HLA-A; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PDIA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-68949 | Orc1 removal from chromatin | 4.34 | 3.54e-09 | 5.17e-08 | 20/48 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-1169091 | Activation of NF-kappaB in B cells | 4.34 | 3.54e-09 | 5.17e-08 | 20/48 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5610780 | Degradation of GLI1 by the proteasome | 4.34 | 3.54e-09 | 5.17e-08 | 20/48 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5668541 | TNFR2 non-canonical NF-kB pathway | 4.34 | 3.54e-09 | 5.17e-08 | 20/48 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-156590 | Glutathione conjugation | 4.34 | 3.56e-03 | 1.46e-02 | 5/12 | GSTM3; MGST3; GSTA4; GSTP1; ESD |
| R-HSA-202131 | Metabolism of nitric oxide | 4.34 | 3.56e-03 | 1.46e-02 | 5/12 | CALM2; DDAH2; CALM1; HSP90AA1; CALM3 |
| R-HSA-203765 | eNOS activation and regulation | 4.34 | 3.56e-03 | 1.46e-02 | 5/12 | CALM2; DDAH2; CALM1; HSP90AA1; CALM3 |
| R-HSA-1168372 | Downstream signaling events of B Cell Receptor (BCR) | 4.31 | 1.07e-10 | 2.48e-09 | 24/58 | PSMA5; PSMD4; CALM2; RPS27A; PSMD6; SKP1; PSMB1; PPIA; PSMC2; PSMD13; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-4641257 | Degradation of AXIN | 4.30 | 1.06e-08 | 1.26e-07 | 19/46 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-1236975 | Antigen processing-Cross presentation | 4.29 | 1.84e-09 | 3.44e-08 | 21/51 | PSMA5; PSMD4; RPS27A; PSMD6; HLA-A; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PDIA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-1236978 | Cross-presentation of soluble exogenous antigens (endosomes) | 4.27 | 1.79e-07 | 1.39e-06 | 16/39 | PSMA5; PSMD4; PSMD6; PSMB1; PSMC2; PSMD13; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; PSMD8; PSMC4 |
| R-HSA-5610783 | Degradation of GLI2 by the proteasome | 4.25 | 5.47e-09 | 7.13e-08 | 20/49 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5610785 | GLI3 is processed to GLI3R by the proteasome | 4.25 | 5.47e-09 | 7.13e-08 | 20/49 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5362768 | Hh mutants that don't undergo autocatalytic processing are degraded by ERAD | 4.21 | 1.62e-08 | 1.81e-07 | 19/47 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5387390 | Hh mutants abrogate ligand secretion | 4.21 | 1.62e-08 | 1.81e-07 | 19/47 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-68867 | Assembly of the pre-replicative complex | 4.21 | 1.62e-08 | 1.81e-07 | 19/47 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5663205 | Infectious disease | 4.20 | 0.00e+00 | 0.00e+00 | 111/275 | RPL22; RPL11; RPS8; RPL5; PSMA5; PSMD4; RPS27; RPS7; CALM2; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; CTNNB1; RPL29; PSMD6; RPL24; AP2M1; RPL35A; RPL9; RPL34; RPS3A; SLC25A4; RPL37; RPS23; SKP1; RPS14; RPS18; RPS10; RPL10A; HSP90AB1; RPS12; PSMB1; PPIA; PSMC2; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; RPL8; RPS6; CHMP5; CLTA; RPL35; RPL12; RPL7A; PSMD13; RPLP2; RPL27A; RPS13; TSG101; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; UBC; RAN; RPL21; PSME2; RPS29; PSMA3; CALM1; HSP90AA1; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; PSMB6; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; PSMC5; KPNA2; RPL38; PSMA7; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; PSMD8; RPS16; PSMC4; RPS19; CALM3; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; CHMP2A; RPL3 |
| R-HSA-70268 | Pyruvate metabolism | 4.16 | 1.78e-03 | 7.80e-03 | 6/15 | PDHB; VDAC1; PDHA1; LDHA; LDHB; BSG |
| R-HSA-9013507 | NOTCH3 Activation and Transmission of Signal to the Nucleus | 4.16 | 1.10e-02 | 4.05e-02 | 4/10 | RPS27A; UBC; UBA52; PSENEN |
| R-HSA-3371568 | Attenuation phase | 4.16 | 1.10e-02 | 4.05e-02 | 4/10 | HSP90AB1; HSPA8; PTGES3; HSP90AA1 |
| R-HSA-4608870 | Asymmetric localization of PCP proteins | 4.12 | 2.45e-08 | 2.63e-07 | 19/48 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69541 | Stabilization of p53 | 4.12 | 2.45e-08 | 2.63e-07 | 19/48 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-8939902 | Regulation of RUNX2 expression and activity | 4.08 | 1.25e-08 | 1.48e-07 | 20/51 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5610787 | Hedgehog 'off' state | 4.07 | 3.29e-11 | 7.87e-10 | 27/69 | PSMA5; PSMD4; RPS27A; TUBA4A; PSMD6; SKP1; TUBB2A; TUBB2B; PSMB1; PSMC2; TUBB4B; PSMD13; TUBA1B; TUBA1A; UBC; PSME2; PSMA3; PSMA4; PSMD7; TUBB3; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-163210 | Formation of ATP by chemiosmotic coupling | 4.05 | 8.87e-04 | 4.17e-03 | 7/18 | ATP5F1; ATP5G3; ATP5C1; ATP5B; ATP5A1; ATP5J; ATP5O |
| R-HSA-3299685 | Detoxification of Reactive Oxygen Species | 4.05 | 8.87e-04 | 4.17e-03 | 7/18 | PRDX1; CYCS; TXN; PRDX5; GSTP1; PRDX2; SOD1 |
| R-HSA-5678895 | Defective CFTR causes cystic fibrosis | 4.04 | 3.65e-08 | 3.70e-07 | 19/49 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-1234176 | Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | 4.00 | 1.86e-08 | 2.05e-07 | 20/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; TCEB1; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5607764 | CLEC7A (Dectin-1) signaling | 3.99 | 1.64e-09 | 3.21e-08 | 23/60 | PSMA5; PSMD4; CALM2; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-5358346 | Hedgehog ligand biogenesis | 3.96 | 5.38e-08 | 5.26e-07 | 19/50 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-437239 | Recycling pathway of L1 | 3.95 | 3.87e-05 | 2.07e-04 | 11/29 | TUBA4A; AP2M1; TUBB2A; TUBB2B; ACTB; CLTA; TUBB4B; TUBA1B; TUBA1A; TUBB3; ACTG1 |
| R-HSA-2871837 | FCERI mediated NF-kB activation | 3.93 | 2.72e-08 | 2.82e-07 | 20/53 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-202424 | Downstream TCR signaling | 3.93 | 2.72e-08 | 2.82e-07 | 20/53 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-422475 | Axon guidance | 3.90 | 0.00e+00 | 0.00e+00 | 123/328 | RPL22; CDC42; RPL11; RPS8; RPL5; PSMA5; PSMD4; RPS27; ARPC5; RPS7; RPS27A; RPL31; RPL37A; ARPC2; TUBA4A; RPL32; RPL15; RPSA; RPL14; RPL29; PSMD6; RPL24; GAP43; AP2M1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; TUBB2A; TUBB2B; CSNK2B; RPS18; RPS10; RPL10A; HSP90AB1; RPS12; PSMB1; ACTB; ARPC1A; PSMC2; RPS4X; RPL36A; RPL39; RPL10; RPS20; SDCBP; RPL7; TCEB1; RPL30; PABPC1; RPL8; RPS6; CLTA; RPL35; RPL12; RPL7A; TUBB4B; PSMD13; RPLP2; RPL27A; RPS13; CFL1; RPS3; RPS25; HSPA8; RPS24; TUBA1B; TUBA1A; RPL41; MYL6; ARPC3; RPL6; RPLP0; MSI1; UBC; RPL21; PSME2; RPS29; PSMA3; HSP90AA1; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; TUBB3; PSMB6; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; PSMC5; RPL38; EIF4A3; ACTG1; MYL12B; PSMA7; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; PSENEN; PSMD8; RPS16; PSMC4; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-5619084 | ABC transporter disorders | 3.88 | 7.83e-08 | 7.41e-07 | 19/51 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-350562 | Regulation of ornithine decarboxylase (ODC) | 3.87 | 8.79e-07 | 5.48e-06 | 16/43 | PSMA5; PSMD4; PSMD6; PSMB1; PSMC2; PSMD13; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; PSMD8; PSMC4 |
| R-HSA-983705 | Signaling by the B Cell Receptor (BCR) | 3.84 | 1.75e-09 | 3.35e-08 | 24/65 | PSMA5; PSMD4; CALM2; RPS27A; PSMD6; SKP1; PSMB1; PPIA; PSMC2; PSMD13; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-5658442 | Regulation of RAS by GAPs | 3.80 | 1.13e-07 | 9.12e-07 | 19/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69002 | DNA Replication Pre-Initiation | 3.80 | 1.13e-07 | 9.12e-07 | 19/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69563 | p53-Dependent G1 DNA Damage Response | 3.80 | 1.13e-07 | 9.12e-07 | 19/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69580 | p53-Dependent G1/S DNA damage checkpoint | 3.80 | 1.13e-07 | 9.12e-07 | 19/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69615 | G1/S DNA Damage Checkpoints | 3.80 | 1.13e-07 | 9.12e-07 | 19/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-174084 | Autodegradation of Cdh1 by Cdh1:APC/C | 3.80 | 1.13e-07 | 9.12e-07 | 19/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5632684 | Hedgehog 'on' state | 3.80 | 1.13e-07 | 9.12e-07 | 19/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-8948751 | Regulation of PTEN stability and activity | 3.79 | 5.67e-08 | 5.48e-07 | 20/55 | PSMA5; PSMD4; RPS27A; PSMD6; CSNK2B; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-9013694 | Signaling by NOTCH4 | 3.77 | 2.85e-08 | 2.92e-07 | 21/58 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSENEN; PSMD8; PSMC4 |
| R-HSA-69017 | CDK-mediated phosphorylation and removal of Cdc6 | 3.73 | 1.60e-07 | 1.26e-06 | 19/53 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-176408 | Regulation of APC/C activators between G1/S and early anaphase | 3.72 | 8.05e-08 | 7.54e-07 | 20/56 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-9020702 | Interleukin-1 signaling | 3.70 | 4.04e-08 | 4.04e-07 | 21/59 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; HMGB1; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-1445148 | Translocation of SLC2A4 (GLUT4) to the plasma membrane | 3.70 | 1.79e-06 | 1.08e-05 | 16/45 | YWHAQ; CALM2; TUBA4A; TUBB2A; TUBB2B; ACTB; RALA; TUBB4B; TUBA1B; TUBA1A; CALM1; TUBB3; YWHAE; ACTG1; YWHAB; CALM3 |
| R-HSA-5339562 | Uptake and actions of bacterial toxins | 3.67 | 3.72e-03 | 1.50e-02 | 6/17 | CALM2; HSP90AB1; CALM1; HSP90AA1; EEF2; CALM3 |
| R-HSA-445355 | Smooth Muscle Contraction | 3.67 | 3.72e-03 | 1.50e-02 | 6/17 | TPM3; CALM2; MYL6; CALM1; MYL12B; CALM3 |
| R-HSA-174154 | APC/C:Cdc20 mediated degradation of Securin | 3.66 | 2.26e-07 | 1.64e-06 | 19/54 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-174178 | APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | 3.66 | 2.26e-07 | 1.64e-06 | 19/54 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-174184 | Cdc20:Phospho-APC/C mediated degradation of Cyclin A | 3.66 | 2.26e-07 | 1.64e-06 | 19/54 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-179419 | APC:Cdc20 mediated degradation of cell cycle proteins prior to satisfation of the cell cycle checkpoint | 3.66 | 2.26e-07 | 1.64e-06 | 19/54 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-6811436 | COPI-independent Golgi-to-ER retrograde traffic | 3.66 | 1.99e-05 | 1.08e-04 | 13/37 | CAPZB; TUBA4A; TUBB2A; TUBB2B; DCTN3; TUBB4B; TUBA1B; TUBA1A; DCTN2; DYNLL1; ACTR10; TUBB3; PAFAH1B3 |
| R-HSA-69202 | Cyclin E associated events during G1/S transition | 3.65 | 1.13e-07 | 9.12e-07 | 20/57 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69656 | Cyclin A:Cdk2-associated events at S phase entry | 3.65 | 1.13e-07 | 9.12e-07 | 20/57 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-174143 | APC/C-mediated degradation of cell cycle proteins | 3.65 | 1.13e-07 | 9.12e-07 | 20/57 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-453276 | Regulation of mitotic cell cycle | 3.65 | 1.13e-07 | 9.12e-07 | 20/57 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-1234174 | Regulation of Hypoxia-inducible Factor (HIF) by oxygen | 3.65 | 1.13e-07 | 9.12e-07 | 20/57 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; TCEB1; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-2262749 | Cellular response to hypoxia | 3.65 | 1.13e-07 | 9.12e-07 | 20/57 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; TCEB1; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-72766 | Translation | 3.65 | 0.00e+00 | 0.00e+00 | 90/257 | RPL22; DDOST; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; SSR4; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; SPCS2; RPS3; RPS25; PPA1; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; TUFM; RPL13; EIF4A1; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; SEC11C; RPN2; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3; MRPS6 |
| R-HSA-162588 | Budding and maturation of HIV virion | 3.64 | 1.82e-03 | 7.97e-03 | 7/20 | RPS27A; PPIA; CHMP5; TSG101; UBC; UBA52; CHMP2A |
| R-HSA-114608 | Platelet degranulation | 3.63 | 5.00e-06 | 2.91e-05 | 15/43 | TAGLN2; CALM2; TUBA4A; PPIA; MAGED2; CFL1; APLP2; PSAP; CD63; CALM1; SCG3; ALDOA; LGALS3BP; CALM3; SOD1 |
| R-HSA-76005 | Response to elevated platelet cytosolic Ca2+ | 3.63 | 5.00e-06 | 2.91e-05 | 15/43 | TAGLN2; CALM2; TUBA4A; PPIA; MAGED2; CFL1; APLP2; PSAP; CD63; CALM1; SCG3; ALDOA; LGALS3BP; CALM3; SOD1 |
| R-HSA-5619115 | Disorders of transmembrane transporters | 3.60 | 3.16e-07 | 2.14e-06 | 19/55 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-176409 | APC/C:Cdc20 mediated degradation of mitotic proteins | 3.60 | 3.16e-07 | 2.14e-06 | 19/55 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-176814 | Activation of APC/C and APC/C:Cdc20 mediated degradation of mitotic proteins | 3.60 | 3.16e-07 | 2.14e-06 | 19/55 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-382556 | ABC-family proteins mediated transport | 3.53 | 4.36e-07 | 2.87e-06 | 19/56 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-8878159 | Transcriptional regulation by RUNX3 | 3.53 | 2.18e-07 | 1.63e-06 | 20/59 | PSMA5; PSMD4; RPS27A; CTNNB1; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69052 | Switching of origins to a post-replicative state | 3.53 | 2.18e-07 | 1.63e-06 | 20/59 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-195253 | Degradation of beta-catenin by the destruction complex | 3.53 | 1.09e-07 | 9.12e-07 | 21/62 | PSMA5; PSMD4; RPS27A; CTNNB1; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-450531 | Regulation of mRNA stability by proteins that bind AU-rich elements | 3.52 | 2.72e-08 | 2.82e-07 | 23/68 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PABPC1; SET; PSMD13; HSPA8; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; YWHAB; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-4086400 | PCP/CE pathway | 3.47 | 1.50e-07 | 1.18e-06 | 21/63 | PSMA5; PSMD4; RPS27A; PSMD6; AP2M1; PSMB1; PSMC2; CLTA; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-8939236 | RUNX1 regulates transcription of genes involved in differentiation of HSCs | 3.47 | 2.99e-07 | 2.08e-06 | 20/60 | PSMA5; PSMD4; H3F3A; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-191273 | Cholesterol biosynthesis | 3.47 | 2.51e-03 | 1.06e-02 | 7/21 | FDPS; ACAT2; EBP; NSDHL; FDFT1; TM7SF2; DHCR7 |
| R-HSA-5205685 | Pink/Parkin Mediated Mitophagy | 3.47 | 1.06e-02 | 3.95e-02 | 5/15 | RPS27A; VDAC1; UBC; MAP1LC3B; UBA52 |
| R-HSA-168643 | Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways | 3.47 | 1.06e-02 | 3.95e-02 | 5/15 | RPS27A; HSP90AB1; TXN; UBC; UBA52 |
| R-HSA-5627123 | RHO GTPases activate PAKs | 3.47 | 5.14e-03 | 2.02e-02 | 6/18 | CDC42; CALM2; MYL6; CALM1; MYL12B; CALM3 |
| R-HSA-5358351 | Signaling by Hedgehog | 3.43 | 3.12e-09 | 4.89e-08 | 27/82 | PSMA5; PSMD4; RPS27A; TUBA4A; PSMD6; SKP1; TUBB2A; TUBB2B; PSMB1; PSMC2; TUBB4B; PSMD13; TUBA1B; TUBA1A; UBC; PSME2; PSMA3; PSMA4; PSMD7; TUBB3; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-202403 | TCR signaling | 3.41 | 4.06e-07 | 2.69e-06 | 20/61 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5621481 | C-type lectin receptors (CLRs) | 3.37 | 6.85e-08 | 6.56e-07 | 23/71 | PSMA5; PSMD4; CALM2; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-5687128 | MAPK6/MAPK4 signaling | 3.36 | 2.74e-07 | 1.94e-06 | 21/65 | CDC42; PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; SEPT7; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-8950505 | Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation | 3.35 | 8.29e-04 | 3.96e-03 | 9/28 | CDC42; HNRNPA2B1; RALA; PPIA; TALDO1; CFL1; RPLP0; PSME2; SOD1 |
| R-HSA-446652 | Interleukin-1 family signaling | 3.31 | 3.67e-07 | 2.45e-06 | 21/66 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; HMGB1; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5689603 | UCH proteinases | 3.17 | 8.48e-07 | 5.33e-06 | 21/69 | PSMA5; PSMD4; RPS27A; PSMD6; UCHL1; PSMB1; ACTB; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-983189 | Kinesins | 3.17 | 4.48e-03 | 1.78e-02 | 7/23 | TUBA4A; TUBB2A; TUBB2B; TUBB4B; TUBA1B; TUBA1A; TUBB3 |
| R-HSA-351202 | Metabolism of polyamines | 3.16 | 1.01e-05 | 5.63e-05 | 17/56 | PSMA5; PSMD4; PSMD6; PSMB1; PSMC2; PSMD13; PSME2; PSMA3; CKB; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; PSMD8; PSMC4 |
| R-HSA-8878166 | Transcriptional regulation by RUNX2 | 3.15 | 1.68e-06 | 1.03e-05 | 20/66 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-69239 | Synthesis of DNA | 3.15 | 1.68e-06 | 1.03e-05 | 20/66 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-9020591 | Interleukin-12 signaling | 3.12 | 1.44e-03 | 6.47e-03 | 9/30 | CDC42; HNRNPA2B1; RALA; PPIA; TALDO1; CFL1; RPLP0; PSME2; SOD1 |
| R-HSA-5205647 | Mitophagy | 3.12 | 9.09e-03 | 3.46e-02 | 6/20 | RPS27A; VDAC1; CSNK2B; UBC; MAP1LC3B; UBA52 |
| R-HSA-3928662 | EPHB-mediated forward signaling | 3.08 | 2.89e-03 | 1.21e-02 | 8/27 | CDC42; ARPC5; ARPC2; ACTB; ARPC1A; CFL1; ARPC3; ACTG1 |
| R-HSA-69481 | G2/M Checkpoints | 3.07 | 4.73e-07 | 3.08e-06 | 23/78 | PSMA5; PSMD4; YWHAQ; RPS27A; SUMO1; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; YWHAE; PSMB6; PSMB3; PSMC5; YWHAB; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-2454202 | Fc epsilon receptor (FCERI) signaling | 3.03 | 6.09e-07 | 3.89e-06 | 23/79 | PSMA5; PSMD4; CALM2; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-3858494 | Beta-catenin independent WNT signaling | 3.03 | 2.00e-07 | 1.54e-06 | 25/86 | PSMA5; PSMD4; CALM2; RPS27A; CTNNB1; PSMD6; AP2M1; PSMB1; PSMC2; CLTA; PSMD13; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-69306 | DNA Replication | 3.02 | 3.65e-06 | 2.17e-05 | 20/69 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-211859 | Biological oxidations | 2.97 | 3.93e-04 | 1.96e-03 | 12/42 | AKR7A2; GSTM3; MGST3; EPHX1; PODXL2; HSP90AB1; GSTA4; GSTP1; PTGES3; ALDH2; ESD; CYB5R3 |
| R-HSA-1266738 | Developmental Biology | 2.95 | 0.00e+00 | 0.00e+00 | 126/445 | RPL22; CDC42; RPL11; RPS8; RPL5; PSMA5; PSMD4; RPS27; ARPC5; H3F3A; RPS7; RPS27A; RPL31; NEUROD1; RPL37A; ARPC2; TUBA4A; RPL32; RPL15; RPSA; RPL14; CTNNB1; RPL29; PSMD6; RPL24; GAP43; AP2M1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; TUBB2A; TUBB2B; CSNK2B; RPS18; RPS10; RPL10A; HSP90AB1; RPS12; PSMB1; ACTB; ARPC1A; PSMC2; RPS4X; RPL36A; RPL39; RPL10; RPS20; SDCBP; RPL7; TCEB1; RPL30; PABPC1; RPL8; RPS6; CLTA; RPL35; RPL12; RPL7A; TUBB4B; PSMD13; RPLP2; RPL27A; RPS13; CFL1; RPS3; RPS25; HSPA8; RPS24; TUBA1B; TUBA1A; RPL41; MYL6; ARPC3; RPL6; RPLP0; MSI1; UBC; RPL21; PSME2; RPS29; PSMA3; HSP90AA1; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; TUBB3; PSMB6; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; PSMC5; RPL38; EIF4A3; ACTG1; MYL12B; PSMA7; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; PSENEN; PSMD8; RPS16; PSMC4; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-69206 | G1/S Transition | 2.93 | 5.93e-06 | 3.38e-05 | 20/71 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-447115 | Interleukin-12 family signaling | 2.93 | 2.38e-03 | 1.01e-02 | 9/32 | CDC42; HNRNPA2B1; RALA; PPIA; TALDO1; CFL1; RPLP0; PSME2; SOD1 |
| R-HSA-69275 | G2/M Transition | 2.90 | 3.77e-09 | 5.41e-08 | 34/122 | PSMA5; PSMD4; RPS27A; TUBA4A; PSMD6; SKP1; TUBB2A; TUBB2B; TUBB; HSP90AB1; PSMB1; PSMC2; DCTN3; TUBB4B; PSMD13; TUBA1B; TUBA1A; DCTN2; DYNLL1; UBC; PSME2; PSMA3; HSP90AA1; PSMA4; PSMD7; TUBB3; YWHAE; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-453274 | Mitotic G2-G2/M phases | 2.88 | 4.76e-09 | 6.40e-08 | 34/123 | PSMA5; PSMD4; RPS27A; TUBA4A; PSMD6; SKP1; TUBB2A; TUBB2B; TUBB; HSP90AB1; PSMB1; PSMC2; DCTN3; TUBB4B; PSMD13; TUBA1B; TUBA1A; DCTN2; DYNLL1; UBC; PSME2; PSMA3; HSP90AA1; PSMA4; PSMD7; TUBB3; YWHAE; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-453279 | Mitotic G1-G1/S phases | 2.88 | 4.84e-06 | 2.86e-05 | 21/76 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; CDKN2D; UBA52; PSMD8; PSMC4 |
| R-HSA-71406 | Pyruvate metabolism and Citric Acid (TCA) cycle | 2.84 | 3.00e-03 | 1.25e-02 | 9/33 | SUCLG1; PDHB; VDAC1; MDH2; PDHA1; LDHA; LDHB; IDH2; BSG |
| R-HSA-109581 | Apoptosis | 2.84 | 2.72e-07 | 1.94e-06 | 27/99 | PSMA5; PSMD4; YWHAQ; RPS27A; CTNNB1; PSMD6; PSMB1; CYCS; PSMC2; BCAP31; PSMD13; DYNLL1; UBC; HMGB1; PSME2; PSMA3; PSMA4; PSMD7; YWHAE; PSMB6; PSMB3; PSMC5; YWHAB; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5357801 | Programmed Cell Death | 2.84 | 2.72e-07 | 1.94e-06 | 27/99 | PSMA5; PSMD4; YWHAQ; RPS27A; CTNNB1; PSMD6; PSMB1; CYCS; PSMC2; BCAP31; PSMD13; DYNLL1; UBC; HMGB1; PSME2; PSMA3; PSMA4; PSMD7; YWHAE; PSMB6; PSMB3; PSMC5; YWHAB; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-162909 | Host Interactions of HIV factors | 2.83 | 8.29e-07 | 5.25e-06 | 25/92 | PSMA5; PSMD4; RPS27A; PSMD6; AP2M1; SLC25A4; SKP1; PSMB1; PPIA; PSMC2; TCEB1; PSMD13; UBC; RAN; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-2029482 | Regulation of actin dynamics for phagocytic cup formation | 2.81 | 1.92e-03 | 8.25e-03 | 10/37 | CDC42; ARPC5; ARPC2; HSP90AB1; ACTB; ARPC1A; CFL1; ARPC3; HSP90AA1; ACTG1 |
| R-HSA-5663213 | RHO GTPases Activate WASPs and WAVEs | 2.80 | 9.34e-03 | 3.53e-02 | 7/26 | CDC42; ARPC5; ARPC2; ACTB; ARPC1A; ARPC3; ACTG1 |
| R-HSA-5620924 | Intraflagellar transport | 2.78 | 5.90e-03 | 2.30e-02 | 8/30 | TUBA4A; TUBB2A; TUBB2B; TUBB4B; TUBA1B; TUBA1A; DYNLL1; TUBB3 |
| R-HSA-2467813 | Separation of Sister Chromatids | 2.74 | 1.37e-07 | 1.09e-06 | 30/114 | NUDC; PSMA5; PSMD4; RPS27; RPS27A; TUBA4A; PSMD6; TUBB2A; TUBB2B; PSMB1; PSMC2; TUBB4B; PSMD13; SMC3; TUBA1B; TUBA1A; DYNLL1; UBC; PSME2; PSMA3; PSMA4; PSMD7; TUBB3; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-1643685 | Disease | 2.71 | 0.00e+00 | 0.00e+00 | 121/464 | RPL22; RPL11; RPS8; PRDX1; RPL5; PSMA5; PSMD4; RPS27; RPS7; CALM2; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; CTNNB1; RPL29; PSMD6; RPL24; AP2M1; RPL35A; RPL9; RPL34; RPS3A; SLC25A4; RPL37; RPS23; SKP1; RPS14; RPS18; RPS10; RPL10A; HSP90AB1; RPS12; PSMB1; ACTB; PPIA; PSMC2; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; RPL8; RPS6; CHMP5; CLTA; RPL35; RPL12; RPL7A; PSMD13; TALDO1; RPLP2; RPL27A; RPS13; TSG101; RPS3; RPS25; RPS24; RPL41; RPL6; PEBP1; RPLP0; UBC; RAN; RPL21; PSME2; RPS29; PSMA3; CALM1; HSP90AA1; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; YWHAE; PSMB6; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; G6PC3; PSMC5; KPNA2; RPL38; ACTG1; YWHAB; PSMA7; RPS21; RPS15; EEF2; RPL36; RPS28; PRDX2; RPL18A; UBA52; PSENEN; PSMD8; RPS16; PSMC4; RPS19; CALM3; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; CHMP2A; RPL3 |
| R-HSA-977225 | Amyloid fiber formation | 2.70 | 1.16e-02 | 4.25e-02 | 7/27 | H3F3A; RPS27A; UBC; ITM2B; CST3; UBA52; PSENEN |
| R-HSA-917937 | Iron uptake and transport | 2.69 | 7.31e-03 | 2.81e-02 | 8/31 | ATP6V0B; RPS27A; SKP1; ATP6AP1; UBC; ATP6V0D1; UBA52; FTL |
| R-HSA-2029480 | Fcgamma receptor (FCGR) dependent phagocytosis | 2.66 | 1.87e-03 | 8.10e-03 | 11/43 | CDC42; ARPC5; ARPC2; HSP90AB1; ACTB; ARPC1A; CFL1; ARPC3; HSP90AA1; ACTG1; PLD3 |
| R-HSA-2682334 | EPH-Ephrin signaling | 2.65 | 4.89e-04 | 2.39e-03 | 14/55 | CDC42; ARPC5; ARPC2; AP2M1; ACTB; ARPC1A; SDCBP; CLTA; CFL1; MYL6; ARPC3; ACTG1; MYL12B; PSENEN |
| R-HSA-2555396 | Mitotic Metaphase and Anaphase | 2.58 | 5.68e-07 | 3.65e-06 | 30/121 | NUDC; PSMA5; PSMD4; RPS27; RPS27A; TUBA4A; PSMD6; TUBB2A; TUBB2B; PSMB1; PSMC2; TUBB4B; PSMD13; SMC3; TUBA1B; TUBA1A; DYNLL1; UBC; PSME2; PSMA3; PSMA4; PSMD7; TUBB3; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-68882 | Mitotic Anaphase | 2.58 | 5.68e-07 | 3.65e-06 | 30/121 | NUDC; PSMA5; PSMD4; RPS27; RPS27A; TUBA4A; PSMD6; TUBB2A; TUBB2B; PSMB1; PSMC2; TUBB4B; PSMD13; SMC3; TUBA1B; TUBA1A; DYNLL1; UBC; PSME2; PSMA3; PSMA4; PSMD7; TUBB3; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5673001 | RAF/MAP kinase cascade | 2.48 | 7.23e-06 | 4.10e-05 | 26/109 | PSMA5; PSMD4; CALM2; RPS27A; PSMD6; PSMB1; ACTB; PSMC2; PSMD13; PEBP1; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; ACTG1; YWHAB; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-5684996 | MAPK1/MAPK3 signaling | 2.46 | 8.65e-06 | 4.87e-05 | 26/110 | PSMA5; PSMD4; CALM2; RPS27A; PSMD6; PSMB1; ACTB; PSMC2; PSMD13; PEBP1; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; ACTG1; YWHAB; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-2565942 | Regulation of PLK1 Activity at G2/M Transition | 2.46 | 1.63e-03 | 7.20e-03 | 13/55 | RPS27A; TUBA4A; SKP1; TUBB; DCTN3; TUBB4B; TUBA1A; DCTN2; DYNLL1; UBC; HSP90AA1; YWHAE; UBA52 |
| R-HSA-380320 | Recruitment of NuMA to mitotic centrosomes | 2.46 | 1.63e-03 | 7.20e-03 | 13/55 | TUBA4A; TUBB2A; TUBB2B; TUBB; DCTN3; TUBB4B; TUBA1B; TUBA1A; DCTN2; DYNLL1; HSP90AA1; TUBB3; YWHAE |
| R-HSA-70326 | Glucose metabolism | 2.44 | 3.99e-03 | 1.61e-02 | 11/47 | ENO1; MDH1; MDH2; PGK1; PGAM1; GAPDH; TPI1; PKM; ALDOA; ENO3; G6PC3 |
| R-HSA-69242 | S Phase | 2.43 | 7.96e-05 | 4.16e-04 | 21/90 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; SMC3; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-5689880 | Ub-specific processing proteases | 2.42 | 3.91e-05 | 2.08e-04 | 23/99 | PSMA5; PSMD4; RPS27A; PSMD6; VDAC1; PSMB1; PSMC2; VDAC3; PSMD13; VDAC2; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; UFD1L |
| R-HSA-6807878 | COPI-mediated anterograde transport | 2.41 | 6.11e-04 | 2.97e-03 | 16/69 | CAPZB; RAB1A; TUBA4A; ARF4; TUBB2A; TUBB2B; DCTN3; TUBB4B; TUBA1B; TUBA1A; DCTN2; DYNLL1; ACTR10; TUBB3; COPE; NAPA |
| R-HSA-2132295 | MHC class II antigen presentation | 2.40 | 9.51e-04 | 4.40e-03 | 15/65 | CAPZB; TUBA4A; RAB7A; AP2M1; TUBB2A; TUBB2B; DCTN3; CLTA; TUBB4B; TUBA1B; TUBA1A; DCTN2; DYNLL1; ACTR10; TUBB3 |
| R-HSA-390466 | Chaperonin-mediated protein folding | 2.36 | 3.59e-03 | 1.47e-02 | 12/53 | CCT7; TUBA4A; CCT5; TUBB2A; TUBB2B; CSNK2B; ACTB; TUBB4B; TUBA1B; TUBA1A; CCT2; TUBB3 |
| R-HSA-391251 | Protein folding | 2.33 | 2.72e-03 | 1.14e-02 | 13/58 | CCT7; TUBA4A; CCT5; TUBB2A; TUBB2B; CSNK2B; ACTB; TUBB4B; TUBA1B; TUBA1A; CCT2; TUBB3; TBCB |
| R-HSA-373760 | L1CAM interactions | 2.30 | 1.56e-03 | 6.94e-03 | 15/68 | TUBA4A; GAP43; AP2M1; TUBB2A; TUBB2B; CSNK2B; ACTB; SDCBP; CLTA; TUBB4B; HSPA8; TUBA1B; TUBA1A; TUBB3; ACTG1 |
| R-HSA-201681 | TCF dependent signaling in response to WNT | 2.29 | 1.38e-04 | 7.07e-04 | 22/100 | PSMA5; PSMD4; H3F3A; RPS27A; CTNNB1; PSMD6; CSNK2B; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-1592230 | Mitochondrial biogenesis | 2.22 | 4.34e-03 | 1.73e-02 | 13/61 | ATP5F1; CALM2; ATP5G3; CYCS; SSBP1; ATP5C1; ATP5B; CALM1; IDH2; ATP5A1; CALM3; ATP5J; ATP5O |
| R-HSA-6798695 | Neutrophil degranulation | 2.19 | 1.06e-06 | 6.54e-06 | 39/185 | DDOST; PSMA5; ILF2; ARPC5; PSMD6; RAB7A; TUBB; CSNK2B; HSP90AB1; DYNLT1; PSMB1; PPIA; PSMC2; PRDX4; ATP6AP2; SDCBP; GGH; TUBB4B; PSMD13; GSTP1; NDUFC2; HSPA8; PSAP; PGAM1; CD63; CCT2; DYNLL1; HMGB1; ACTR10; HSP90AA1; SRP14; PKM; ALDOA; PSMD7; VAPA; CST3; EEF2; FTL; CYB5R3 |
| R-HSA-5683057 | MAPK family signaling cascades | 2.19 | 3.96e-05 | 2.09e-04 | 28/133 | CDC42; PSMA5; PSMD4; CALM2; RPS27A; PSMD6; PSMB1; ACTB; SEPT7; PSMC2; PSMD13; PEBP1; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; ACTG1; YWHAB; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | 2.18 | 2.23e-05 | 1.21e-04 | 30/143 | ATP5F1; SUCLG1; ATP5G3; UQCRC1; PDHB; VDAC1; NDUFA4; CYCS; MDH2; PDHA1; NDUFB11; COX6C; NDUFB6; LDHA; NDUFV1; NDUFC2; ATP5C1; NDUFB8; LDHB; ATP5B; COX5A; IDH2; UQCRC2; NDUFV2; ATP5A1; BSG; NDUFA13; ETFB; ATP5J; ATP5O |
| R-HSA-2262752 | Cellular responses to stress | 2.18 | 4.60e-08 | 4.55e-07 | 49/234 | CAPZB; PRDX1; PSMA5; PSMD4; H3F3A; RPS27A; TUBA4A; PSMD6; TUBB2A; TUBB2B; HSP90AB1; PSMB1; CYCS; PSMC2; TCEB1; DNAJA1; DCTN3; TXN; TUBB4B; PSMD13; PRDX5; GSTP1; C11orf73; HSPA8; TUBA1B; TUBA1A; PTGES3; DCTN2; DYNLL1; UBC; PSME2; ACTR10; PSMA3; HSP90AA1; PSMA4; PSMD7; TERF2IP; TUBB3; YWHAE; PSMB6; PSMB3; PSMC5; PSMA7; CDKN2D; PRDX2; UBA52; PSMD8; PSMC4; SOD1 |
| R-HSA-449147 | Signaling by Interleukins | 2.18 | 1.25e-05 | 6.92e-05 | 32/153 | CDC42; PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; HNRNPA2B1; RALA; PPIA; PSMC2; PSMD13; TALDO1; CFL1; HSPA8; HSP90B1; RPLP0; UBC; HMGB1; PSME2; PSMA3; HSP90AA1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; SOD1 |
| R-HSA-8878171 | Transcriptional regulation by RUNX1 | 2.14 | 3.86e-04 | 1.93e-03 | 22/107 | PSMA5; PSMD4; H3F3A; RPS27A; PSMD6; CSNK2B; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; PRMT1 |
| R-HSA-8856688 | Golgi-to-ER retrograde transport | 2.13 | 1.84e-03 | 8.02e-03 | 17/83 | CAPZB; RAB1A; TUBA4A; ARF4; TUBB2A; TUBB2B; DCTN3; TUBB4B; TUBA1B; TUBA1A; DCTN2; DYNLL1; ACTR10; TUBB3; COPE; PAFAH1B3; NAPA |
| R-HSA-157118 | Signaling by NOTCH | 2.12 | 4.43e-04 | 2.18e-03 | 22/108 | PSMA5; PSMD4; H3F3A; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSENEN; PSMD8; PSMC4 |
| R-HSA-983231 | Factors involved in megakaryocyte development and platelet production | 2.12 | 1.19e-02 | 4.33e-02 | 11/54 | CAPZB; CDC42; H3F3A; TUBA4A; TUBB2A; TUBB2B; ACTB; TUBB4B; TUBA1B; TUBA1A; TUBB3 |
| R-HSA-6811434 | COPI-dependent Golgi-to-ER retrograde traffic | 2.12 | 1.19e-02 | 4.33e-02 | 11/54 | RAB1A; TUBA4A; ARF4; TUBB2A; TUBB2B; TUBB4B; TUBA1B; TUBA1A; TUBB3; COPE; NAPA |
| R-HSA-71387 | Metabolism of carbohydrates | 2.10 | 8.98e-04 | 4.20e-03 | 20/99 | ENO1; CALM2; RPS27A; MDH1; TKT; MDH2; AKR1B1; PGK1; TALDO1; PGAM1; GAPDH; TPI1; UBC; CALM1; PKM; ALDOA; ENO3; G6PC3; UBA52; CALM3 |
| R-HSA-6807070 | PTEN Regulation | 2.08 | 7.71e-04 | 3.71e-03 | 21/105 | PSMA5; PSMD4; RPS27A; PSMD6; CSNK2B; PSMB1; PSMC2; RRAGA; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-8856828 | Clathrin-mediated endocytosis | 2.05 | 4.89e-03 | 1.93e-02 | 15/76 | ARPC5; RPS27A; ARPC2; COPS8; AP2M1; ACTB; ARPC1A; COPS6; CLTA; HSPA8; ARPC3; UBC; COPS3; ACTG1; UBA52 |
| R-HSA-68886 | M Phase | 2.04 | 5.30e-06 | 3.06e-05 | 40/204 | NUDC; PSMA5; PSMD4; RPS27; H3F3A; RPS27A; RAB1A; TUBA4A; PSMD6; TUBB2A; TUBB2B; TUBB; CSNK2B; PSMB1; PSMC2; RAB2A; DCTN3; SET; TUBB4B; PSMD13; SMC3; TUBA1B; TUBA1A; DCTN2; DYNLL1; UBC; PSME2; PSMA3; HSP90AA1; PSMA4; PSMD7; TUBB3; YWHAE; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4 |
| R-HSA-195721 | Signaling by WNT | 2.03 | 1.30e-04 | 6.71e-04 | 29/149 | PSMA5; PSMD4; H3F3A; CALM2; RPS27A; CTNNB1; PSMD6; AP2M1; SKP1; CSNK2B; PSMB1; PSMC2; CLTA; PSMD13; VPS29; UBC; PSME2; PSMA3; CALM1; PSMA4; PSMD7; PSMB6; PSMB3; PSMC5; PSMA7; UBA52; PSMD8; PSMC4; CALM3 |
| R-HSA-195258 | RHO GTPase Effectors | 2.02 | 1.73e-04 | 8.83e-04 | 28/144 | CDC42; NUDC; RPS27; ARPC5; H3F3A; YWHAQ; CALM2; ARPC2; TUBA4A; CTNNB1; TUBB2A; TUBB2B; ACTB; ARPC1A; TUBB4B; CFL1; TUBA1B; TUBA1A; MYL6; ARPC3; DYNLL1; CALM1; TUBB3; YWHAE; ACTG1; MYL12B; YWHAB; CALM3 |
| R-HSA-5663220 | RHO GTPases Activate Formins | 2.02 | 9.89e-03 | 3.72e-02 | 13/67 | CDC42; NUDC; RPS27; TUBA4A; TUBB2A; TUBB2B; ACTB; TUBB4B; TUBA1B; TUBA1A; DYNLL1; TUBB3; ACTG1 |
| R-HSA-8957322 | Metabolism of steroids | 2.01 | 1.32e-02 | 4.78e-02 | 12/62 | FDPS; HSD17B11; ACAT2; AKR1B1; EBP; NSDHL; FDFT1; TM7SF2; DHCR7; AKR1C1; RAN; SUMO2 |
Factor 3 : 33 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-192823 | Viral mRNA Translation | 40.80 | 0 | 0 | 66/83 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-156902 | Peptide chain elongation | 40.30 | 0 | 0 | 66/84 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-2408557 | Selenocysteine synthesis | 39.80 | 0 | 0 | 66/85 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 39.40 | 0 | 0 | 66/86 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72764 | Eukaryotic Translation Termination | 39.40 | 0 | 0 | 66/86 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 38.50 | 0 | 0 | 66/88 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 35.60 | 0 | 0 | 66/95 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-2408522 | Selenoamino acid metabolism | 34.20 | 0 | 0 | 66/99 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 32.50 | 0 | 0 | 66/104 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 32.20 | 0 | 0 | 66/105 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 32.20 | 0 | 0 | 66/105 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 32.20 | 0 | 0 | 66/105 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 31.90 | 0 | 0 | 66/106 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 30.00 | 0 | 0 | 66/113 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 30.00 | 0 | 0 | 66/113 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 30.00 | 0 | 0 | 66/113 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-168255 | Influenza Life Cycle | 28.00 | 0 | 0 | 66/121 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 27.80 | 0 | 0 | 26/48 | RPS8; RPS27; RPS7; RPS27A; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS17; RPS15A; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-168254 | Influenza Infection | 26.80 | 0 | 0 | 67/128 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72649 | Translation initiation complex formation | 24.20 | 0 | 0 | 26/55 | RPS8; RPS27; RPS7; RPS27A; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS17; RPS15A; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 24.20 | 0 | 0 | 26/55 | RPS8; RPS27; RPS7; RPS27A; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS17; RPS15A; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 23.80 | 0 | 0 | 26/56 | RPS8; RPS27; RPS7; RPS27A; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS17; RPS15A; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 22.70 | 0 | 0 | 66/149 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 22.00 | 0 | 0 | 66/154 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 21.20 | 0 | 0 | 66/160 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72312 | rRNA processing | 20.60 | 0 | 0 | 66/164 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-376176 | Signaling by ROBO receptors | 19.00 | 0 | 0 | 66/178 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 16.20 | 0 | 0 | 66/209 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72766 | Translation | 13.20 | 0 | 0 | 66/257 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-5663205 | Infectious disease | 12.50 | 0 | 0 | 67/275 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-422475 | Axon guidance | 10.30 | 0 | 0 | 66/328 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-1266738 | Developmental Biology | 7.61 | 0 | 0 | 66/445 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-1643685 | Disease | 7.41 | 0 | 0 | 67/464 | RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23; RPL19; RPL38; RPS21; RPS15; RPL36; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
Factor 4 : 48 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-5576892 | Phase 0 - rapid depolarisation | 7.97 | 2.17e-04 | 9.34e-03 | 5/12 | SCN3A; SCN9A; CAMK2B; CALM1 |
| R-HSA-373080 | Class B/2 (Secretin family receptors) | 7.36 | 3.38e-04 | 1.12e-02 | 5/13 | GNG5; GNG4; FZD1; GNG8; PTH2 |
| R-HSA-442729 | CREB phosphorylation through the activation of CaMKII | 6.96 | 1.79e-03 | 3.63e-02 | 4/11 | CAMK2B; NEFL; CALM1 |
| R-HSA-442982 | Ras activation upon Ca2+ influx through NMDA receptor | 6.96 | 1.79e-03 | 3.63e-02 | 4/11 | CAMK2B; NEFL; CALM1 |
| R-HSA-500792 | GPCR ligand binding | 6.38 | 4.97e-06 | 1.03e-03 | 9/27 | GNG5; AGT; GNG4; GRM4; FZD1; GAL; SSTR2; GNG8; PTH2 |
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 6.38 | 2.06e-04 | 9.34e-03 | 6/18 | TUBB2B; ACTB; CALM1; ACTG1; TUBB4A |
| R-HSA-445095 | Interaction between L1 and Ankyrins | 6.38 | 7.23e-04 | 2.07e-02 | 5/15 | SCN3A; SCN9A; ACTB; ANK3; ACTG1 |
| R-HSA-392170 | ADP signalling through P2Y purinoceptor 12 | 6.38 | 2.57e-03 | 4.51e-02 | 4/12 | GNG5; GNG4; GNAI2; GNG8 |
| R-HSA-400042 | Adrenaline,noradrenaline inhibits insulin secretion | 6.38 | 2.57e-03 | 4.51e-02 | 4/12 | GNG5; GNG4; GNAI2; GNG8 |
| R-HSA-451326 | Activation of kainate receptors upon glutamate binding | 5.98 | 1.01e-03 | 2.63e-02 | 5/16 | GNG5; GNG4; CALM1; GNG8 |
| R-HSA-5578775 | Ion homeostasis | 5.82 | 1.14e-04 | 6.56e-03 | 7/23 | ATP1B1; RYR2; CAMK2B; SRI; CALM1; FXYD7 |
| R-HSA-445355 | Smooth Muscle Contraction | 5.63 | 1.37e-03 | 3.10e-02 | 5/17 | CALD1; MYL6; CALM1; MYL12A |
| R-HSA-5576891 | Cardiac conduction | 5.47 | 7.00e-06 | 1.03e-03 | 10/35 | ATP1B1; RYR2; SCN3A; SCN9A; CAMK2B; SRI; AKAP9; CALM1; FXYD7 |
| R-HSA-190828 | Gap junction trafficking | 5.31 | 1.81e-03 | 3.63e-02 | 5/18 | AP2M1; TUBB2B; ACTB; ACTG1; TUBB4A |
| R-HSA-397014 | Muscle contraction | 5.22 | 6.38e-08 | 5.49e-05 | 15/55 | ATP1B1; RYR2; SCN3A; SCN9A; MYL1; CAMK2B; SRI; AKAP9; CALD1; MYL6; CALM1; ACTC1; MYL12A; FXYD7 |
| R-HSA-157858 | Gap junction trafficking and regulation | 5.03 | 2.36e-03 | 4.32e-02 | 5/19 | AP2M1; TUBB2B; ACTB; ACTG1; TUBB4A |
| R-HSA-2672351 | Stimuli-sensing channels | 4.78 | 1.15e-03 | 2.67e-02 | 6/24 | RYR2; RPS27A; SRI; CALM1; TTYH2 |
| R-HSA-936837 | Ion transport by P-type ATPases | 4.78 | 1.15e-03 | 2.67e-02 | 6/24 | ATP1B1; CAMK2B; SRI; CALM1; FXYD7 |
| R-HSA-418555 | G alpha (s) signalling events | 4.78 | 1.15e-03 | 2.67e-02 | 6/24 | GNG5; GNG4; REEP1; GNAI2; GNG8; PTH2 |
| R-HSA-437239 | Recycling pathway of L1 | 4.62 | 5.57e-04 | 1.71e-02 | 7/29 | AP2M1; TUBB2B; EZR; ACTB; DPYSL2; ACTG1; TUBB4A |
| R-HSA-418346 | Platelet homeostasis | 4.46 | 6.95e-04 | 2.06e-02 | 7/30 | GNG5; GNG4; SRI; CALM1; GNG8; PDE9A |
| R-HSA-112314 | Neurotransmitter receptors and postsynaptic signal transmission | 4.29 | 3.01e-05 | 3.24e-03 | 11/49 | GNG5; GNG4; CHRNA1; GNAI2; AP2M1; GRIA2; CAMK2B; NEFL; CALM1; GNG8 |
| R-HSA-373755 | Semaphorin interactions | 3.94 | 1.54e-03 | 3.39e-02 | 7/34 | RHOC; DPYSL3; HSP90AB1; DPYSL2; CFL1; MYL6; MYL12A |
| R-HSA-418594 | G alpha (i) signalling events | 3.53 | 1.02e-04 | 6.56e-03 | 12/65 | GNG5; RGS16; AGT; GNG4; GNAI2; GRM4; GAL; RGS10; CALM1; SSTR2; GNG8 |
| R-HSA-373760 | L1CAM interactions | 3.38 | 1.61e-04 | 7.70e-03 | 12/68 | SCN3A; SCN9A; NRP2; ALCAM; AP2M1; TUBB2B; EZR; ACTB; DPYSL2; ANK3; ACTG1; TUBB4A |
| R-HSA-156842 | Eukaryotic Translation Elongation | 3.34 | 2.77e-05 | 3.24e-03 | 15/86 | RPS27A; EEF1B2; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-112315 | Transmission across Chemical Synapses | 3.23 | 7.61e-05 | 6.55e-03 | 14/83 | GNG5; GNG4; CHRNA1; GNAI2; AP2M1; GRIA2; CAMK2B; STX1A; NEFL; CALM1; CACNA1A; RAB3A; GNG8 |
| R-HSA-192823 | Viral mRNA Translation | 3.23 | 7.61e-05 | 6.55e-03 | 14/83 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-112316 | Neuronal System | 3.22 | 7.17e-06 | 1.03e-03 | 18/107 | PTPRF; GNG5; GNG4; NRXN1; CHRNA1; GNAI2; AP2M1; GRIA2; CAMK2B; STX1A; NEFL; SYT7; CALM1; CACNA1A; RAB3A; HOMER3; GNG8 |
| R-HSA-156902 | Peptide chain elongation | 3.19 | 8.71e-05 | 6.56e-03 | 14/84 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-2408557 | Selenocysteine synthesis | 3.15 | 9.96e-05 | 6.56e-03 | 14/85 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-72764 | Eukaryotic Translation Termination | 3.11 | 1.14e-04 | 6.56e-03 | 14/86 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 3.04 | 1.47e-04 | 7.43e-03 | 14/88 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 2.82 | 3.37e-04 | 1.12e-02 | 14/95 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 2.73 | 2.93e-04 | 1.07e-02 | 15/105 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; EIF4A3; RPS11; RPL28 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 2.73 | 2.93e-04 | 1.07e-02 | 15/105 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; EIF4A3; RPS11; RPL28 |
| R-HSA-372790 | Signaling by GPCR | 2.71 | 1.27e-04 | 6.86e-03 | 17/120 | GNG5; RHOC; RGS16; AGT; GNG4; REEP1; GNAI2; GRM4; FZD1; GAL; RGS10; CALM1; SSTR2; PREX1; GNG8; PTH2 |
| R-HSA-2408522 | Selenoamino acid metabolism | 2.71 | 5.21e-04 | 1.66e-02 | 14/99 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-388396 | GPCR downstream signalling | 2.64 | 2.78e-04 | 1.07e-02 | 16/116 | GNG5; RHOC; RGS16; AGT; GNG4; REEP1; GNAI2; GRM4; GAL; RGS10; CALM1; SSTR2; PREX1; GNG8; PTH2 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 2.58 | 8.65e-04 | 2.40e-02 | 14/104 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 2.55 | 9.53e-04 | 2.56e-02 | 14/105 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 2.53 | 1.05e-03 | 2.65e-02 | 14/106 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 2.37 | 1.97e-03 | 3.68e-02 | 14/113 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 2.37 | 1.97e-03 | 3.68e-02 | 14/113 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 2.37 | 1.97e-03 | 3.68e-02 | 14/113 | RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; RPS11; RPL28 |
| R-HSA-109582 | Hemostasis | 2.26 | 2.98e-04 | 1.07e-02 | 21/178 | PRKCZ; GNG5; TAGLN2; ATP1B1; GNG4; GYPC; GNAI2; TUBB2B; HDAC2; ACTB; SRI; TMSB4X; CFL1; CD63; CALM1; KIF19; BSG; TUBB4A; GNG8; PDE9A |
| R-HSA-422475 | Axon guidance | 2.22 | 1.17e-06 | 5.02e-04 | 38/328 | RHOC; LHX9; RPS27A; SCN3A; SCN9A; NRP2; RPL29; ALCAM; AP2M1; RPS3A; RPL37; RGMB; DPYSL3; PDLIM7; TUBB2B; HSP90AB1; EZR; ACTB; RPS4X; RPL39; DPYSL2; RPL7; RPL12; RPL27A; CFL1; RPS3; ANK3; MYL6; RPLP0; RPLP1; EFNB3; EIF4A3; ACTG1; MYL12A; PSMA7; TUBB4A; RPS11; RPL28 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 2.18 | 1.67e-03 | 3.60e-02 | 17/149 | LHX9; RPS27A; RPL29; RPS3A; RPL37; RPS4X; RPL39; RPL7; RPL12; RPL27A; RPS3; RPLP0; RPLP1; EIF4A3; PSMA7; RPS11; RPL28 |
Factor 5 : 152 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-8978934 | Metabolism of cofactors | 6.14 | 7.34e-04 | 4.45e-03 | 5/12 | COQ5; CALM1; HSP90AA1; COQ9 |
| R-HSA-3371568 | Attenuation phase | 5.90 | 3.13e-03 | 1.76e-02 | 4/10 | HSP90AB1; HSF1; HSPA8; HSP90AA1 |
| R-HSA-450408 | AUF1 (hnRNP D0) binds and destabilizes mRNA | 5.33 | 3.40e-09 | 1.48e-06 | 17/47 | PSMD6; PSMD2; PSMB1; HSPB1; PSMC2; PSMB7; PSMD13; HSPA8; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-1236978 | Cross-presentation of soluble exogenous antigens (endosomes) | 5.29 | 9.79e-08 | 3.24e-06 | 14/39 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-180534 | Vpu mediated degradation of CD4 | 5.14 | 5.23e-08 | 2.82e-06 | 15/43 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-211733 | Regulation of activated PAK-2p34 by proteasome mediated degradation | 5.14 | 5.23e-08 | 2.82e-06 | 15/43 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69601 | Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | 5.14 | 5.23e-08 | 2.82e-06 | 15/43 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69610 | p53-Independent DNA Damage Response | 5.14 | 5.23e-08 | 2.82e-06 | 15/43 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69613 | p53-Independent G1/S DNA damage checkpoint | 5.14 | 5.23e-08 | 2.82e-06 | 15/43 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69229 | Ubiquitin-dependent degradation of Cyclin D1 | 5.03 | 7.47e-08 | 3.21e-06 | 15/44 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-75815 | Ubiquitin-dependent degradation of Cyclin D | 5.03 | 7.47e-08 | 3.21e-06 | 15/44 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-169911 | Regulation of Apoptosis | 5.03 | 7.47e-08 | 3.21e-06 | 15/44 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-174113 | SCF-beta-TrCP mediated degradation of Emi1 | 5.03 | 7.47e-08 | 3.21e-06 | 15/44 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-187577 | SCF(Skp2)-mediated degradation of p27/p21 | 5.02 | 2.75e-08 | 2.49e-06 | 16/47 | CKS1B; PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5607761 | Dectin-1 mediated noncanonical NF-kB signaling | 5.02 | 2.75e-08 | 2.49e-06 | 16/47 | PSMD6; UBA3; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5676590 | NIK-->noncanonical NF-kB signaling | 5.02 | 2.75e-08 | 2.49e-06 | 16/47 | PSMD6; UBA3; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5668541 | TNFR2 non-canonical NF-kB pathway | 4.92 | 3.88e-08 | 2.82e-06 | 16/48 | PSMD6; UBA3; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-349425 | Autodegradation of the E3 ubiquitin ligase COP1 | 4.92 | 1.05e-07 | 3.24e-06 | 15/45 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-4641258 | Degradation of DVL | 4.92 | 1.05e-07 | 3.24e-06 | 15/45 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-68827 | CDT1 association with the CDC6:ORC:origin complex | 4.92 | 1.05e-07 | 3.24e-06 | 15/45 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-8854050 | FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | 4.92 | 1.05e-07 | 3.24e-06 | 15/45 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-8941858 | Regulation of RUNX3 expression and activity | 4.92 | 1.05e-07 | 3.24e-06 | 15/45 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-8852276 | The role of GTSE1 in G2/M progression after G2 checkpoint | 4.83 | 7.35e-09 | 1.58e-06 | 18/55 | PSMD6; PSMD2; HSP90AB1; PSMB1; PSMC2; PSMB7; PSMD13; TUBA1A; UBC; PSMC6; PSMC1; HSP90AA1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-180585 | Vif-mediated degradation of APOBEC3G | 4.81 | 1.47e-07 | 3.51e-06 | 15/46 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-4641257 | Degradation of AXIN | 4.81 | 1.47e-07 | 3.51e-06 | 15/46 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-350562 | Regulation of ornithine decarboxylase (ODC) | 4.80 | 3.98e-07 | 5.80e-06 | 14/43 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5362768 | Hh mutants that don't undergo autocatalytic processing are degraded by ERAD | 4.71 | 2.03e-07 | 4.06e-06 | 15/47 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5387390 | Hh mutants abrogate ligand secretion | 4.71 | 2.03e-07 | 4.06e-06 | 15/47 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-68867 | Assembly of the pre-replicative complex | 4.71 | 2.03e-07 | 4.06e-06 | 15/47 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-9604323 | Negative regulation of NOTCH4 signaling | 4.71 | 2.03e-07 | 4.06e-06 | 15/47 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-389957 | Prefoldin mediated transfer of substrate to CCT/TriC | 4.69 | 4.27e-04 | 2.72e-03 | 7/22 | PFDN2; CCT7; CCT5; TCP1; ACTB; VBP1; TUBA1A |
| R-HSA-5607764 | CLEC7A (Dectin-1) signaling | 4.67 | 5.15e-09 | 1.48e-06 | 19/60 | PSMD6; UBA3; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; CALM1; PDPK1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-68949 | Orc1 removal from chromatin | 4.61 | 2.77e-07 | 4.68e-06 | 15/48 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-1169091 | Activation of NF-kappaB in B cells | 4.61 | 2.77e-07 | 4.68e-06 | 15/48 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-4608870 | Asymmetric localization of PCP proteins | 4.61 | 2.77e-07 | 4.68e-06 | 15/48 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5610780 | Degradation of GLI1 by the proteasome | 4.61 | 2.77e-07 | 4.68e-06 | 15/48 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69541 | Stabilization of p53 | 4.61 | 2.77e-07 | 4.68e-06 | 15/48 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-1168372 | Downstream signaling events of B Cell Receptor (BCR) | 4.58 | 1.91e-08 | 2.49e-06 | 18/58 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; CALM1; PSMB6; PSMD11; PSMB3; PSMD3; FKBP1A; PSMA7; PSMD8 |
| R-HSA-69563 | p53-Dependent G1 DNA Damage Response | 4.54 | 1.39e-07 | 3.51e-06 | 16/52 | PCBP4; PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69580 | p53-Dependent G1/S DNA damage checkpoint | 4.54 | 1.39e-07 | 3.51e-06 | 16/52 | PCBP4; PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69615 | G1/S DNA Damage Checkpoints | 4.54 | 1.39e-07 | 3.51e-06 | 16/52 | PCBP4; PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-1234176 | Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | 4.54 | 1.39e-07 | 3.51e-06 | 16/52 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; EGLN2 |
| R-HSA-5610783 | Degradation of GLI2 by the proteasome | 4.51 | 3.76e-07 | 5.58e-06 | 15/49 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5610785 | GLI3 is processed to GLI3R by the proteasome | 4.51 | 3.76e-07 | 5.58e-06 | 15/49 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5678895 | Defective CFTR causes cystic fibrosis | 4.51 | 3.76e-07 | 5.58e-06 | 15/49 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-389958 | Cooperation of Prefoldin and TriC/CCT in actin and tubulin folding | 4.49 | 5.79e-04 | 3.53e-03 | 7/23 | PFDN2; CCT7; CCT5; TCP1; ACTB; VBP1; TUBA1A |
| R-HSA-2871837 | FCERI mediated NF-kB activation | 4.45 | 1.88e-07 | 4.06e-06 | 16/53 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PDPK1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-202424 | Downstream TCR signaling | 4.45 | 1.88e-07 | 4.06e-06 | 16/53 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PDPK1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-1236974 | ER-Phagosome pathway | 4.42 | 5.05e-07 | 7.13e-06 | 15/50 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5358346 | Hedgehog ligand biogenesis | 4.42 | 5.05e-07 | 7.13e-06 | 15/50 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-174154 | APC/C:Cdc20 mediated degradation of Securin | 4.37 | 2.51e-07 | 4.68e-06 | 16/54 | PSMD6; PSMD2; PTTG1; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-174178 | APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | 4.37 | 2.51e-07 | 4.68e-06 | 16/54 | PSMD6; PSMD2; PTTG1; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5619084 | ABC transporter disorders | 4.34 | 6.72e-07 | 8.15e-06 | 15/51 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-1236975 | Antigen processing-Cross presentation | 4.34 | 6.72e-07 | 8.15e-06 | 15/51 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-8939902 | Regulation of RUNX2 expression and activity | 4.34 | 6.72e-07 | 8.15e-06 | 15/51 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5339562 | Uptake and actions of bacterial toxins | 4.34 | 4.33e-03 | 2.32e-02 | 5/17 | HSP90AB1; STX1A; CALM1; HSP90AA1 |
| R-HSA-5619115 | Disorders of transmembrane transporters | 4.29 | 3.33e-07 | 5.30e-06 | 16/55 | TPR; PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-176409 | APC/C:Cdc20 mediated degradation of mitotic proteins | 4.29 | 3.33e-07 | 5.30e-06 | 16/55 | PSMD6; PSMD2; PTTG1; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-176814 | Activation of APC/C and APC/C:Cdc20 mediated degradation of mitotic proteins | 4.29 | 3.33e-07 | 5.30e-06 | 16/55 | PSMD6; PSMD2; PTTG1; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5658442 | Regulation of RAS by GAPs | 4.25 | 8.88e-07 | 1.02e-05 | 15/52 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69002 | DNA Replication Pre-Initiation | 4.25 | 8.88e-07 | 1.02e-05 | 15/52 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-174084 | Autodegradation of Cdh1 by Cdh1:APC/C | 4.25 | 8.88e-07 | 1.02e-05 | 15/52 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5632684 | Hedgehog 'on' state | 4.25 | 8.88e-07 | 1.02e-05 | 15/52 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-8939236 | RUNX1 regulates transcription of genes involved in differentiation of HSCs | 4.18 | 2.15e-07 | 4.21e-06 | 17/60 | PSMD6; PSMD2; H2AFZ; HIST1H4C; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69017 | CDK-mediated phosphorylation and removal of Cdc6 | 4.17 | 1.17e-06 | 1.30e-05 | 15/53 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69202 | Cyclin E associated events during G1/S transition | 4.14 | 5.72e-07 | 7.35e-06 | 16/57 | CKS1B; PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69656 | Cyclin A:Cdk2-associated events at S phase entry | 4.14 | 5.72e-07 | 7.35e-06 | 16/57 | CKS1B; PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-174143 | APC/C-mediated degradation of cell cycle proteins | 4.14 | 5.72e-07 | 7.35e-06 | 16/57 | PSMD6; PSMD2; PTTG1; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-453276 | Regulation of mitotic cell cycle | 4.14 | 5.72e-07 | 7.35e-06 | 16/57 | PSMD6; PSMD2; PTTG1; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-1234174 | Regulation of Hypoxia-inducible Factor (HIF) by oxygen | 4.14 | 5.72e-07 | 7.35e-06 | 16/57 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; EGLN2 |
| R-HSA-2262749 | Cellular response to hypoxia | 4.14 | 5.72e-07 | 7.35e-06 | 16/57 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; EGLN2 |
| R-HSA-174184 | Cdc20:Phospho-APC/C mediated degradation of Cyclin A | 4.10 | 1.52e-06 | 1.64e-05 | 15/54 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-179419 | APC:Cdc20 mediated degradation of cell cycle proteins prior to satisfation of the cell cycle checkpoint | 4.10 | 1.52e-06 | 1.64e-05 | 15/54 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 4.10 | 5.67e-03 | 3.00e-02 | 5/18 | ACTB; TUBA1A; CALM1; ACTG1 |
| R-HSA-163210 | Formation of ATP by chemiosmotic coupling | 4.10 | 5.67e-03 | 3.00e-02 | 5/18 | ATP5G3; ATP5B; ATP5A1; ATP5E; ATP5O |
| R-HSA-983705 | Signaling by the B Cell Receptor (BCR) | 4.08 | 1.37e-07 | 3.51e-06 | 18/65 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; CALM1; PSMB6; PSMD11; PSMB3; PSMD3; FKBP1A; PSMA7; PSMD8 |
| R-HSA-195253 | Degradation of beta-catenin by the destruction complex | 4.04 | 3.63e-07 | 5.58e-06 | 17/62 | PSMD6; PSMD2; PPP2CA; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-8948751 | Regulation of PTEN stability and activity | 4.02 | 1.96e-06 | 1.99e-05 | 15/55 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-9020702 | Interleukin-1 signaling | 4.00 | 9.58e-07 | 1.09e-05 | 16/59 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; HMGB1; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-382556 | ABC-family proteins mediated transport | 3.95 | 2.52e-06 | 2.46e-05 | 15/56 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-176408 | Regulation of APC/C activators between G1/S and early anaphase | 3.95 | 2.52e-06 | 2.46e-05 | 15/56 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5621481 | C-type lectin receptors (CLRs) | 3.95 | 1.11e-07 | 3.29e-06 | 19/71 | PSMD6; UBA3; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; CALM1; PDPK1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-202403 | TCR signaling | 3.87 | 1.57e-06 | 1.66e-05 | 16/61 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PDPK1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-9013694 | Signaling by NOTCH4 | 3.81 | 4.07e-06 | 3.77e-05 | 15/58 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-8878159 | Transcriptional regulation by RUNX3 | 3.75 | 5.14e-06 | 4.56e-05 | 15/59 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69052 | Switching of origins to a post-replicative state | 3.75 | 5.14e-06 | 4.56e-05 | 15/59 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-4086400 | PCP/CE pathway | 3.74 | 2.50e-06 | 2.46e-05 | 16/63 | PSMD6; AP2M1; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69206 | G1/S Transition | 3.74 | 5.90e-07 | 7.47e-06 | 18/71 | CKS1B; PSMD6; PSMD2; PPP2CA; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-450531 | Regulation of mRNA stability by proteins that bind AU-rich elements | 3.69 | 1.52e-06 | 1.64e-05 | 17/68 | PSMD6; PSMD2; PSMB1; HSPB1; PSMC2; PSMB7; PSMD13; HSPA8; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-351202 | Metabolism of polyamines | 3.69 | 1.32e-05 | 1.08e-04 | 14/56 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5687128 | MAPK6/MAPK4 signaling | 3.63 | 3.91e-06 | 3.66e-05 | 16/65 | PSMD6; PSMD2; PSMB1; HSPB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-446652 | Interleukin-1 family signaling | 3.57 | 4.85e-06 | 4.40e-05 | 16/66 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; HMGB1; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 3.49 | 3.79e-09 | 1.48e-06 | 27/114 | NDUFS2; COX5B; ATP5G3; SDHA; NDUFS6; COX7C; NDUFB11; NDUFS8; NDUFC2; SDHD; ATP5B; NDUFA12; COX6A1; NDUFB10; TRAP1; UQCRC2; NDUFAB1; ATP5A1; ATP5E; ATP5O; MT-ND1; MT-ND2; MT-CO1; MT-CO2; MT-CO3; MT-ND4; MT-CYB |
| R-HSA-453279 | Mitotic G1-G1/S phases | 3.49 | 1.75e-06 | 1.82e-05 | 18/76 | CKS1B; PSMD6; PSMD2; PPP2CA; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-8949613 | Cristae formation | 3.44 | 3.19e-03 | 1.78e-02 | 7/30 | ATP5G3; CHCHD3; ATP5B; ATP5A1; ATP5E; SAMM50; ATP5O |
| R-HSA-5610787 | Hedgehog 'off' state | 3.42 | 9.01e-06 | 7.65e-05 | 16/69 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; TUBA1A; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5689603 | UCH proteinases | 3.42 | 9.01e-06 | 7.65e-05 | 16/69 | PSMD6; PSMD2; PSMB1; ACTB; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-611105 | Respiratory electron transport | 3.41 | 1.79e-07 | 4.06e-06 | 22/95 | NDUFS2; COX5B; SDHA; NDUFS6; COX7C; NDUFB11; NDUFS8; NDUFC2; SDHD; NDUFA12; COX6A1; NDUFB10; TRAP1; UQCRC2; NDUFAB1; MT-ND1; MT-ND2; MT-CO1; MT-CO2; MT-CO3; MT-ND4; MT-CYB |
| R-HSA-69481 | G2/M Checkpoints | 3.40 | 2.62e-06 | 2.53e-05 | 18/78 | YWHAQ; PSMD6; PSMD2; HIST1H4C; PSMB1; PSMC2; PSMB7; PSMD13; KAT5; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-2454202 | Fc epsilon receptor (FCERI) signaling | 3.36 | 3.19e-06 | 3.02e-05 | 18/79 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; CALM1; PDPK1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-8878166 | Transcriptional regulation by RUNX2 | 3.35 | 2.24e-05 | 1.77e-04 | 15/66 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69239 | Synthesis of DNA | 3.35 | 2.24e-05 | 1.77e-04 | 15/66 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-3858494 | Beta-catenin independent WNT signaling | 3.26 | 2.76e-06 | 2.64e-05 | 19/86 | PSMD6; AP2M1; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; GNG3; UBC; PSMC6; PSMC1; CALM1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-69306 | DNA Replication | 3.21 | 3.94e-05 | 3.00e-04 | 15/69 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-3371497 | HSP90 chaperone cycle for steroid hormone receptors (SHR) | 3.19 | 2.74e-03 | 1.56e-02 | 8/37 | HSP90AB1; CAPZA2; DCTN3; HSPA8; TUBA1A; DCTN2; ACTR10; HSP90AA1 |
| R-HSA-69275 | G2/M Transition | 3.14 | 7.85e-08 | 3.22e-06 | 26/122 | CENPF; PSMD6; PSMD2; PPP2CA; TUBB; HSP90AB1; PSMB1; PSMC2; DCTN3; PSMB7; PSMD13; TUBA1A; DCTN2; PHLDA1; UBC; PSMC6; PSMC1; HSP90AA1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; RAB8A; PSMD8; PPP2R1A |
| R-HSA-6799198 | Complex I biogenesis | 3.12 | 5.55e-04 | 3.47e-03 | 11/52 | NDUFS2; NDUFS6; NDUFB11; NDUFS8; NDUFC2; NDUFA12; NDUFB10; NDUFAB1; MT-ND1; MT-ND2; MT-ND4 |
| R-HSA-453274 | Mitotic G2-G2/M phases | 3.12 | 9.37e-08 | 3.24e-06 | 26/123 | CENPF; PSMD6; PSMD2; PPP2CA; TUBB; HSP90AB1; PSMB1; PSMC2; DCTN3; PSMB7; PSMD13; TUBA1A; DCTN2; PHLDA1; UBC; PSMC6; PSMC1; HSP90AA1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; RAB8A; PSMD8; PPP2R1A |
| R-HSA-201681 | TCF dependent signaling in response to WNT | 3.10 | 1.95e-06 | 1.99e-05 | 21/100 | PSMD6; PSMD2; H2AFZ; PPP2CA; SOX4; HIST1H4C; PSMB1; PSMC2; PSMB7; PSMD13; KAT5; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | 3.09 | 1.06e-08 | 1.82e-06 | 30/143 | NDUFS2; FH; COX5B; ATP5G3; PDHB; SDHA; NDUFS6; COX7C; NDUFB11; NDUFS8; NDUFC2; SDHD; ATP5B; NDUFA12; COX6A1; SUCLA2; NDUFB10; TRAP1; UQCRC2; NDUFAB1; ATP5A1; ATP5E; ATP5O; MT-ND1; MT-ND2; MT-CO1; MT-CO2; MT-CO3; MT-ND4; MT-CYB |
| R-HSA-162909 | Host Interactions of HIV factors | 3.05 | 7.98e-06 | 7.01e-05 | 19/92 | TPR; ARF1; PSMD6; AP2M1; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; RAE1; PSMA7; PSMD8 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 2.97 | 2.90e-08 | 2.49e-06 | 30/149 | RPL5; RBM8A; RPL31; PSMD6; PSMD2; RPL34; RPS3A; PSMB1; PSMC2; RPS4X; RPL10; RPL7; PSMB7; PSMD13; RPS24; RPLP0; UBC; RPS29; PSMC6; PSMC1; RNPS1; PSMB6; PSMD11; PSMB3; RPL19; PSMD3; EIF4A3; PSMA7; PSMD8; RPL3 |
| R-HSA-5358351 | Signaling by Hedgehog | 2.88 | 8.82e-05 | 6.33e-04 | 16/82 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; TUBA1A; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5673001 | RAF/MAP kinase cascade | 2.84 | 8.36e-06 | 7.27e-05 | 21/109 | PSMD6; PSMD2; PPP2CA; PSMB1; ACTB; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; CALM1; PSMB6; PSMD11; PSMB3; PSMD3; ACTG1; PSMA7; PSMD8; PPP2R1A |
| R-HSA-109581 | Apoptosis | 2.83 | 2.43e-05 | 1.87e-04 | 19/99 | YWHAQ; PSMD6; PSMD2; HMGB2; PSMB1; PSMC2; FNTA; PSMB7; PSMD13; UBC; HMGB1; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5357801 | Programmed Cell Death | 2.83 | 2.43e-05 | 1.87e-04 | 19/99 | YWHAQ; PSMD6; PSMD2; HMGB2; PSMB1; PSMC2; FNTA; PSMB7; PSMD13; UBC; HMGB1; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-5684996 | MAPK1/MAPK3 signaling | 2.82 | 9.71e-06 | 8.12e-05 | 21/110 | PSMD6; PSMD2; PPP2CA; PSMB1; ACTB; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; CALM1; PSMB6; PSMD11; PSMB3; PSMD3; ACTG1; PSMA7; PSMD8; PPP2R1A |
| R-HSA-72649 | Translation initiation complex formation | 2.68 | 3.30e-03 | 1.82e-02 | 10/55 | EIF4A2; EIF4E; RPS3A; RPS4X; EIF3E; RPS24; RPS29; EIF3J; EIF4A1; EIF3D |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 2.68 | 3.30e-03 | 1.82e-02 | 10/55 | EIF4A2; EIF4E; RPS3A; RPS4X; EIF3E; RPS24; RPS29; EIF3J; EIF4A1; EIF3D |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 2.63 | 3.78e-03 | 2.06e-02 | 10/56 | EIF4A2; EIF4E; RPS3A; RPS4X; EIF3E; RPS24; RPS29; EIF3J; EIF4A1; EIF3D |
| R-HSA-69242 | S Phase | 2.62 | 2.77e-04 | 1.81e-03 | 16/90 | CKS1B; PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-8951664 | Neddylation | 2.61 | 1.36e-05 | 1.11e-04 | 23/130 | PSMD6; UBA3; PSMD2; COPS4; PSMB1; PSMC2; COPS5; DCAF13; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; COPS3; PSMD11; PSMB3; PSMD3; PSMA7; FBXL12; KEAP1; PSMD8; FBXO7 |
| R-HSA-157118 | Signaling by NOTCH | 2.59 | 8.57e-05 | 6.20e-04 | 19/108 | PSMD6; PSMD2; H2AFZ; HIST1H4C; PSMB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; NCOR1; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; MFNG |
| R-HSA-2467813 | Separation of Sister Chromatids | 2.59 | 5.78e-05 | 4.29e-04 | 20/114 | CENPF; PSMD6; PSMD2; PPP2CA; PTTG1; PSMB1; PSMC2; PSMB7; PSMD13; TUBA1A; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-5689880 | Ub-specific processing proteases | 2.53 | 2.76e-04 | 1.81e-03 | 17/99 | PSMD6; PSMD2; PSMB1; PSMC2; PSMB7; PSMD13; SUDS3; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; KEAP1; PSMD8 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 2.53 | 1.86e-04 | 1.26e-03 | 18/105 | RPL5; RPL31; EIF4A2; EIF4E; RPL34; RPS3A; RPS4X; RPL10; RPL7; EIF3E; RPS24; RPLP0; RPS29; EIF3J; EIF4A1; RPL19; EIF3D; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 2.50 | 2.11e-04 | 1.42e-03 | 18/106 | RPL5; RPL31; EIF4A2; EIF4E; RPL34; RPS3A; RPS4X; RPL10; RPL7; EIF3E; RPS24; RPLP0; RPS29; EIF3J; EIF4A1; RPL19; EIF3D; RPL3 |
| R-HSA-376176 | Signaling by ROBO receptors | 2.49 | 1.75e-06 | 1.82e-05 | 30/178 | RPL5; RBM8A; RPL31; PSMD6; PSMD2; RPL34; RPS3A; PSMB1; PSMC2; RPS4X; RPL10; RPL7; PSMB7; PSMD13; RPS24; RPLP0; UBC; RPS29; PSMC6; PSMC1; RNPS1; PSMB6; PSMD11; PSMB3; RPL19; PSMD3; EIF4A3; PSMA7; PSMD8; RPL3 |
| R-HSA-8878171 | Transcriptional regulation by RUNX1 | 2.48 | 2.38e-04 | 1.59e-03 | 18/107 | PSMD6; PSMD2; H2AFZ; HIST1H4C; PSMB1; PSMC2; PSMB7; PSMD13; YAF2; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-195721 | Signaling by WNT | 2.47 | 1.49e-05 | 1.20e-04 | 25/149 | PSMD6; AP2M1; PSMD2; H2AFZ; PPP2CA; SOX4; HIST1H4C; PSMB1; PSMC2; PSMB7; PSMD13; GNG3; KAT5; UBC; PSMC6; PSMC1; CALM1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-5683057 | MAPK family signaling cascades | 2.44 | 6.24e-05 | 4.59e-04 | 22/133 | PSMD6; PSMD2; PPP2CA; PSMB1; ACTB; HSPB1; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; CALM1; PSMB6; PSMD11; PSMB3; PSMD3; ACTG1; PSMA7; PSMD8; PPP2R1A |
| R-HSA-2555396 | Mitotic Metaphase and Anaphase | 2.44 | 1.38e-04 | 9.57e-04 | 20/121 | CENPF; PSMD6; PSMD2; PPP2CA; PTTG1; PSMB1; PSMC2; PSMB7; PSMD13; TUBA1A; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-68882 | Mitotic Anaphase | 2.44 | 1.38e-04 | 9.57e-04 | 20/121 | CENPF; PSMD6; PSMD2; PPP2CA; PTTG1; PSMB1; PSMC2; PSMB7; PSMD13; TUBA1A; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-1592230 | Mitochondrial biogenesis | 2.42 | 7.10e-03 | 3.68e-02 | 10/61 | ATP5G3; CHCHD3; ATP5B; CALM1; NCOR1; ATP5A1; ATP5E; SAMM50; ATP5O |
| R-HSA-449147 | Signaling by Interleukins | 2.41 | 2.39e-05 | 1.87e-04 | 25/153 | ARF1; PSMD6; PSMD2; PPP2CA; TCP1; PSMB1; STX1A; PSMC2; PSMB7; PSMD13; HSPA8; RPLP0; UBC; HMGB1; PSMC6; PSMC1; HSP90AA1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; JUNB; PSMD8; PPP2R1A |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 2.39 | 5.66e-04 | 3.48e-03 | 17/105 | RPL5; RBM8A; RPL31; RPL34; RPS3A; PPP2CA; RPS4X; RPL10; RPL7; RPS24; RPLP0; RPS29; RNPS1; RPL19; EIF4A3; PPP2R1A; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 2.39 | 5.66e-04 | 3.48e-03 | 17/105 | RPL5; RBM8A; RPL31; RPL34; RPS3A; PPP2CA; RPS4X; RPL10; RPL7; RPS24; RPLP0; RPS29; RNPS1; RPL19; EIF4A3; PPP2R1A; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 2.35 | 4.79e-04 | 3.01e-03 | 18/113 | RPL5; RPL31; EIF4A2; EIF4E; RPL34; RPS3A; RPS4X; RPL10; RPL7; EIF3E; RPS24; RPLP0; RPS29; EIF3J; EIF4A1; RPL19; EIF3D; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 2.35 | 4.79e-04 | 3.01e-03 | 18/113 | RPL5; RPL31; EIF4A2; EIF4E; RPL34; RPS3A; RPS4X; RPL10; RPL7; EIF3E; RPS24; RPLP0; RPS29; EIF3J; EIF4A1; RPL19; EIF3D; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 2.33 | 1.55e-03 | 9.10e-03 | 15/95 | RPL5; RPL31; RPL34; RPS3A; RPS4X; RPL10; RPL7; EIF3E; RPS24; RPLP0; RPS29; EIF3J; RPL19; EIF3D; RPL3 |
| R-HSA-69620 | Cell Cycle Checkpoints | 2.32 | 1.38e-04 | 9.57e-04 | 22/140 | CENPF; YWHAQ; PCBP4; PSMD6; PSMD2; PPP2CA; HIST1H4C; PSMB1; PSMC2; PSMB7; PSMD13; KAT5; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-6807070 | PTEN Regulation | 2.25 | 1.61e-03 | 9.35e-03 | 16/105 | PSMD6; PSMD2; PSMB1; PSMC2; MAF1; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8 |
| R-HSA-1280215 | Cytokine Signaling in Immune system | 2.23 | 9.06e-06 | 7.65e-05 | 32/212 | TPR; ARF1; PSMD6; UBA3; PSMD2; EIF4A2; EIF4E; PPP2CA; TCP1; PSMB1; STX1A; PSMC2; PSMB7; PSMD13; HSPA8; RPLP0; UBC; HMGB1; PSMC6; PSMC1; HSP90AA1; PSMB6; EIF4A1; PSMD11; PSMB3; PSMD3; EIF4A3; RAE1; PSMA7; JUNB; PSMD8; PPP2R1A |
| R-HSA-2262752 | Cellular responses to stress | 2.14 | 1.09e-05 | 9.02e-05 | 34/234 | TPR; PSMD6; PSMD2; H2AFZ; HIST1H4C; HSP90AB1; PSMB1; PSMC2; CAPZA2; HSF1; DCTN3; PSMB7; PSMD13; KAT5; GSTP1; HSPA8; PRDX3; TUBA1A; DCTN2; UBC; PSMC6; ACTR10; PSMC1; HSP90AA1; TERF2IP; PSMB6; PSMD11; PSMB3; PSMD3; RAE1; PSMA7; PRDX2; PSMD8; EGLN2 |
| R-HSA-192823 | Viral mRNA Translation | 2.13 | 9.25e-03 | 4.74e-02 | 12/83 | RPL5; RPL31; RPL34; RPS3A; RPS4X; RPL10; RPL7; RPS24; RPLP0; RPS29; RPL19; RPL3 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 2.12 | 4.81e-05 | 3.64e-04 | 30/209 | RPL5; RPL31; PDHB; PSMD6; PSMD2; RPL34; RPS3A; PSMB1; PSMC2; RPS4X; RPL10; RPL7; PSMB7; PSMD13; SLC3A2; RPS24; RPLP0; RPS29; PSMC6; PSMC1; NDUFAB1; PSMB6; PSMD11; PSMB3; RPL19; PSMD3; PSMA7; PSMD8; MPST; RPL3 |
| R-HSA-68886 | M Phase | 2.10 | 7.80e-05 | 5.69e-04 | 29/204 | TPR; CENPF; PSMD6; PSMD2; H2AFZ; PPP2CA; PTTG1; HIST1H4C; TUBB; PSMB1; PSMC2; RAB2A; DCTN3; PSMB7; PSMD13; TUBA1A; DCTN2; UBC; PSMC6; PSMC1; HSP90AA1; PSMB6; PSMD11; PSMB3; PSMD3; RAE1; PSMA7; PSMD8; PPP2R1A |
| R-HSA-5663205 | Infectious disease | 2.09 | 4.12e-06 | 3.78e-05 | 39/275 | RPL5; TPR; ARF1; RPL31; PSMD6; AP2M1; PSMD2; RPL34; RPS3A; HSP90AB1; PSMB1; STX1A; TAF6; PSMC2; RPS4X; RPL10; RPL7; PSMB7; PSMD13; RPS24; RPLP0; UBC; RPS29; PSMC6; PSMC1; CALM1; HSP90AA1; POLR2C; PSMB6; PSMD11; PSMB3; RPL19; PSMD3; RAE1; NELFCD; PSMA7; PSMD8; RPL3 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 2.09 | 3.48e-03 | 1.91e-02 | 16/113 | RPL5; TPR; RPL31; RPL34; RPS3A; RPS4X; RPL10; RPL7; RPS24; RPLP0; RPS29; HSP90AA1; POLR2C; RPL19; RAE1; RPL3 |
| R-HSA-162906 | HIV Infection | 2.05 | 7.98e-04 | 4.80e-03 | 22/158 | TPR; ARF1; PSMD6; AP2M1; PSMD2; PSMB1; TAF6; PSMC2; PSMB7; PSMD13; UBC; PSMC6; PSMC1; POLR2C; PSMB6; PSMD11; PSMB3; PSMD3; RAE1; NELFCD; PSMA7; PSMD8 |
| R-HSA-9006925 | Intracellular signaling by second messengers | 2.04 | 1.16e-03 | 6.91e-03 | 21/152 | PSMD6; PSMD2; PPP2CA; PSMB1; PSMC2; MAF1; PSMB7; PSMD13; UBC; PSMC6; PSMC1; CALM1; PDPK1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
| R-HSA-1257604 | PIP3 activates AKT signaling | 2.00 | 2.46e-03 | 1.41e-02 | 19/140 | PSMD6; PSMD2; PPP2CA; PSMB1; PSMC2; MAF1; PSMB7; PSMD13; UBC; PSMC6; PSMC1; PDPK1; PSMB6; PSMD11; PSMB3; PSMD3; PSMA7; PSMD8; PPP2R1A |
Factor 6 : 2 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-983170 | Antigen Presentation: Folding, assembly and peptide loading of class I MHC | 14.10 | 1.26e-05 | 0.0108 | 5/11 | CANX; HLA-A; HSPA5; PDIA3; CALR |
| R-HSA-168255 | Influenza Life Cycle | 3.34 | 1.06e-04 | 0.0457 | 13/121 | RPS7; RPL22L1; CANX; RPS18; RPL10A; RPS20; RPL12; RPS25; RAN; HSP90AA1; RPS21; CALR; RPS9 |
Factor 7 : 54 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-156902 | Peptide chain elongation | 7.87 | 0.00e+00 | 0.00e+00 | 57/84 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; EEF1A1; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; EEF2; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 7.69 | 0.00e+00 | 0.00e+00 | 57/86 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; EEF1A1; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; EEF2; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-192823 | Viral mRNA Translation | 7.69 | 0.00e+00 | 0.00e+00 | 55/83 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-72764 | Eukaryotic Translation Termination | 7.56 | 0.00e+00 | 0.00e+00 | 56/86 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; TRMT112; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-2408557 | Selenocysteine synthesis | 7.51 | 0.00e+00 | 0.00e+00 | 55/85 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 7.45 | 0.00e+00 | 0.00e+00 | 61/95 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL35; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; EIF3K; RPS19; RPL18; RPL13A; RPS11; EIF3L; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 7.25 | 0.00e+00 | 0.00e+00 | 55/88 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-163210 | Formation of ATP by chemiosmotic coupling | 7.09 | 3.01e-08 | 6.65e-07 | 11/18 | ATP5G3; ATP5I; ATP5J2; ATP5L; ATP5G2; ATP5G1; ATP5H; ATP5A1; ATP5E; ATP5J; ATP5O |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 6.85 | 0.00e+00 | 0.00e+00 | 62/105 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; EIF4A2; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL35; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; EIF3K; RPS19; RPL18; RPL13A; RPS11; EIF3L; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 6.79 | 0.00e+00 | 0.00e+00 | 62/106 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; EIF4A2; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL35; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; EIF3K; RPS19; RPL18; RPL13A; RPS11; EIF3L; RPL3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 6.77 | 0.00e+00 | 0.00e+00 | 28/48 | RPS8; RPS27; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS26; RPS29; EIF3J; RPS17; RPS2; RPS15A; EIF3K; RPS19; RPS11; EIF3L |
| R-HSA-8876725 | Protein methylation | 6.77 | 1.79e-05 | 3.58e-04 | 7/12 | CALM2; EEF1A1; CALM1; RPS2; EEF2; CALM3; ETFB |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 6.69 | 0.00e+00 | 0.00e+00 | 60/104 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; SPCS1; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SEC61G; RPS4X; RPL39; SSR4; RPL10; RPS20; RPL7; RPL30; RPS6; SEC61B; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 6.47 | 0.00e+00 | 0.00e+00 | 63/113 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; EIF4A2; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL35; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; EIF3K; RPS19; RPL18; RPL13A; RPS11; EIF3L; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 6.47 | 0.00e+00 | 0.00e+00 | 63/113 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; EIF4A2; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL35; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; EIF3K; RPS19; RPL18; RPL13A; RPS11; EIF3L; RPL3 |
| R-HSA-2408522 | Selenoamino acid metabolism | 6.45 | 0.00e+00 | 0.00e+00 | 55/99 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 6.26 | 0.00e+00 | 0.00e+00 | 61/113 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; POLR2J; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPS6; RPL35; RPL7A; POLR2L; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; HSP90AA1; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; POLR2I; RPS19; RPL18; RPL13A; RPS11; POLR2F; RPL3 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 6.22 | 0.00e+00 | 0.00e+00 | 30/56 | RPS8; RPS27; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; EIF4EBP1; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS26; RPS29; EIF3J; RPS17; RPS2; RPS15A; EIF3K; RPS19; RPS11; EIF3L |
| R-HSA-72649 | Translation initiation complex formation | 6.12 | 0.00e+00 | 0.00e+00 | 29/55 | RPS8; RPS27; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS26; RPS29; EIF3J; RPS17; RPS2; RPS15A; EIF3K; RPS19; RPS11; EIF3L |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 6.12 | 0.00e+00 | 0.00e+00 | 29/55 | RPS8; RPS27; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS26; RPS29; EIF3J; RPS17; RPS2; RPS15A; EIF3K; RPS19; RPS11; EIF3L |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 6.08 | 0.00e+00 | 0.00e+00 | 55/105 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 6.08 | 0.00e+00 | 0.00e+00 | 55/105 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-6799198 | Complex I biogenesis | 6.02 | 4.44e-16 | 1.09e-14 | 27/52 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFC1; NDUFS6; NDUFA2; NDUFB2; NDUFB11; NDUFA1; NDUFB9; NDUFB6; NDUFS8; NDUFC2; NDUFB8; NDUFB1; NDUFB10; NDUFAB1; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND3; MT-ND4 |
| R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 6.01 | 0.00e+00 | 0.00e+00 | 59/114 | NDUFS5; UQCRH; COX20; COX5B; ATP5G3; NDUFB3; NDUFAF3; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; UQCRQ; NDUFA2; NDUFA4; CYCS; ATP5J2; NDUFB2; NDUFB11; COX7B; NDUFA1; UQCRB; COX6C; NDUFB9; NDUFB6; COX8A; NDUFS8; NDUFC2; ATP5L; NDUFB8; COX14; ATP5G2; COX6A1; COX16; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; ETFB; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| R-HSA-168255 | Influenza Life Cycle | 5.95 | 0.00e+00 | 0.00e+00 | 62/121 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; POLR2J; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPS6; RPL35; RPL7A; POLR2L; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RAN; RPL21; RPS29; RPL36AL; HSP90AA1; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; POLR2I; RPS19; RPL18; RPL13A; RPS11; POLR2F; RPL3 |
| R-HSA-611105 | Respiratory electron transport | 5.86 | 0.00e+00 | 0.00e+00 | 48/95 | NDUFS5; UQCRH; COX20; COX5B; NDUFB3; NDUFAF3; NDUFB4; NDUFC1; NDUFS6; COX7C; UQCRQ; NDUFA2; NDUFA4; CYCS; NDUFB2; NDUFB11; COX7B; NDUFA1; UQCRB; COX6C; NDUFB9; NDUFB6; COX8A; NDUFS8; NDUFC2; NDUFB8; COX14; COX6A1; COX16; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; NDUFS7; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; ETFB; NDUFA3; UQCR10; NDUFA6; MT-CO1; MT-ND3; MT-ND4 |
| R-HSA-3299685 | Detoxification of Reactive Oxygen Species | 5.80 | 5.65e-06 | 1.19e-04 | 9/18 | PRDX1; GPX1; ATOX1; CYCS; TXN; PRDX5; GSTP1; PRDX2; SOD1 |
| R-HSA-168254 | Influenza Infection | 5.71 | 0.00e+00 | 0.00e+00 | 63/128 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; POLR2J; SLC25A6; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPS6; RPL35; RPL7A; POLR2L; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RAN; RPL21; RPS29; RPL36AL; HSP90AA1; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; POLR2I; RPS19; RPL18; RPL13A; RPS11; POLR2F; RPL3 |
| R-HSA-5601884 | PIWI-interacting RNA (piRNA) biogenesis | 5.36 | 3.98e-04 | 7.13e-03 | 6/13 | POLR2J; POLR2K; POLR2L; HSP90AA1; POLR2I; POLR2F |
| R-HSA-3371511 | HSF1 activation | 5.27 | 1.38e-03 | 2.32e-02 | 5/11 | EEF1A1; RPA3; HSP90AA1; HSBP1; YWHAE |
| R-HSA-430039 | mRNA decay by 5' to 3' exoribonuclease | 5.27 | 1.38e-03 | 2.32e-02 | 5/11 | LSM3; LSM6; LSM5; LSM7; LSM4 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 4.83 | 0.00e+00 | 0.00e+00 | 62/149 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SHFM1; RPS4X; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; RPL26; RPL23A; PSMB3; RPL23; RPL19; PSMA7; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3; RBX1 |
| R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | 4.79 | 0.00e+00 | 0.00e+00 | 59/143 | NDUFS5; UQCRH; COX20; COX5B; ATP5G3; NDUFB3; NDUFAF3; NDUFB4; ATP5I; NDUFC1; NDUFS6; COX7C; UQCRQ; NDUFA2; NDUFA4; CYCS; ATP5J2; NDUFB2; NDUFB11; COX7B; NDUFA1; UQCRB; COX6C; NDUFB9; NDUFB6; COX8A; NDUFS8; NDUFC2; ATP5L; NDUFB8; COX14; ATP5G2; COX6A1; COX16; NDUFB1; COX5A; NDUFB10; NDUFAB1; COX4I1; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; ETFB; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-CO1; MT-ND3; MT-ND4 |
| R-HSA-8949613 | Cristae formation | 4.64 | 2.84e-06 | 6.10e-05 | 12/30 | MINOS1; ATP5G3; ATP5I; ATP5J2; ATP5L; ATP5G2; ATP5G1; ATP5H; ATP5A1; ATP5E; ATP5J; ATP5O |
| R-HSA-451326 | Activation of kainate receptors upon glutamate binding | 4.35 | 1.48e-03 | 2.46e-02 | 6/16 | GNG5; CALM2; GNG3; CALM1; CALM3; GNG8 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 4.28 | 0.00e+00 | 0.00e+00 | 59/160 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; TRMT112; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; NOP10; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3; SNU13 |
| R-HSA-72312 | rRNA processing | 4.25 | 0.00e+00 | 0.00e+00 | 60/164 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS18; RPL10A; RPS12; HSD17B10; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; TRMT112; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; NOP10; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3; SNU13 |
| R-HSA-5628897 | TP53 Regulates Metabolic Genes | 4.22 | 2.60e-10 | 5.90e-09 | 24/66 | PRDX1; LAMTOR5; LAMTOR2; COX20; COX5B; COX7C; NDUFA4; CYCS; LAMTOR4; COX7B; COX6C; YWHAZ; TXN; COX8A; PRDX5; COX14; COX6A1; COX16; COX5A; COX4I1; YWHAE; PRDX2; COX6B1; MT-CO1 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 4.22 | 0.00e+00 | 0.00e+00 | 56/154 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3; SNU13 |
| R-HSA-376176 | Signaling by ROBO receptors | 4.11 | 0.00e+00 | 0.00e+00 | 63/178 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SHFM1; RPS4X; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; PFN1; RPL26; RPL23A; PSMB3; RPL23; RPL19; PSMA7; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3; RBX1 |
| R-HSA-5339562 | Uptake and actions of bacterial toxins | 4.10 | 2.13e-03 | 3.46e-02 | 6/17 | CALM2; CALM1; HSP90AA1; VAMP2; EEF2; CALM3 |
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 3.87 | 2.97e-03 | 4.65e-02 | 6/18 | CALM2; TUBB2B; TUBA1A; CALM1; ACTG1; CALM3 |
| R-HSA-72766 | Translation | 3.61 | 0.00e+00 | 0.00e+00 | 80/257 | AURKAIP1; MRPL20; RPL11; RPS8; RPL5; MRPS21; RPS27; MRPL33; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; SPCS1; RPL22L1; EIF4A2; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; EEF1A1; RPS12; SEC61G; RPS4X; RPL39; SSR4; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; SEC61B; RPL35; RPL7A; MRPL41; EIF3F; RPL27A; RPS13; TRMT112; RPS3; CHCHD1; RPS24; MRPL51; RPS26; RPL41; RPL6; RPLP0; RPL21; MRPL52; RPS29; RPL36AL; SRP14; EIF3J; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; EEF2; RPL18A; EIF3K; RPS19; RPL18; RPL13A; RPS11; EIF3L; RPL3; MRPS6 |
| R-HSA-5663205 | Infectious disease | 3.50 | 0.00e+00 | 0.00e+00 | 83/275 | RPL11; RPS8; RPL5; RPS27; CALM2; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; SKP1; RPS14; RPS18; RPL10A; RPS12; GTF2H5; PPIA; SHFM1; POLR2J; SLC25A6; RPS4X; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; POLR2K; RPS6; RPL35; RPL7A; POLR2L; RPL27A; RPS13; BANF1; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; UBC; RAN; RPL21; RPS29; RPL36AL; CALM1; HSP90AA1; GTF2A2; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; VAMP2; RPL26; RPL23A; PSMB3; RPL23; RPL19; PSMA7; EEF2; RPL18A; POLR2I; RPS19; CALM3; AP2S1; RPL18; RPL13A; RPS11; CHMP2A; RANBP1; POLR2F; RPL3; RBX1 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 3.39 | 0.00e+00 | 0.00e+00 | 61/209 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SHFM1; HSD17B10; RPS4X; RPL39; RPL10; RPS20; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPS17; RPS2; RPS15A; NDUFAB1; RPL13; RPL26; RPL23A; PSMB3; RPL23; RPL19; PSMA7; OAZ1; RPL18A; RPS19; RPL18; RPL13A; RPS11; RPL3 |
| R-HSA-6782210 | Gap-filling DNA repair synthesis and ligation in TC-NER | 3.27 | 3.02e-04 | 5.54e-03 | 11/39 | RPS27A; POLE4; GTF2H5; RPA3; POLR2J; POLR2K; POLR2L; UBC; POLR2I; POLR2F; RBX1 |
| R-HSA-1592230 | Mitochondrial biogenesis | 3.23 | 8.25e-06 | 1.69e-04 | 17/61 | MINOS1; CALM2; ATP5G3; ATP5I; CYCS; ATP5J2; SSBP1; ATP5L; ATP5G2; CALM1; ATP5G1; ATP5H; ATP5A1; ATP5E; CALM3; ATP5J; ATP5O |
| R-HSA-72165 | mRNA Splicing - Minor Pathway | 3.19 | 6.35e-05 | 1.24e-03 | 14/51 | SNRPE; SF3B6; SNRPG; SF3B5; POLR2J; POLR2K; POLR2L; SF3B2; SNRPF; SNRPD1; POLR2I; SNRPD2; POLR2F; SNU13 |
| R-HSA-6782135 | Dual incision in TC-NER | 3.11 | 4.87e-04 | 8.56e-03 | 11/41 | RPS27A; POLE4; GTF2H5; RPA3; POLR2J; POLR2K; POLR2L; UBC; POLR2I; POLR2F; RBX1 |
| R-HSA-422475 | Axon guidance | 2.65 | 0.00e+00 | 0.00e+00 | 75/328 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; GAP43; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; TUBB2B; RPS18; RPL10A; CD24; RPS12; SHFM1; RPS4X; RPL39; RPL10; DPYSL2; RPS20; RPL7; TCEB1; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; CFL1; RPS3; RPS24; TUBA1A; RPS26; RPL41; MYL6; RPL6; RPLP0; UBC; RPL21; RPS29; RPL36AL; HSP90AA1; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; PFN1; RPL26; RPL23A; PSMB3; RPL23; RPL19; ACTG1; MYL12B; PSMA7; RPL18A; PSENEN; RPS19; AP2S1; RPL18; RPL13A; RPS11; RPL3; RBX1 |
| R-HSA-6781827 | Transcription-Coupled Nucleotide Excision Repair (TC-NER) | 2.55 | 2.85e-03 | 4.54e-02 | 11/50 | RPS27A; POLE4; GTF2H5; RPA3; POLR2J; POLR2K; POLR2L; UBC; POLR2I; POLR2F; RBX1 |
| R-HSA-1643685 | Disease | 2.25 | 1.55e-15 | 3.72e-14 | 90/464 | RPL11; RPS8; PRDX1; RPL5; RPS27; CALM2; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; RPL22L1; RPL9; RPL34; RPS3A; RPL37; RPS23; SKP1; RPS14; RPS18; RPL10A; RPS12; GTF2H5; PPIA; SHFM1; POLR2J; SLC25A6; RPS4X; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; POLR2K; RPS6; RPL35; RPL7A; POLR2L; RPL27A; RPS13; BANF1; RPS3; RPS24; RPS26; RPL41; RPL6; PEBP1; RPLP0; UBC; RAN; RPL21; RPS29; RPL36AL; CALM1; HSP90AA1; GTF2A2; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; YWHAE; VAMP2; RPL26; RPL23A; PSMB3; RPL23; RPL19; PHB; ACTG1; PSMA7; EEF2; PRDX2; RPL18A; PSENEN; POLR2I; RPS19; CALM3; AP2S1; RPL18; RPL13A; RPS11; CHMP2A; RANBP1; POLR2F; RPL3; RBX1 |
| R-HSA-1266738 | Developmental Biology | 2.14 | 9.52e-13 | 2.22e-11 | 82/445 | RPL11; RPS8; RPL5; RPS27; H3F3A; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL29; GAP43; RPL22L1; RPL9; H2AFZ; RPL34; RPS3A; RPL37; RPS23; RPS14; TUBB2B; RPS18; RPL10A; CD24; RPS12; SHFM1; POLR2J; RPS4X; RPL39; RPL10; DPYSL2; RPS20; RPL7; TCEB1; RPL30; POLR2K; RPS6; RPL35; RPL7A; POLR2L; RPL27A; RPS13; CFL1; RPS3; RPS24; TUBA1A; RPS26; RPL41; MYL6; RPL6; RPLP0; UBC; RPL21; RPS29; RPL36AL; HSP90AA1; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; PFN1; RPL26; RPL23A; PSMB3; RPL23; RPL19; ACTG1; MYL12B; PSMA7; RPL18A; PSENEN; POLR2I; RPS19; AP2S1; RPL18; RPL13A; RPS11; POLR2F; RPL3; RBX1 |
| R-HSA-1852241 | Organelle biogenesis and maintenance | 2.01 | 1.52e-04 | 2.84e-03 | 29/167 | MINOS1; HSPB11; CALM2; DYNC1I2; ATP5G3; TCTEX1D2; ATP5I; TUBB2B; TUBB; CYCS; ATP5J2; SSBP1; DCTN3; ATP5L; TUBA1A; ATP5G2; DCTN2; DYNLL1; CALM1; HSP90AA1; YWHAE; ATP5G1; ATP5H; ATP5A1; DYNLRB1; ATP5E; CALM3; ATP5J; ATP5O |
Factor 8 : 37 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-445355 | Smooth Muscle Contraction | 9.34 | 3.79e-06 | 1.26e-04 | 7/17 | CALM2; ANXA6; TPM2; MYL6B; MYL6; CALM1 |
| R-HSA-2514856 | The phototransduction cascade | 9.07 | 6.20e-04 | 1.54e-02 | 4/10 | CALM2; METAP2; CALM1 |
| R-HSA-2514859 | Inactivation, recovery and regulation of the phototransduction cascade | 9.07 | 6.20e-04 | 1.54e-02 | 4/10 | CALM2; METAP2; CALM1 |
| R-HSA-5218921 | VEGFR2 mediated cell proliferation | 9.07 | 6.20e-04 | 1.54e-02 | 4/10 | CALM2; KRAS; CALM1 |
| R-HSA-5576892 | Phase 0 - rapid depolarisation | 7.56 | 1.36e-03 | 3.26e-02 | 4/12 | CALM2; CACNB3; CALM1 |
| R-HSA-451326 | Activation of kainate receptors upon glutamate binding | 7.09 | 4.59e-04 | 1.28e-02 | 5/16 | GNG4; CALM2; CALM1; GNG8 |
| R-HSA-5339562 | Uptake and actions of bacterial toxins | 6.67 | 6.28e-04 | 1.54e-02 | 5/17 | CALM2; SYT1; CALM1; VAMP2 |
| R-HSA-192823 | Viral mRNA Translation | 6.01 | 2.38e-12 | 9.01e-10 | 22/83 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-156902 | Peptide chain elongation | 5.94 | 3.11e-12 | 9.01e-10 | 22/84 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-2408557 | Selenocysteine synthesis | 5.87 | 4.04e-12 | 9.01e-10 | 22/85 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 5.80 | 5.23e-12 | 9.01e-10 | 22/86 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-72764 | Eukaryotic Translation Termination | 5.80 | 5.23e-12 | 9.01e-10 | 22/86 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 5.67 | 8.66e-12 | 1.24e-09 | 22/88 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-2408522 | Selenoamino acid metabolism | 5.27 | 1.45e-11 | 1.79e-09 | 23/99 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; MARS; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 5.25 | 4.51e-11 | 4.59e-09 | 22/95 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 4.97 | 5.33e-11 | 4.59e-09 | 23/105 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; MAGOHB; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 4.97 | 5.33e-11 | 4.59e-09 | 23/105 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; MAGOHB; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 4.80 | 3.01e-10 | 2.36e-08 | 22/104 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 4.75 | 3.67e-10 | 2.63e-08 | 22/105 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 4.71 | 4.46e-10 | 2.96e-08 | 22/106 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-418346 | Platelet homeostasis | 4.54 | 1.64e-03 | 3.72e-02 | 6/30 | GNG4; CALM2; ATP2B1; CALM1; GNG8 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 4.42 | 1.63e-09 | 7.82e-08 | 22/113 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 4.42 | 1.63e-09 | 7.82e-08 | 22/113 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 4.42 | 1.63e-09 | 7.82e-08 | 22/113 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-422356 | Regulation of insulin secretion | 4.39 | 1.96e-03 | 4.33e-02 | 6/31 | GNG4; MARCKS; SLC25A6; CACNB3; VAMP2; GNG8 |
| R-HSA-168255 | Influenza Life Cycle | 4.31 | 1.10e-09 | 6.32e-08 | 23/121 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RAN; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-168254 | Influenza Infection | 4.25 | 6.16e-10 | 3.79e-08 | 24/128 | RPL32; RPL24; RPL34; RPS18; SLC25A6; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RAN; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-397014 | Muscle contraction | 4.12 | 1.09e-04 | 3.14e-03 | 10/55 | CALM2; ANXA6; TPM2; KCNIP2; CACNB3; MYL6B; MYL6; ATP2B1; CALM1 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 3.65 | 1.51e-08 | 6.85e-07 | 24/149 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; MAGOHB; RPS26; RPL41; RPL6; RPLP0; MSI1; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-376176 | Signaling by ROBO receptors | 3.31 | 2.94e-08 | 1.26e-06 | 26/178 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; MAGOHB; RPS26; RPL41; SRGAP1; RPL6; RPLP0; MSI1; RPS17; RPL13; RPL26; RPL23A; RPL23; DCC; RPL36; RPL28 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 3.24 | 5.78e-07 | 2.37e-05 | 22/154 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 3.12 | 1.14e-06 | 4.07e-05 | 22/160 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-72312 | rRNA processing | 3.04 | 1.74e-06 | 6.01e-05 | 22/164 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 2.82 | 8.01e-07 | 3.13e-05 | 26/209 | RPL32; RPL24; RPL34; CDO1; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; BCAT1; RPS26; RPL41; MARS; RPL6; RPLP0; CKB; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-5663205 | Infectious disease | 2.39 | 5.53e-06 | 1.76e-04 | 29/275 | CALM2; RPL32; RPL24; RPL34; RPS18; SLC25A6; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; RPS26; RPL41; SYT1; RPL6; RPLP0; RAN; CALM1; RPS17; RPL13; VAMP2; RPL26; RPL23A; RPL23; RPL36; RPL28 |
| R-HSA-422475 | Axon guidance | 2.35 | 1.07e-06 | 4.01e-05 | 34/328 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; PTK2; RPL8; RPL12; RPL27A; RPS13; RPS3; MAGOHB; KRAS; COL2A1; CACNB3; TUBA1B; RPS26; RPL41; MYL6; SRGAP1; ARPC3; RPL6; RPLP0; MSI1; RPS17; RPL13; RPL26; RPL23A; RPL23; DCC; RPL36; NCAN; RPL28 |
| R-HSA-72766 | Translation | 2.30 | 3.84e-05 | 1.18e-03 | 26/257 | RPL32; RPL24; RPL34; RPS18; RPL36A; RPL10; RPL8; RPL12; RPL27A; RPS13; RPS3; MRPL51; RPS26; RPL41; MARS; TSFM; MRPL42; RPL6; RPLP0; RPS17; RPL13; RPL26; RPL23A; RPL23; RPL36; RPL28 |
Factor 9 : 36 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-390522 | Striated Muscle Contraction | 9.19 | 5.90e-04 | 0.02539 | 4/10 | TPM2; ACTC1; TNNC2; TPM4 |
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 8.94 | 5.48e-06 | 0.00157 | 7/18 | TUBA4A; CTNNB1; ACTB; TUBA1B; CALM1; ACTG1; CALM3 |
| R-HSA-373076 | Class A/1 (Rhodopsin-like receptors) | 8.36 | 8.96e-04 | 0.03085 | 4/11 | SST; GAL; NMB; SSTR2 |
| R-HSA-2871809 | FCERI mediated Ca+2 mobilization | 8.36 | 8.96e-04 | 0.03085 | 4/11 | SHC1; VAV2; CALM1; CALM3 |
| R-HSA-445355 | Smooth Muscle Contraction | 8.11 | 5.12e-05 | 0.00722 | 6/17 | TPM2; VCL; MYL6; CALM1; TPM4; CALM3 |
| R-HSA-418990 | Adherens junctions interactions | 7.66 | 1.30e-03 | 0.03726 | 4/12 | CTNNB1; ACTB; CADM1; ACTG1 |
| R-HSA-5576892 | Phase 0 - rapid depolarisation | 7.66 | 1.30e-03 | 0.03726 | 4/12 | CAMK2D; CACNA2D1; CALM1; CALM3 |
| R-HSA-375170 | CDO in myogenesis | 7.07 | 1.81e-03 | 0.04459 | 4/13 | CTNNB1; MEF2C; TCF12; TCF4 |
| R-HSA-525793 | Myogenesis | 7.07 | 1.81e-03 | 0.04459 | 4/13 | CTNNB1; MEF2C; TCF12; TCF4 |
| R-HSA-190828 | Gap junction trafficking | 6.38 | 7.91e-04 | 0.02959 | 5/18 | TUBA4A; ACTB; DNM1; TUBA1B; ACTG1 |
| R-HSA-6794361 | Neurexins and neuroligins | 6.27 | 2.57e-04 | 0.01847 | 6/22 | EPB41; NRXN1; SYT7; SYT1; EPB41L3; HOMER3 |
| R-HSA-157858 | Gap junction trafficking and regulation | 6.05 | 1.04e-03 | 0.03245 | 5/19 | TUBA4A; ACTB; DNM1; TUBA1B; ACTG1 |
| R-HSA-5578775 | Ion homeostasis | 5.99 | 3.36e-04 | 0.02066 | 6/23 | CAMK2D; FXYD6; ATP2B1; CALM1; FXYD1; CALM3 |
| R-HSA-500792 | GPCR ligand binding | 5.96 | 1.10e-04 | 0.00945 | 7/27 | GNG5; SST; GAL; NMB; SSTR2; GNG8; PTH2 |
| R-HSA-936837 | Ion transport by P-type ATPases | 5.75 | 4.32e-04 | 0.02188 | 6/24 | CAMK2D; FXYD6; ATP2B1; CALM1; FXYD1; CALM3 |
| R-HSA-4086398 | Ca2+ pathway | 5.75 | 4.32e-04 | 0.02188 | 6/24 | GNG5; CTNNB1; PRKG2; CALM1; CALM3; GNG8 |
| R-HSA-446728 | Cell junction organization | 5.52 | 5.48e-04 | 0.02539 | 6/25 | CTNNB1; ACTB; ILK; CADM1; RSU1; ACTG1 |
| R-HSA-5218920 | VEGFR2 mediated vascular permeability | 5.47 | 1.69e-03 | 0.04459 | 5/21 | CTNNB1; VAV2; CALM1; CALM3; PRR5 |
| R-HSA-397014 | Muscle contraction | 5.43 | 3.60e-07 | 0.00031 | 13/55 | CAMK2D; CACNA2D1; TPM2; FXYD6; VCL; MYL6; ATP2B1; CALM1; ACTC1; TNNC2; TPM4; FXYD1; CALM3 |
| R-HSA-8986944 | Transcriptional Regulation by MECP2 | 5.19 | 2.81e-04 | 0.01858 | 7/31 | SST; CAMK2D; GRIA2; MEF2C; HDAC2; CALM1; CALM3 |
| R-HSA-114608 | Platelet degranulation | 4.81 | 7.00e-05 | 0.00722 | 9/43 | TUBA4A; WDR1; SPARC; TMSB4X; CFL1; VCL; CALM1; PFN1; CALM3 |
| R-HSA-76005 | Response to elevated platelet cytosolic Ca2+ | 4.81 | 7.00e-05 | 0.00722 | 9/43 | TUBA4A; WDR1; SPARC; TMSB4X; CFL1; VCL; CALM1; PFN1; CALM3 |
| R-HSA-5576891 | Cardiac conduction | 4.60 | 6.19e-04 | 0.02540 | 7/35 | CAMK2D; CACNA2D1; FXYD6; ATP2B1; CALM1; FXYD1; CALM3 |
| R-HSA-418346 | Platelet homeostasis | 4.60 | 1.53e-03 | 0.04262 | 6/30 | GNG5; PRKG2; ATP2B1; CALM1; CALM3; GNG8 |
| R-HSA-1500931 | Cell-Cell communication | 4.23 | 1.04e-03 | 0.03245 | 7/38 | CTNNB1; CD47; ACTB; ILK; CADM1; RSU1; ACTG1 |
| R-HSA-112314 | Neurotransmitter receptors and postsynaptic signal transmission | 3.75 | 1.06e-03 | 0.03245 | 8/49 | GNG5; CHRNA1; CAMK2D; GRIA2; TSPAN7; CALM1; CALM3; GNG8 |
| R-HSA-373760 | L1CAM interactions | 3.72 | 1.33e-04 | 0.01038 | 11/68 | CNTN2; TUBA4A; ALCAM; ANK2; ACTB; DNM1; VAV2; NCAM1; ANK3; TUBA1B; ACTG1 |
| R-HSA-76002 | Platelet activation, signaling and aggregation | 3.43 | 7.55e-05 | 0.00722 | 13/87 | GNG5; SHC1; TUBA4A; WDR1; SPARC; TMSB4X; VAV2; CFL1; VCL; CALM1; PFN1; CALM3; GNG8 |
| R-HSA-194138 | Signaling by VEGF | 3.40 | 2.02e-03 | 0.04710 | 8/54 | CTNNB1; ACTB; VAV2; PGF; CALM1; ACTG1; CALM3; PRR5 |
| R-HSA-112316 | Neuronal System | 3.22 | 4.49e-05 | 0.00722 | 15/107 | EPB41; GNG5; NRXN1; CHRNA1; CAMK2D; GRIA2; CACNA2D1; TSPAN7; SYT7; SYT1; CALM1; EPB41L3; HOMER3; CALM3; GNG8 |
| R-HSA-418594 | G alpha (i) signalling events | 3.18 | 1.75e-03 | 0.04459 | 9/65 | GNG5; RGS16; RBP1; SST; GAL; CALM1; SSTR2; CALM3; GNG8 |
| R-HSA-388396 | GPCR downstream signalling | 2.77 | 4.14e-04 | 0.02188 | 14/116 | GNG5; RHOC; RGS16; RBP1; SST; VAV2; GAL; CALM1; NMB; SSTR2; PREX1; CALM3; GNG8; PTH2 |
| R-HSA-109582 | Hemostasis | 2.71 | 1.96e-05 | 0.00422 | 21/178 | GNG5; SHC1; GYPC; TUBA4A; CD47; WDR1; PRKG2; SPARC; HDAC2; ACTB; TMSB4X; TSPAN7; VAV2; CFL1; VCL; TUBA1B; ATP2B1; CALM1; PFN1; CALM3; GNG8 |
| R-HSA-372790 | Signaling by GPCR | 2.68 | 5.88e-04 | 0.02539 | 14/120 | GNG5; RHOC; RGS16; RBP1; SST; VAV2; GAL; CALM1; NMB; SSTR2; PREX1; CALM3; GNG8; PTH2 |
| R-HSA-194315 | Signaling by Rho GTPases | 2.16 | 1.89e-03 | 0.04524 | 17/181 | RHOC; ARPC5; TUBA4A; SRGAP3; CTNNB1; FAM13A; ACTB; VAV2; CFL1; TUBA1B; MYL6; DYNLL1; CALM1; PFN1; ACTG1; PREX1; CALM3 |
| R-HSA-1266738 | Developmental Biology | 2.07 | 2.93e-06 | 0.00126 | 40/445 | JUN; RHOC; SHC1; PBX1; ARPC5; LHX9; CNTN2; NCOA1; PKP4; NEUROD1; RPL37A; TUBA4A; SRGAP3; CTNNB1; ROBO1; ALCAM; ANK2; MEF2C; RGMB; DPYSL3; EBF1; PDLIM7; CAP2; ACTB; DNM1; VAV2; CFL1; RPS3; NCAM1; ANK3; TUBA1B; MYL6; TCF12; PFN1; ADAM11; ACTG1; DCC; TCF4; INSM1; LGI4 |
Factor 11 : 62 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-381033 | ATF6 (ATF6-alpha) activates chaperones | 17.70 | 5.11e-04 | 1.18e-02 | 3/10 | DDIT3; XBP1; ATF4 |
| R-HSA-427359 | SIRT1 negatively regulates rRNA expression | 14.70 | 9.14e-04 | 1.87e-02 | 3/12 | H2AFZ; HIST1H4C; HIST1H2AC |
| R-HSA-5334118 | DNA methylation | 14.70 | 9.14e-04 | 1.87e-02 | 3/12 | H2AFZ; HIST1H4C; HIST1H2AC |
| R-HSA-73728 | RNA Polymerase I Promoter Opening | 14.70 | 9.14e-04 | 1.87e-02 | 3/12 | H2AFZ; HIST1H4C; HIST1H2AC |
| R-HSA-171306 | Packaging Of Telomere Ends | 13.60 | 1.17e-03 | 2.20e-02 | 3/13 | H2AFZ; HIST1H4C; HIST1H2AC |
| R-HSA-5625886 | Activated PKN1 stimulates transcription of AR (androgen receptor) regulated genes KLK2 and KLK3 | 13.60 | 1.17e-03 | 2.20e-02 | 3/13 | H2AFZ; HIST1H4C; HIST1H2AC |
| R-HSA-389977 | Post-chaperonin tubulin folding pathway | 13.60 | 1.17e-03 | 2.20e-02 | 3/13 | TUBB2A; TUBB2B; TUBA1A |
| R-HSA-190828 | Gap junction trafficking | 13.10 | 1.91e-04 | 4.85e-03 | 4/18 | TUBB2A; TUBB2B; ACTB; TUBA1A |
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 13.10 | 1.91e-04 | 4.85e-03 | 4/18 | TUBB2A; TUBB2B; ACTB; TUBA1A |
| R-HSA-8955332 | Carboxyterminal post-translational modifications of tubulin | 12.60 | 1.48e-03 | 2.49e-02 | 3/14 | TUBB2A; TUBB2B; TUBA1A |
| R-HSA-157858 | Gap junction trafficking and regulation | 12.40 | 2.39e-04 | 5.89e-03 | 4/19 | TUBB2A; TUBB2B; ACTB; TUBA1A |
| R-HSA-2299718 | Condensation of Prophase Chromosomes | 11.10 | 2.22e-03 | 3.41e-02 | 3/16 | H2AFZ; HIST1H4C; HIST1H2AC |
| R-HSA-389960 | Formation of tubulin folding intermediates by CCT/TriC | 11.10 | 2.22e-03 | 3.41e-02 | 3/16 | TUBB2A; TUBB2B; TUBA1A |
| R-HSA-389957 | Prefoldin mediated transfer of substrate to CCT/TriC | 10.70 | 4.35e-04 | 1.04e-02 | 4/22 | TUBB2A; TUBB2B; ACTB; TUBA1A |
| R-HSA-912446 | Meiotic recombination | 10.40 | 2.66e-03 | 3.95e-02 | 3/17 | H2AFZ; HIST1H4C; HIST1H2AC |
| R-HSA-389958 | Cooperation of Prefoldin and TriC/CCT in actin and tubulin folding | 10.30 | 5.20e-04 | 1.18e-02 | 4/23 | TUBB2A; TUBB2B; ACTB; TUBA1A |
| R-HSA-2408557 | Selenocysteine synthesis | 9.71 | 5.08e-11 | 1.21e-08 | 14/85 | SARS; RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-192823 | Viral mRNA Translation | 9.24 | 5.29e-10 | 6.51e-08 | 13/83 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-156902 | Peptide chain elongation | 9.13 | 6.19e-10 | 6.66e-08 | 13/84 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 8.92 | 8.39e-10 | 7.22e-08 | 13/86 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-72764 | Eukaryotic Translation Termination | 8.92 | 8.39e-10 | 7.22e-08 | 13/86 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 8.71 | 1.13e-09 | 8.09e-08 | 13/88 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 8.60 | 1.21e-05 | 3.46e-04 | 7/48 | RPS10; RPS12; RPS24; RPS2; EIF2S2; RPS19; RPS11 |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 8.58 | 2.77e-06 | 8.51e-05 | 8/55 | RPS10; RPS12; RPS24; EIF5; RPS2; EIF2S2; RPS19; RPS11 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 8.43 | 3.19e-06 | 9.46e-05 | 8/56 | RPS10; RPS12; EIF4EBP1; RPS24; RPS2; EIF2S2; RPS19; RPS11 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 8.35 | 1.86e-11 | 8.01e-09 | 16/113 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; EIF4EBP1; RPL12; RPS24; RPL21; EIF5; RPS2; RPL27; EIF2S2; RPS19; RPS11 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 8.35 | 1.86e-11 | 8.01e-09 | 16/113 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; EIF4EBP1; RPL12; RPS24; RPL21; EIF5; RPS2; RPL27; EIF2S2; RPS19; RPS11 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 8.35 | 8.98e-11 | 1.55e-08 | 15/106 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; EIF5; RPS2; RPL27; EIF2S2; RPS19; RPS11 |
| R-HSA-2408522 | Selenoamino acid metabolism | 8.34 | 4.26e-10 | 6.12e-08 | 14/99 | SARS; RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-3214858 | RMTs methylate histone arginines | 8.14 | 1.29e-03 | 2.27e-02 | 4/29 | H2AFZ; HIST1H4C; HIST1H2AC; RPS2 |
| R-HSA-437239 | Recycling pathway of L1 | 8.14 | 1.29e-03 | 2.27e-02 | 4/29 | TUBB2A; TUBB2B; ACTB; TUBA1A |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 8.07 | 3.00e-09 | 1.99e-07 | 13/95 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-5250924 | B-WICH complex positively regulates rRNA expression | 7.86 | 1.47e-03 | 2.49e-02 | 4/30 | H2AFZ; HIST1H4C; HIST1H2AC; ACTB |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 7.86 | 9.55e-10 | 7.48e-08 | 14/105 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; EIF2S2; RPS19; RPS11 |
| R-HSA-72649 | Translation initiation complex formation | 7.51 | 3.03e-05 | 8.41e-04 | 7/55 | RPS10; RPS12; RPS24; RPS2; EIF2S2; RPS19; RPS11 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 7.37 | 9.39e-09 | 5.69e-07 | 13/104 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 7.30 | 1.06e-08 | 5.69e-07 | 13/105 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 7.30 | 1.06e-08 | 5.69e-07 | 13/105 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 6.79 | 2.63e-08 | 1.26e-06 | 13/113 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-3371497 | HSP90 chaperone cycle for steroid hormone receptors (SHR) | 6.38 | 3.25e-03 | 4.51e-02 | 4/37 | TUBB2A; TUBB2B; TUBA1A; DYNC1H1 |
| R-HSA-6811436 | COPI-independent Golgi-to-ER retrograde traffic | 6.38 | 3.25e-03 | 4.51e-02 | 4/37 | TUBB2A; TUBB2B; TUBA1A; DYNC1H1 |
| R-HSA-168255 | Influenza Life Cycle | 6.34 | 6.07e-08 | 2.75e-06 | 13/121 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-168254 | Influenza Infection | 5.99 | 1.20e-07 | 4.91e-06 | 13/128 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 5.94 | 1.19e-08 | 6.03e-07 | 15/149 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; PSMB7; RPL12; RPS24; RPL21; PSMB5; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 5.64 | 5.61e-11 | 1.21e-08 | 20/209 | SARS; PHGDH; RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; PSAT1; PSMB7; RPL12; SLC3A2; RPS24; RPL21; PSMB5; RPS2; RPL27; OAZ1; RPS19; RPS11 |
| R-HSA-380320 | Recruitment of NuMA to mitotic centrosomes | 5.36 | 2.15e-03 | 3.41e-02 | 5/55 | TUBB2A; TUBB2B; TUBB; TUBA1A; DYNC1H1 |
| R-HSA-8852276 | The role of GTSE1 in G2/M progression after G2 checkpoint | 5.36 | 2.15e-03 | 3.41e-02 | 5/55 | TUBB2A; TUBB2B; PSMB7; TUBA1A; PSMB5 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 4.98 | 1.06e-06 | 3.65e-05 | 13/154 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-376176 | Signaling by ROBO receptors | 4.97 | 1.35e-07 | 5.29e-06 | 15/178 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; PSMB7; RPL12; RPS24; RPL21; PSMB5; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-8939236 | RUNX1 regulates transcription of genes involved in differentiation of HSCs | 4.92 | 3.16e-03 | 4.51e-02 | 5/60 | H2AFZ; HIST1H4C; HIST1H2AC; PSMB7; PSMB5 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 4.79 | 1.65e-06 | 5.46e-05 | 13/160 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-72312 | rRNA processing | 4.68 | 2.18e-06 | 6.97e-05 | 13/164 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; RPL12; RPS24; RPL21; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-72766 | Translation | 4.13 | 1.01e-07 | 4.36e-06 | 18/257 | SARS; RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; EIF4EBP1; RPL12; CARS; RPS24; RPL21; EIF5; RPS2; RPL27; EIF2S2; RPS19; RPS11 |
| R-HSA-69275 | G2/M Transition | 3.87 | 9.01e-04 | 1.87e-02 | 8/122 | TUBB2A; TUBB2B; TUBB; PSMB7; TUBA1A; PHLDA1; PSMB5; DYNC1H1 |
| R-HSA-453274 | Mitotic G2-G2/M phases | 3.84 | 9.51e-04 | 1.90e-02 | 8/123 | TUBB2A; TUBB2B; TUBB; PSMB7; TUBA1A; PHLDA1; PSMB5; DYNC1H1 |
| R-HSA-422475 | Axon guidance | 3.42 | 8.37e-07 | 3.00e-05 | 19/328 | RPL37A; RPL35A; RPL37; TUBB2A; TUBB2B; RPS10; RPS12; ACTB; RPL39; PSMB7; RPL12; RPS24; TUBA1A; RPL21; PSMB5; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-195258 | RHO GTPase Effectors | 3.28 | 2.63e-03 | 3.95e-02 | 8/144 | H2AFZ; TUBB2A; TUBB2B; HIST1H4C; HIST1H2AC; ACTB; TUBA1A; DYNC1H1 |
| R-HSA-5663205 | Infectious disease | 3.22 | 3.37e-05 | 9.08e-04 | 15/275 | RPL37A; RPL35A; RPL37; RPS10; RPS12; RPL39; PSMB7; RPL12; RPS24; RPL21; PSMB5; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-1266738 | Developmental Biology | 3.05 | 2.97e-07 | 1.11e-05 | 23/445 | NEUROD1; RPL37A; RPL35A; H2AFZ; RPL37; TUBB2A; TUBB2B; HIST1H4C; HIST1H2AC; RPS10; RPS12; ACTB; RPL39; PSMB7; RPL12; RPS24; TUBA1A; RPL21; PSMB5; RPS2; RPL27; RPS19; RPS11 |
| R-HSA-68886 | M Phase | 2.89 | 1.93e-03 | 3.20e-02 | 10/204 | H2AFZ; TUBB2A; TUBB2B; HIST1H4C; HIST1H2AC; TUBB; PSMB7; TUBA1A; PSMB5; DYNC1H1 |
| R-HSA-69278 | Cell Cycle, Mitotic | 2.56 | 2.96e-03 | 4.31e-02 | 11/253 | H2AFZ; TUBB2A; TUBB2B; HIST1H4C; HIST1H2AC; TUBB; PSMB7; TUBA1A; PHLDA1; PSMB5; DYNC1H1 |
| R-HSA-1643685 | Disease | 2.16 | 1.27e-03 | 2.27e-02 | 17/464 | PRDX1; RPL37A; RPL35A; RPL37; RPS10; RPS12; ACTB; RPL39; PSMB7; RPL12; RPS24; RPL21; PSMB5; RPS2; RPL27; RPS19; RPS11 |
Factor 13 : 1 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 42.1 | 3.89e-05 | 0.0335 | 3/18 | CALM2; ACTB; ACTG1 |
Factor 15 : 1 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-397014 | Muscle contraction | 7.26 | 3.77e-05 | 0.0324 | 7/55 | ATP2B4; CALM2; SCN9A; SRI; MYL6; MYL12A; FXYD7 |
Factor 16 : 38 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-442729 | CREB phosphorylation through the activation of CaMKII | 14.60 | 1.03e-04 | 0.0119 | 4/11 | CALM2; NEFL; CALM1 |
| R-HSA-442982 | Ras activation upon Ca2+ influx through NMDA receptor | 14.60 | 1.03e-04 | 0.0119 | 4/11 | CALM2; NEFL; CALM1 |
| R-HSA-8876725 | Protein methylation | 13.40 | 1.52e-04 | 0.0119 | 4/12 | CALM2; CALM1; RPS2 |
| R-HSA-5576892 | Phase 0 - rapid depolarisation | 13.40 | 1.52e-04 | 0.0119 | 4/12 | CALM2; SCN9A; CALM1 |
| R-HSA-2514856 | The phototransduction cascade | 12.10 | 1.57e-03 | 0.0423 | 3/10 | CALM2; CALM1 |
| R-HSA-2514859 | Inactivation, recovery and regulation of the phototransduction cascade | 12.10 | 1.57e-03 | 0.0423 | 3/10 | CALM2; CALM1 |
| R-HSA-5218921 | VEGFR2 mediated cell proliferation | 12.10 | 1.57e-03 | 0.0423 | 3/10 | CALM2; CALM1 |
| R-HSA-983695 | Antigen activates B Cell Receptor (BCR) leading to generation of second messengers | 12.10 | 1.57e-03 | 0.0423 | 3/10 | CALM2; CALM1 |
| R-HSA-70221 | Glycogen breakdown (glycogenolysis) | 12.10 | 1.57e-03 | 0.0423 | 3/10 | CALM2; CALM1 |
| R-HSA-2871809 | FCERI mediated Ca+2 mobilization | 11.00 | 2.12e-03 | 0.0499 | 3/11 | CALM2; CALM1 |
| R-HSA-111933 | Calmodulin induced events | 11.00 | 2.12e-03 | 0.0499 | 3/11 | CALM2; CALM1 |
| R-HSA-111997 | CaM pathway | 11.00 | 2.12e-03 | 0.0499 | 3/11 | CALM2; CALM1 |
| R-HSA-451326 | Activation of kainate receptors upon glutamate binding | 10.10 | 5.17e-04 | 0.0262 | 4/16 | CALM2; GNG3; CALM1 |
| R-HSA-442742 | CREB phosphorylation through the activation of Ras | 10.10 | 5.17e-04 | 0.0262 | 4/16 | CALM2; NEFL; CALM1 |
| R-HSA-5339562 | Uptake and actions of bacterial toxins | 9.46 | 6.63e-04 | 0.0301 | 4/17 | CALM2; SYT1; CALM1 |
| R-HSA-445355 | Smooth Muscle Contraction | 9.46 | 6.63e-04 | 0.0301 | 4/17 | CALM2; CALD1; CALM1 |
| R-HSA-70263 | Gluconeogenesis | 8.47 | 1.04e-03 | 0.0389 | 4/19 | GAPDH; TPI1; ALDOA; ENO3 |
| R-HSA-425393 | Transport of inorganic cations/anions and amino acids/oligopeptides | 8.47 | 1.04e-03 | 0.0389 | 4/19 | CALM2; SLC17A6; CALM1 |
| R-HSA-2672351 | Stimuli-sensing channels | 8.38 | 2.48e-04 | 0.0145 | 5/24 | CALM2; ASIC4; ASIC1; CALM1 |
| R-HSA-418346 | Platelet homeostasis | 8.04 | 7.32e-05 | 0.0119 | 6/30 | CALM2; GNG3; ATP2B1; CALM1; PDE9A |
| R-HSA-438064 | Post NMDA receptor activation events | 8.04 | 1.28e-03 | 0.0422 | 4/20 | CALM2; NEFL; CALM1 |
| R-HSA-442755 | Activation of NMDA receptors and postsynaptic events | 7.66 | 1.55e-03 | 0.0423 | 4/21 | CALM2; NEFL; CALM1 |
| R-HSA-5578775 | Ion homeostasis | 6.99 | 2.20e-03 | 0.0499 | 4/23 | CALM2; ATP2B1; CALM1 |
| R-HSA-5576891 | Cardiac conduction | 6.89 | 1.81e-04 | 0.0120 | 6/35 | CALM2; SCN9A; KCNIP2; ATP2B1; CALM1 |
| R-HSA-2151201 | Transcriptional activation of mitochondrial biogenesis | 6.70 | 7.40e-04 | 0.0319 | 5/30 | CALM2; MEF2C; ATP5B; CALM1 |
| R-HSA-114608 | Platelet degranulation | 6.55 | 7.20e-05 | 0.0119 | 7/43 | TAGLN2; CALM2; TMSB4X; SERPING1; CALM1; ALDOA |
| R-HSA-76005 | Response to elevated platelet cytosolic Ca2+ | 6.55 | 7.20e-05 | 0.0119 | 7/43 | TAGLN2; CALM2; TMSB4X; SERPING1; CALM1; ALDOA |
| R-HSA-8986944 | Transcriptional Regulation by MECP2 | 6.49 | 8.65e-04 | 0.0355 | 5/31 | CALM2; SST; MEF2C; CALM1 |
| R-HSA-425407 | SLC-mediated transmembrane transport | 6.09 | 1.16e-03 | 0.0406 | 5/33 | CALM2; SLC17A6; CALM1; SLCO3A1 |
| R-HSA-397014 | Muscle contraction | 5.85 | 4.97e-05 | 0.0119 | 8/55 | CALM2; SCN9A; MYL1; CALD1; KCNIP2; ATP2B1; CALM1 |
| R-HSA-418594 | G alpha (i) signalling events | 4.95 | 1.68e-04 | 0.0120 | 8/65 | RGS16; CALM2; SST; GNG3; GAL; RGS10; CALM1 |
| R-HSA-112314 | Neurotransmitter receptors and postsynaptic signal transmission | 4.92 | 1.18e-03 | 0.0406 | 6/49 | CALM2; CHRNA1; NEFL; GNG3; CALM1 |
| R-HSA-76002 | Platelet activation, signaling and aggregation | 4.16 | 2.52e-04 | 0.0145 | 9/87 | TAGLN2; CALM2; ABHD6; TMSB4X; SERPING1; GNG3; CALM1; ALDOA |
| R-HSA-388396 | GPCR downstream signalling | 3.81 | 1.11e-04 | 0.0119 | 11/116 | RHOC; RGS16; CALM2; ABHD6; SST; GNG3; GAL; RGS10; CALM1; PTH2 |
| R-HSA-372790 | Signaling by GPCR | 3.69 | 1.51e-04 | 0.0119 | 11/120 | RHOC; RGS16; CALM2; ABHD6; SST; GNG3; GAL; RGS10; CALM1; PTH2 |
| R-HSA-611105 | Respiratory electron transport | 3.39 | 2.20e-03 | 0.0499 | 8/95 | COX7B; NDUFB9; COX8A; COX6A1; COX5A; NDUFB10; UQCR11; MT-CO3 |
| R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 3.17 | 1.84e-03 | 0.0479 | 9/114 | COX7B; NDUFB9; COX8A; ATP5B; COX6A1; COX5A; NDUFB10; UQCR11; MT-CO3 |
| R-HSA-109582 | Hemostasis | 3.16 | 9.66e-05 | 0.0119 | 14/178 | TAGLN2; CALM2; GYPC; ABHD6; CD47; TMSB4X; SERPING1; GNG3; ATP2B1; CALM1; ALDOA; H3F3B; PDE9A |
Factor 17 : 7 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 15.90 | 1.01e-05 | 0.00868 | 5/18 | CALM2; ACTB; TUBA1B; TUBA1A; ACTG1 |
| R-HSA-190828 | Gap junction trafficking | 12.70 | 2.18e-04 | 0.04131 | 4/18 | ACTB; TUBA1B; TUBA1A; ACTG1 |
| R-HSA-157858 | Gap junction trafficking and regulation | 12.00 | 2.72e-04 | 0.04131 | 4/19 | ACTB; TUBA1B; TUBA1A; ACTG1 |
| R-HSA-1445148 | Translocation of SLC2A4 (GLUT4) to the plasma membrane | 7.61 | 1.08e-04 | 0.04131 | 6/45 | CALM2; ACTB; TUBA1B; TUBA1A; YWHAE; ACTG1 |
| R-HSA-2565942 | Regulation of PLK1 Activity at G2/M Transition | 6.23 | 3.36e-04 | 0.04131 | 6/55 | SKP1; TUBB; DCTN3; TUBA1A; UBC; YWHAE |
| R-HSA-109582 | Hemostasis | 3.53 | 2.05e-04 | 0.04131 | 11/178 | CAP1; GNG5; H3F3A; CALM2; HDAC2; ACTB; TMSB4X; MAGED2; TUBA1B; TUBA1A; CD63 |
| R-HSA-69278 | Cell Cycle, Mitotic | 2.93 | 3.26e-04 | 0.04131 | 13/253 | CKS1B; CENPF; H3F3A; SMC4; SKP1; PTTG1; HIST1H4C; TUBB; DCTN3; TUBA1B; TUBA1A; UBC; YWHAE |
Factor 18 : 33 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-156902 | Peptide chain elongation | 33.60 | 0 | 0 | 79/84 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 33.30 | 0 | 0 | 80/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; EEF1B2; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-192823 | Viral mRNA Translation | 33.20 | 0 | 0 | 77/83 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-2408557 | Selenocysteine synthesis | 32.40 | 0 | 0 | 77/85 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72764 | Eukaryotic Translation Termination | 32.00 | 0 | 0 | 77/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 31.30 | 0 | 0 | 77/88 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 29.00 | 0 | 0 | 77/95 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-2408522 | Selenoamino acid metabolism | 27.80 | 0 | 0 | 77/99 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 26.50 | 0 | 0 | 77/104 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 26.20 | 0 | 0 | 77/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 26.20 | 0 | 0 | 77/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 26.20 | 0 | 0 | 77/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 26.00 | 0 | 0 | 77/106 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 24.40 | 0 | 0 | 77/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 24.40 | 0 | 0 | 77/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 24.40 | 0 | 0 | 77/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-168255 | Influenza Life Cycle | 22.70 | 0 | 0 | 77/121 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 22.30 | 0 | 0 | 30/48 | RPS8; RPS27; RPS7; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-168254 | Influenza Infection | 21.80 | 0 | 0 | 78/128 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72649 | Translation initiation complex formation | 19.50 | 0 | 0 | 30/55 | RPS8; RPS27; RPS7; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 19.50 | 0 | 0 | 30/55 | RPS8; RPS27; RPS7; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 19.20 | 0 | 0 | 30/56 | RPS8; RPS27; RPS7; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; RPS6; RPS13; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 18.50 | 0 | 0 | 77/149 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 17.90 | 0 | 0 | 77/154 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 17.20 | 0 | 0 | 77/160 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72312 | rRNA processing | 16.80 | 0 | 0 | 77/164 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-376176 | Signaling by ROBO receptors | 15.50 | 0 | 0 | 77/178 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 13.20 | 0 | 0 | 77/209 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72766 | Translation | 11.10 | 0 | 0 | 80/257 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; EEF1B2; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-5663205 | Infectious disease | 10.30 | 0 | 0 | 79/275 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-422475 | Axon guidance | 8.50 | 0 | 0 | 78/328 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; MYL6; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-1266738 | Developmental Biology | 6.27 | 0 | 0 | 78/445 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; MYL6; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-1643685 | Disease | 6.09 | 0 | 0 | 79/464 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
Factor 19 : 37 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-72689 | Formation of a pool of free 40S subunits | 15.10 | 0.00e+00 | 0.00e+00 | 32/95 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| R-HSA-156902 | Peptide chain elongation | 14.90 | 0.00e+00 | 0.00e+00 | 28/84 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; EEF1A1; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; EEF2; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 14.90 | 9.99e-16 | 2.97e-14 | 16/48 | RPS7; RPSA; RPS3A; RPS4X; RPS20; EIF3E; EIF3H; EIF3F; EIF3M; RPS3; RPS29; RPS2; RPS28; RPS9; EIF3D; EIF3L |
| R-HSA-156842 | Eukaryotic Translation Elongation | 14.60 | 0.00e+00 | 0.00e+00 | 28/86 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; EEF1A1; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; EEF2; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 14.10 | 0.00e+00 | 0.00e+00 | 33/105 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; EIF4A2; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| R-HSA-192823 | Viral mRNA Translation | 14.00 | 0.00e+00 | 0.00e+00 | 26/83 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 13.90 | 0.00e+00 | 0.00e+00 | 33/106 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; EIF4A2; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| R-HSA-72649 | Translation initiation complex formation | 13.80 | 4.44e-16 | 1.47e-14 | 17/55 | RPS7; RPSA; EIF4A2; RPS3A; RPS4X; RPS20; EIF3E; EIF3H; EIF3F; EIF3M; RPS3; RPS29; RPS2; RPS28; RPS9; EIF3D; EIF3L |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 13.80 | 4.44e-16 | 1.47e-14 | 17/55 | RPS7; RPSA; EIF4A2; RPS3A; RPS4X; RPS20; EIF3E; EIF3H; EIF3F; EIF3M; RPS3; RPS29; RPS2; RPS28; RPS9; EIF3D; EIF3L |
| R-HSA-2408557 | Selenocysteine synthesis | 13.70 | 0.00e+00 | 0.00e+00 | 26/85 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 13.60 | 6.66e-16 | 2.12e-14 | 17/56 | RPS7; RPSA; EIF4A2; RPS3A; RPS4X; RPS20; EIF3E; EIF3H; EIF3F; EIF3M; RPS3; RPS29; RPS2; RPS28; RPS9; EIF3D; EIF3L |
| R-HSA-72764 | Eukaryotic Translation Termination | 13.50 | 0.00e+00 | 0.00e+00 | 26/86 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 13.20 | 0.00e+00 | 0.00e+00 | 26/88 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 13.10 | 0.00e+00 | 0.00e+00 | 33/113 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; EIF4A2; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 13.10 | 0.00e+00 | 0.00e+00 | 33/113 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; EIF4A2; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| R-HSA-2408522 | Selenoamino acid metabolism | 12.20 | 0.00e+00 | 0.00e+00 | 27/99 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; QARS; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 11.60 | 0.00e+00 | 0.00e+00 | 27/104 | RPL5; SRP9; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-8876725 | Protein methylation | 11.20 | 2.04e-03 | 4.74e-02 | 3/12 | EEF1A1; RPS2; EEF2 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 11.10 | 0.00e+00 | 0.00e+00 | 26/105 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 11.10 | 0.00e+00 | 0.00e+00 | 26/105 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 10.70 | 0.00e+00 | 0.00e+00 | 27/113 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; HSP90AA1; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-191273 | Cholesterol biosynthesis | 10.70 | 7.51e-05 | 1.90e-03 | 5/21 | FDPS; ACAT2; EBP; SQLE; DHCR7 |
| R-HSA-168255 | Influenza Life Cycle | 10.00 | 0.00e+00 | 0.00e+00 | 27/121 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; HSP90AA1; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-168254 | Influenza Infection | 9.80 | 0.00e+00 | 0.00e+00 | 28/128 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; SLC25A6; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; HSP90AA1; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 7.82 | 0.00e+00 | 0.00e+00 | 26/149 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 7.56 | 0.00e+00 | 0.00e+00 | 26/154 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 7.28 | 1.11e-16 | 4.16e-15 | 26/160 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-72312 | rRNA processing | 7.10 | 2.22e-16 | 7.97e-15 | 26/164 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-376176 | Signaling by ROBO receptors | 6.54 | 1.55e-15 | 4.46e-14 | 26/178 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-72766 | Translation | 6.45 | 0.00e+00 | 0.00e+00 | 37/257 | RPL5; SRP9; RPS7; RPL37A; RPL15; RPSA; RPL14; QARS; EIF4A2; RPS3A; RPL10A; EEF1A1; RPS4X; RPL10; RPS20; RPL7; EIF3E; EIF3H; RPL35; RPL7A; EIF3F; EIF3M; RPS3; RPL41; RPL6; RPS29; RPL4; RPS2; RPL23; EEF2; RPL36; RPS28; RPL13A; RPS9; EIF3D; EIF3L; RPL3 |
| R-HSA-2029482 | Regulation of actin dynamics for phagocytic cup formation | 6.05 | 1.22e-03 | 2.96e-02 | 5/37 | ARPC5; ACTB; CFL1; HSP90AA1; ACTG1 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 6.00 | 8.88e-16 | 2.73e-14 | 28/209 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; QARS; RPS3A; RPL10A; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; CKB; RPL4; RPS2; RPL23; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-5663205 | Infectious disease | 5.05 | 2.44e-15 | 6.78e-14 | 31/275 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; NPM1; HMGA1; RPL10A; SLC25A6; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; HSP90AA1; RPL4; RPS2; RPL23; EEF2; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-2682334 | EPH-Ephrin signaling | 4.89 | 1.24e-03 | 2.96e-02 | 6/55 | ARPC5; ACTB; CFL1; MYL6; ACTG1; MYL12A |
| R-HSA-422475 | Axon guidance | 4.78 | 1.11e-16 | 4.16e-15 | 35/328 | RPL5; ARPC5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; PDLIM7; RPL10A; ACTB; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; CFL1; RPS3; TUBA1A; RPL41; MYL6; RPL6; RPS29; HSP90AA1; RPL4; RPS2; RPL23; ACTG1; MYL12A; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-1266738 | Developmental Biology | 3.72 | 3.59e-14 | 9.65e-13 | 37/445 | RPL5; ARPC5; H3F3A; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; PDLIM7; RPL10A; ACTB; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; CFL1; RPS3; TUBA1A; RPL41; MYL6; RPL6; RPS29; HSP90AA1; RPL4; RPS2; RPL23; H3F3B; ACTG1; MYL12A; RPL36; RPS28; RPL13A; RPS9; RPL3 |
| R-HSA-1643685 | Disease | 3.28 | 2.79e-11 | 7.27e-10 | 34/464 | RPL5; RPS7; RPL37A; RPL15; RPSA; RPL14; RPS3A; NPM1; HMGA1; RPL10A; ACTB; SLC25A6; RPS4X; RPL10; RPS20; RPL7; RPL35; RPL7A; RPS3; RPL41; RPL6; RPS29; HSP90AA1; RPL4; RPS2; RPL23; ACTG1; FKBP1A; EEF2; RPL36; RPS28; RPL13A; RPS9; RPL3 |
Factor 20 : 14 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-5576892 | Phase 0 - rapid depolarisation | 15.90 | 7.68e-05 | 0.006616 | 4/12 | SCN9A; CAMK2B; CALM1 |
| R-HSA-5578775 | Ion homeostasis | 12.50 | 5.19e-06 | 0.001253 | 6/23 | ATP2B4; RYR2; CAMK2B; FXYD6; CALM1 |
| R-HSA-500792 | GPCR ligand binding | 12.40 | 8.34e-07 | 0.000718 | 7/27 | GNG5; SST; GABBR2; GNG3; GAL; SSTR2; PTH2 |
| R-HSA-451326 | Activation of kainate receptors upon glutamate binding | 12.00 | 2.65e-04 | 0.019021 | 4/16 | GNG5; GNG3; CALM1 |
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 10.60 | 4.32e-04 | 0.028598 | 4/18 | ACTB; TUBA1A; CALM1 |
| R-HSA-936837 | Ion transport by P-type ATPases | 9.96 | 1.09e-04 | 0.008512 | 5/24 | ATP2B4; CAMK2B; FXYD6; CALM1 |
| R-HSA-418346 | Platelet homeostasis | 9.56 | 2.72e-05 | 0.003170 | 6/30 | GNG5; ATP2B4; GNG3; CALM1; PDE9A |
| R-HSA-5576891 | Cardiac conduction | 9.56 | 5.52e-06 | 0.001253 | 7/35 | ATP2B4; RYR2; SCN9A; CAMK2B; FXYD6; CALM1 |
| R-HSA-397014 | Muscle contraction | 6.96 | 1.38e-05 | 0.002373 | 8/55 | ATP2B4; RYR2; SCN9A; MYL1; CAMK2B; FXYD6; CALM1 |
| R-HSA-418594 | G alpha (i) signalling events | 6.62 | 5.82e-06 | 0.001253 | 9/65 | GNG5; RGS16; SST; GABBR2; GNG3; GAL; CALM1; SSTR2 |
| R-HSA-112314 | Neurotransmitter receptors and postsynaptic signal transmission | 5.86 | 4.67e-04 | 0.028728 | 6/49 | GNG5; CAMK2B; GABBR2; GNG3; CALM1 |
| R-HSA-388396 | GPCR downstream signalling | 4.54 | 2.13e-05 | 0.003061 | 11/116 | GNG5; RGS16; RGS2; SST; GABBR2; GNG3; GAL; CALM1; SSTR2; PTH2 |
| R-HSA-372790 | Signaling by GPCR | 4.38 | 2.95e-05 | 0.003170 | 11/120 | GNG5; RGS16; RGS2; SST; GABBR2; GNG3; GAL; CALM1; SSTR2; PTH2 |
| R-HSA-109582 | Hemostasis | 3.49 | 5.99e-05 | 0.005732 | 13/178 | PRKCZ; GNG5; ATP2B4; H3F3A; ACTB; TMSB4X; GNG3; TUBA1A; CALM1; KIF19; LGALS3BP; PDE9A |
| Factor | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Signif_GO_terms | 165 | 217 | 33 | 48 | 152 | 2 | 54 | 37 | 36 | 0 |
| Factor | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| Signif_GO_terms | 62 | 0 | 1 | 0 | 1 | 38 | 7 | 33 | 37 | 14 |
6 Gene Set Enrichment Analysis by Perturbation
Target genes: Genes w/ LFSR < 0.05 under each perturbation marker gene;
Backgroud genes: all 6000 genes used in factor analysis;
Statistical test: hypergeometric test (over-representation test);
Only GO terms/pathways that satisfy fold change \(\geq\) 2 and test FDR \(<\) 0.05 are shown below.
6.1 GO Slim Over-Representation Analysis
Gene sets: Gene ontology "Biological Process" (non-redundant).
ADNP : 25 significant GO terms| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0007218 | neuropeptide signaling pathway | 4.48 | 7.36e-04 | 2.25e-02 | 6/10 | PCSK1N; GAL; SCG5; NMB; SSTR2; PTH2 |
| GO:0070972 | protein localization to endoplasmic reticulum | 4.14 | 0.00e+00 | 0.00e+00 | 66/119 | RPL22; RPL11; RPS8; RPL5; RYR2; RTN4; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; ANK2; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; BCAP31; RPL10; RPL7; RPL30; GPAA1; RPS6; RPL35; HSPA5; RPL7A; RPL27A; RPS13; SPCS2; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; VAPA; DDRGK1; RPS15; RPS28; RPL18A; RPS16; RPS19; KDELR1; RPL18; RPL13A; RPS5; RPL3 |
| GO:0070670 | response to interleukin-4 | 4.07 | 1.44e-03 | 3.64e-02 | 6/11 | HSPA5; TUBA1B; CDK4; CORO1A; XBP1; RPL3 |
| GO:0060191 | regulation of lipase activity | 3.14 | 1.93e-03 | 4.42e-02 | 8/19 | PRKCZ; HPCA; RGS2; AGT; ARF4; SNCA; NTF3; BICD1 |
| GO:0002576 | platelet degranulation | 2.90 | 1.15e-04 | 5.29e-03 | 14/36 | TAGLN2; TUBA4A; MANF; WDR1; SPARC; MAGED2; APLP2; PSAP; VCL; CD63; SCG3; ALDOA; LGALS3BP; SOD1 |
| GO:0008037 | cell recognition | 2.90 | 1.15e-04 | 5.29e-03 | 14/36 | CNTN2; RTN4; CCT7; ROBO1; GAP43; CASP3; CCT5; TCP1; CADM1; VDAC2; ALDOA; CORO1A; EFNB3; BSG |
| GO:0006413 | translational initiation | 2.80 | 7.77e-15 | 1.90e-12 | 59/157 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; EIF4EBP1; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; PPP1CA; RPS24; RPS26; RPL41; RPL6; RPLP0; DENR; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; EIF2S2; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| GO:0090150 | establishment of protein localization to membrane | 2.71 | 0.00e+00 | 0.00e+00 | 74/204 | RPL22; RPL11; HPCA; RPS8; RPL5; ATP1B1; YWHAQ; RPS27A; REEP1; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; CPLX1; NSG1; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; SDCBP; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; ANK3; RPS24; CACNB3; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; HSP90AA1; SRP14; RAB11A; RPL4; RPLP1; RPS17; RPS2; RAB26; RPS15A; ARL6IP1; RPL13; YWHAE; RPL26; RPL23A; RPL19; RPL27; SNAP25; YWHAB; RPS15; RPS28; RPL18A; NDUFA13; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3; WRB |
| GO:0014074 | response to purine-containing compound | 2.68 | 3.11e-04 | 1.20e-02 | 14/39 | SEPN1; JUN; RYR2; IGFBP5; SPARC; HDAC2; EZR; AKAP9; HSPA5; RAPGEF1; LDHA; HSP90B1; BSG; SOD1 |
| GO:0048857 | neural nucleus development | 2.65 | 1.53e-03 | 3.73e-02 | 11/31 | CDC42; ATP5F1; YWHAQ; G6PD; NFIB; HSPA5; SUDS3; DYNLL1; CKB; YWHAE; ATP5J |
| GO:0046683 | response to organophosphorus | 2.63 | 9.92e-04 | 2.60e-02 | 12/34 | JUN; IGFBP5; SPARC; EZR; AKAP9; HSPA5; RAPGEF1; LDHA; AKR1C1; HSP90B1; BSG; SOD1 |
| GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 2.61 | 4.21e-04 | 1.47e-02 | 14/40 | PTPRF; CNTN2; NRXN1; PTPRG; ROBO1; ALCAM; MDGA1; CADM1; PCDH17; PCDH9; PKD1; CBLN1; BSG; PVRL2 |
| GO:0002181 | cytoplasmic translation | 2.45 | 1.38e-05 | 1.01e-03 | 24/73 | RPL11; RPL31; RPL32; RPL15; RPL35A; RPL9; RPS23; RPL36A; EIF4EBP1; RPL30; ZNF385A; RPS26; RPL41; RPL6; RPLP0; DENR; RPS29; RPLP1; RPL26; RPL19; RPS28; RPL18A; RPL18; RPL13A |
| GO:0010959 | regulation of metal ion transport | 2.41 | 7.85e-07 | 7.20e-05 | 32/99 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; GNAI2; ATP1B3; SNCA; ANK2; CAMK2D; GLRX; TSPAN13; CAMK2B; SRI; AKAP9; G6PD; CHD7; GAL; FXYD6; KIF5B; ANK3; KCNIP2; CACNB3; CD63; JPH4; B2M; CORO1A; YWHAE; NKAIN4; FXYD1; FXYD7 |
| GO:0031099 | regeneration | 2.36 | 2.90e-04 | 1.18e-02 | 18/57 | SEPN1; PTPRF; JUN; GPX1; GAP43; CSNK2B; MTPN; NEFL; NINJ1; TNC; GSTP1; CDK4; PGF; PKM; ENO3; RPL19; RPS16; RTN4R |
| GO:0003012 | muscle system process | 2.33 | 5.94e-07 | 6.56e-05 | 35/112 | ENO1; TPM3; NOS1AP; ATP1B1; RGS2; AGT; RYR2; CHRNA1; IGFBP5; ANK2; CAMK2D; SORBS2; COL4A3BP; ANXA6; HDAC2; CAMK2B; SRI; CALD1; MTPN; G6PD; TPM2; VCL; KCNIP2; MYL6B; MYL6; ACTC1; ALDOA; SSTR2; MYL12A; MYL12B; TNNC2; TPM4; FXYD1; SOD1; PDE9A |
| GO:0034765 | regulation of ion transmembrane transport | 2.30 | 6.25e-07 | 6.56e-05 | 36/117 | SEPN1; HPCA; NOS1AP; ATP1B1; RGS2; AGT; RYR2; YWHAQ; SCN3A; ARL6IP5; ATP1B3; WDR1; SNCA; ANK2; CAMK2D; GLRX; CLIC1; TSPAN13; SRI; AKAP9; G6PD; CHD7; GAL; FXYD6; KIF5B; ANK3; KCNIP2; CACNB3; CD63; JPH4; ARL6IP1; CORO1A; YWHAE; CACNA1A; FXYD1; FXYD7 |
| GO:0072524 | pyridine-containing compound metabolic process | 2.17 | 9.02e-04 | 2.45e-02 | 18/62 | ENO1; MDH1; TKT; PDHB; MDH2; PDHA1; PGK1; G6PD; TALDO1; LDHA; PGAM1; TPI1; PKM; IDH2; QPRT; ALDOA; ENO3; DCXR |
| GO:0034764 | positive regulation of transmembrane transport | 2.15 | 7.34e-04 | 2.25e-02 | 19/66 | NOS1AP; ATP1B1; AGT; RYR2; CAPN10; ATP1B3; SNCA; ANK2; GLRX; SRI; AKAP9; G6PD; GAL; KIF5B; ANK3; KCNIP2; CACNB3; ARL6IP1; FXYD1 |
| GO:0032409 | regulation of transporter activity | 2.12 | 2.62e-04 | 1.13e-02 | 23/81 | SEPN1; HPCA; NOS1AP; ATP1B1; RYR2; ATP1B3; SNCA; ANK2; CAMK2D; GLRX; NDFIP1; SRI; AKAP9; GAL; FXYD6; KIF5B; ANK3; KCNIP2; CACNB3; JPH4; YWHAE; FXYD1; FXYD7 |
| GO:0009410 | response to xenobiotic stimulus | 2.09 | 5.91e-04 | 1.97e-02 | 21/75 | SEPN1; HPCA; GSTM3; PHGDH; MGST3; RGS2; EPHX1; RYR2; GPX1; SNCA; MGST2; HDAC2; EIF4EBP1; FBXO32; HSPA5; GSTP1; AKR1C1; RGS10; ASIC1; SERPINF1; SOD1 |
| GO:0006401 | RNA catabolic process | 2.09 | 1.02e-09 | 1.49e-07 | 66/236 | RPL22; RPL11; YBX1; RPS8; RPL5; RPS27A; RPL31; SSB; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; SET; RPL7A; RNH1; RPL27A; RPS13; RNASEH2C; HSPA8; RPS24; MAGOHB; RPS26; RPL41; CNOT2; RPL6; RPLP0; UBC; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; EIF4A3; YWHAB; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| GO:0042391 | regulation of membrane potential | 2.05 | 2.65e-05 | 1.77e-03 | 33/120 | PRKCZ; JUN; NOS1AP; ATP1B1; RYR2; NRXN1; SCN3A; CHRNA1; ARL6IP5; ATP1B3; WDR1; SNCA; ANK2; CAMK2D; GLRX; CLIC1; HEBP2; STX1A; SRI; AKAP9; KIF5B; ANK3; KCNIP2; CACNB3; ASIC1; CBLN1; YWHAE; EIF4A3; BAIAP2; CELF4; CACNA1A; FXYD1; SOD1 |
| GO:0008544 | epidermis development | 2.04 | 1.99e-03 | 4.42e-02 | 18/66 | POU3F1; CRABP2; PKP4; KLF7; IGFBP5; SATB1; CASP3; H2AFY; FAM65B; HDAC2; NSDHL; AGPAT2; TSG101; GAL; PSAP; PPHLN1; CAPNS1; SOD1 |
| GO:0006605 | protein targeting | 2.03 | 8.60e-10 | 1.49e-07 | 71/261 | RPL22; RPL11; HPCA; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RAB7A; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; SLC25A6; RPS4X; RPL36A; RPL10; SDCBP; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; SPCS2; HSPA8; ANK3; RPS24; CACNB3; RPS26; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; HSP90AA1; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; ARL6IP1; UQCRC2; RPL13; RPL26; RPL23A; RPL19; RPL27; YWHAB; RPS15; RPS28; RPL18A; NDUFA13; ECH1; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
ARID1B : 20 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 7.10 | 0.00e+00 | 0.00e+00 | 51/119 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; SPCS1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SEC61G; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; SEC61B; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| GO:0010257 | NADH dehydrogenase complex assembly | 6.62 | 1.11e-14 | 8.15e-13 | 24/60 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFA2; NDUFB2; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFS8; NDUFC2; NDUFB8; NDUFB1; NDUFB10; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND3 |
| GO:0006413 | translational initiation | 5.91 | 0.00e+00 | 0.00e+00 | 56/157 | RPL11; RPS8; RPL5; RPS27; DDX1; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; MCTS1; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; EIF1; RPL18A; RPS19; RPL18; RPL13A; EIF3L; RPL3; ATF4 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 5.90 | 0.00e+00 | 0.00e+00 | 31/87 | NDUFS5; COX20; NDUFB3; NDUFAF3; COX17; NDUFB4; NDUFA2; NDUFB2; NDUFB11; NDUFA1; NDUFB9; TMEM261; NDUFB6; NDUFS8; NDUFC2; NDUFB8; COX14; COX16; NDUFB1; NDUFB10; COA3; NDUFS7; NDUFA11; PET100; NDUFA7; NDUFB7; NDUFA13; NDUFA3; UQCR10; NDUFA6; MT-ND3 |
| GO:0002181 | cytoplasmic translation | 5.44 | 1.79e-12 | 1.01e-10 | 24/73 | RPL11; RPL32; RPL15; TMA7; RPL9; RPS23; RPL10A; RPL36A; RPL39; MCTS1; EIF4EBP1; RPL30; EIF3E; EIF3H; EIF3F; RPS26; RPL6; RPLP0; RPL26; RPL19; RPL18A; RPL18; RPL13A; EIF3L |
| GO:1902600 | proton transmembrane transport | 5.38 | 8.32e-13 | 5.09e-11 | 25/77 | PARK7; COX5B; ATP5G3; COX17; ATP5I; COX7C; COX7A2; NDUFA4; ATP5J2; ATP6V1F; TMSB4X; COX7B; COX6C; COX8A; ATP5L; ATP5G2; COX5A; COX4I1; ATP5G1; ATP5H; ATP5E; NDUFS7; COX6B1; ATP5J; MT-CO1 |
| GO:0090150 | establishment of protein localization to membrane | 4.71 | 0.00e+00 | 0.00e+00 | 58/204 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL9; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPL10A; CD24; RPS12; FIS1; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; YWHAZ; RPS6; SEC61B; RPL7A; RPL27A; RPS13; TIMM10; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; CALM1; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; ROMO1; RPL18A; NDUFA13; PDCD5; RPS19; RPL18; RPL13A; RPL3 |
| GO:0017004 | cytochrome complex assembly | 4.14 | 1.07e-03 | 4.15e-02 | 7/28 | COX20; COX17; COX14; COX16; COA3; PET100; UQCR10 |
| GO:0009141 | nucleoside triphosphate metabolic process | 4.12 | 0.00e+00 | 0.00e+00 | 47/189 | PARK7; ATPIF1; NDUFS5; UQCRH; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; COX7C; UQCRQ; NDUFA2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; NDUFB9; NDUFB6; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5G2; DNAJC15; NDUFB1; COX5A; NDUFB10; DCTPP1; COX4I1; ATP5G1; NME1; ATP5H; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; MT-CO1; MT-ND3 |
| GO:0006605 | protein targeting | 3.93 | 0.00e+00 | 0.00e+00 | 62/261 | RPL11; ATPIF1; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; SPCS1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SEC61G; FIS1; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; YWHAZ; RPS6; SEC61B; RPL7A; RPL27A; RPS13; IMMP1L; TIMM10; RPS3; TIMM8B; RPS24; RPS26; RPL6; RPLP0; RPL21; DNAJC15; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; ROMO1; UBL5; RPL18A; NDUFA13; PDCD5; RPS19; RPL18; RPL13A; RPL3 |
| GO:0009123 | nucleoside monophosphate metabolic process | 3.91 | 1.11e-16 | 9.05e-15 | 46/195 | PARK7; ATPIF1; NDUFS5; UQCRH; COX5B; ATP5G3; NDUFB3; NDUFB4; ATP5I; COX7C; UQCRQ; NDUFA2; COX7A2; HDDC2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; NDUFB9; NDUFB6; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5G2; DNAJC15; NDUFB1; COX5A; NDUFB10; COX4I1; ATP5G1; ATP5H; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; MT-CO1; MT-ND3 |
| GO:0006401 | RNA catabolic process | 3.86 | 0.00e+00 | 0.00e+00 | 55/236 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL9; LSM6; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; LSM5; LINC01420; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; YWHAZ; EIF3E; RPS6; RPL7A; RPL27A; RPS13; RNASEH2C; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; DDX5; RPL18A; LSM4; RPS19; RPL18; RPL13A; RPL3 |
| GO:0006091 | generation of precursor metabolites and energy | 3.54 | 0.00e+00 | 0.00e+00 | 53/248 | PARK7; SH3BGRL3; ATPIF1; NDUFS5; UQCRH; COX20; COX5B; NDUFB3; COX17; NDUFB4; ATP5I; COX7C; GLRX; UQCRQ; NDUFA2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; COX7B; NDUFA1; COX6C; NDUFB9; NDUFB6; COX8A; NDUFS8; NDUFC2; ATP5L; CISD1; NDUFB8; TPI1; BLOC1S1; HMGB1; DNAJC15; NDUFB1; COX5A; NDUFB10; COX4I1; ATP5H; GNAS; ATP5E; NDUFS7; UQCR11; NDUFA7; NDUFB7; COX6B1; NDUFA3; UQCR10; NDUFA6; ATP5J; MT-CO1; MT-ND3 |
| GO:0007006 | mitochondrial membrane organization | 3.27 | 1.06e-05 | 5.54e-04 | 17/86 | ATPIF1; ATP5G3; ATP5I; DYNLT1; ATP5J2; YWHAZ; TIMM10; ATP5L; ATP5G2; CALM1; ATP5G1; ATP5H; ROMO1; ATP5E; NDUFA13; PDCD5; ATP5J |
| GO:0009259 | ribonucleotide metabolic process | 3.09 | 4.32e-13 | 2.88e-11 | 48/257 | PARK7; ATPIF1; NDUFS5; UQCRH; COX5B; DBI; ATP5G3; NDUFB3; NDUFB4; ATP5I; COX7C; HINT1; UQCRQ; NDUFA2; COX7A2; NDUFA4; CYCS; ATP5J2; NDUFB2; TMSB4X; NDUFA1; NDUFB9; NDUFB6; AK1; NDUFS8; NDUFC2; ATP5L; NDUFB8; TPI1; ATP5G2; DNAJC15; NDUFB1; COX5A; NDUFB10; COX4I1; ATP5G1; NME1; ATP5H; ATP5E; NDUFS7; NDUFA7; NDUFB7; NDUFA3; UQCR10; NDUFA6; ATP5J; MT-CO1; MT-ND3 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 2.42 | 1.30e-05 | 5.94e-04 | 27/185 | MRPL20; RPL11; RPL5; RPS27; SNRPE; DDX1; SNRPG; RPSA; RPS23; RPS14; LINC01420; MCTS1; EIF3E; EIF3H; EIF3F; SNRPF; RPL6; RPLP0; RPL23A; SNRPD1; LSM4; RPS19; SNRPD2; RPL13A; EIF3L; RPL3; SNU13 |
| GO:0006839 | mitochondrial transport | 2.25 | 4.62e-04 | 2.00e-02 | 20/147 | ATPIF1; COX5B; ATP5I; DYNLT1; ATP5J2; FIS1; YWHAZ; IMMP1L; TIMM10; TIMM8B; ATP5L; DNAJC15; ATP5H; ROMO1; ATP5E; UBL5; NDUFA13; PDCD5; SMDT1; ATP5J |
| GO:0006979 | response to oxidative stress | 2.04 | 1.01e-03 | 4.10e-02 | 22/179 | PARK7; PRDX1; TXNIP; SELK; NDUFB4; ATOX1; GNB2L1; CYCS; TXN; PRDX5; GSTP1; NDUFS8; RPS3; PEBP1; ROMO1; GPX4; PRDX2; ATF4; NDUFA6; SOD1; MT-CO1; MT-ND3 |
| GO:0016072 | rRNA metabolic process | 2.03 | 1.32e-03 | 4.84e-02 | 21/171 | RPL11; RPS8; RPL5; RPS27; NCL; RPSA; LSM6; RPS14; NHP2; RPL10A; GTF2H5; RPL7; RPS6; RPL7A; RPS24; NOP10; RPS17; RPS2; RPL26; RPS19; SNU13 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 2.01 | 1.17e-05 | 5.73e-04 | 40/330 | MRPL20; RPL11; RPS8; RPL5; RPS27; SNRPE; DDX1; SNRPG; RPSA; LSM6; RPS23; RPS14; NHP2; RPL10A; GTF2H5; LINC01420; MCTS1; RPL7; EIF3E; EIF3H; RPS6; RPL7A; EIF3F; RPS24; SNRPF; RPL6; RPLP0; NOP10; RPS17; RPS2; RPL26; RPL23A; SNRPD1; LSM4; RPS19; SNRPD2; RPL13A; EIF3L; RPL3; SNU13 |
ASH1L : 33 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0010737 | protein kinase A signaling | 7.76 | 2.29e-04 | 1.29e-02 | 5/11 | RAB13; AKAP12; EZR; GAL; AKAP6 |
| GO:0007218 | neuropeptide signaling pathway | 6.83 | 1.83e-03 | 4.19e-02 | 4/10 | PCSK1N; GAL; SSTR2; PTH2 |
| GO:0010644 | cell communication by electrical coupling | 6.57 | 5.80e-04 | 2.36e-02 | 5/13 | ATP1B1; RYR2; SRI; ANK3; CALM1 |
| GO:0015844 | monoamine transport | 6.03 | 2.73e-04 | 1.34e-02 | 6/17 | AGT; SNCA; STX1A; GPM6B; SYT7; SYT1 |
| GO:0060191 | regulation of lipase activity | 5.39 | 5.43e-04 | 2.34e-02 | 6/19 | PRKCZ; HPCA; RHOC; AGT; SNCA; NTF3 |
| GO:0031644 | regulation of neurological system process | 4.55 | 2.37e-04 | 1.29e-02 | 8/30 | PRKCZ; AGT; NRXN1; STX1A; AKAP9; EIF4A3; BAIAP2; CELF4 |
| GO:0014074 | response to purine-containing compound | 3.94 | 3.18e-04 | 1.46e-02 | 9/39 | SEPN1; JUN; RYR2; IGFBP5; HDAC2; EZR; AKAP9; RAPGEF1; AKAP6 |
| GO:0099565 | chemical synaptic transmission, postsynaptic | 3.90 | 7.36e-04 | 2.54e-02 | 8/35 | PRKCZ; NRXN1; CHRNA1; SNCA; STX1A; EIF4A3; BAIAP2; CELF4 |
| GO:0010959 | regulation of metal ion transport | 3.79 | 3.29e-08 | 2.42e-05 | 22/99 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; GNAI2; SNCA; TSPAN13; CAMK2B; SRI; AKAP9; G6PD; CHD7; GAL; ANK3; JPH4; AKAP6; CALM1; NKAIN4; HOMER3; FXYD7 |
| GO:0034764 | positive regulation of transmembrane transport | 3.62 | 1.98e-05 | 2.08e-03 | 14/66 | NOS1AP; ATP1B1; AGT; RYR2; CAPN10; SNCA; SRI; AKAP9; TMSB4X; G6PD; GAL; ANK3; AKAP6; CALM1 |
| GO:0035637 | multicellular organismal signaling | 3.57 | 6.89e-04 | 2.54e-02 | 9/43 | ATP1B1; AGT; RYR2; SCN3A; CHRNA1; SRI; AKAP9; ANK3; CALM1 |
| GO:0034765 | regulation of ion transmembrane transport | 3.36 | 1.78e-07 | 6.55e-05 | 23/117 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; SCN3A; SNCA; CLIC1; TSPAN13; SRI; AKAP9; TMSB4X; G6PD; CHD7; GAL; ANK3; JPH4; AKAP6; CALM1; CACNA1A; FXYD7; KCNJ6 |
| GO:0043270 | positive regulation of ion transport | 3.26 | 1.23e-04 | 8.22e-03 | 13/68 | NOS1AP; ATP1B1; AGT; RYR2; SNCA; SRI; AKAP9; TMSB4X; G6PD; GAL; ANK3; AKAP6; CALM1 |
| GO:0061351 | neural precursor cell proliferation | 3.22 | 8.34e-04 | 2.66e-02 | 10/53 | GNG5; ID2; SIX3; GNAI2; RAPGEF1; ILK; EMX2; NEUROD4; NAP1L1; INSM1 |
| GO:0032409 | regulation of transporter activity | 3.16 | 5.49e-05 | 5.04e-03 | 15/81 | SEPN1; HPCA; NOS1AP; ATP1B1; RYR2; SNCA; SRI; AKAP9; TMSB4X; GAL; ANK3; JPH4; AKAP6; CALM1; FXYD7 |
| GO:0072511 | divalent inorganic cation transport | 3.00 | 7.40e-06 | 9.05e-04 | 20/114 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; GNAI2; SNCA; GPM6A; TSPAN13; CAMK2B; SRI; G6PD; CHD7; ASIC1; JPH4; AKAP6; CALM1; CACNA1A; HOMER3 |
| GO:0042391 | regulation of membrane potential | 2.99 | 4.53e-06 | 6.65e-04 | 21/120 | PRKCZ; JUN; NOS1AP; ATP1B1; RYR2; NRXN1; SCN3A; CHRNA1; SNCA; GNB2L1; CLIC1; STX1A; SRI; AKAP9; ANK3; ASIC1; AKAP6; EIF4A3; BAIAP2; CELF4; CACNA1A |
| GO:0099177 | regulation of trans-synaptic signaling | 2.92 | 5.02e-07 | 1.23e-04 | 26/152 | PRKCZ; AGT; NRXN1; GNAI2; ABHD6; NSG1; SNCA; CPLX2; GRM4; AKAP12; CAMK2B; STX1A; AKAP9; SYT7; NTF3; ASIC1; SYT1; JPH4; CALM1; RAB26; EFNB3; EIF4A3; BAIAP2; CELF4; CACNA1A; RAB3A |
| GO:0019932 | second-messenger-mediated signaling | 2.78 | 2.46e-04 | 1.29e-02 | 15/92 | SEPN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; NEUROD1; GNAI2; SRI; GAL; JPH4; AKAP6; CALM1; HOMER3; APLP1 |
| GO:0009410 | response to xenobiotic stimulus | 2.73 | 1.21e-03 | 3.42e-02 | 12/75 | SEPN1; HPCA; RYR2; GPX1; SNCA; NR3C1; HSP90AB1; GSTA4; HDAC2; GSTP1; ASIC1; CALM1 |
| GO:0009914 | hormone transport | 2.52 | 1.64e-03 | 4.01e-02 | 13/88 | AGT; SOX11; NEUROD1; KLF7; CAPN10; STX1A; SRI; SLC25A6; CHD7; SYT7; GAL; CACNA1A; SELM |
| GO:0001503 | ossification | 2.52 | 1.12e-03 | 3.29e-02 | 14/95 | PBX1; H3F3A; SOX11; ID2; IGFBP5; PDLIM7; CLIC1; FZD1; GPM6B; RBMX; CTHRC1; TNC; ILK; TP53INP2 |
| GO:0060560 | developmental growth involved in morphogenesis | 2.49 | 1.82e-03 | 4.19e-02 | 13/89 | PRKCZ; CRABP2; MAP2; ALCAM; HSP90AB1; AUTS2; DPYSL2; TNC; ILK; GAL; SYT1; RND2; RTN4R |
| GO:0061564 | axon development | 2.47 | 2.55e-06 | 4.67e-04 | 30/207 | STMN1; PTPRF; JUN; S100A6; CRABP2; LHX9; NRXN1; KLF7; MAP2; ALCAM; SPON2; CASP3; PDLIM7; HSP90AB1; EZR; AUTS2; NEFM; NEFL; DPYSL2; TNC; ILK; FEZ1; ANK3; MYCBP2; EFNB3; RND2; NGFR; BAIAP2; RAB3A; RTN4R |
| GO:0003012 | muscle system process | 2.44 | 7.16e-04 | 2.54e-02 | 16/112 | NOS1AP; ATP1B1; AGT; RYR2; CHRNA1; IGFBP5; HDAC2; CAMK2B; SRI; CALD1; G6PD; AKAP6; CALM1; ALDOA; SSTR2; EEF2 |
| GO:0051047 | positive regulation of secretion | 2.44 | 1.05e-03 | 3.20e-02 | 15/105 | PRKCZ; WLS; AGT; SOX11; CAPN10; GNAI2; SNCA; EZR; STX1A; SRI; SYT7; GAL; GAPDH; SYT1; RAB3A |
| GO:0048880 | sensory system development | 2.44 | 1.53e-03 | 4.00e-02 | 14/98 | HPCA; JUN; SOX11; SIX3; NEUROD1; IMPDH2; MAB21L2; GPM6A; HDAC2; CHD7; ZEB1; NEUROD4; MAB21L1; CELF4 |
| GO:0023061 | signal release | 2.36 | 1.20e-04 | 8.22e-03 | 22/159 | WLS; AGT; SOX11; NRXN1; NEUROD1; KLF7; CAPN10; SNCA; CPLX2; GRM4; STX1A; SRI; SLC25A6; CHD7; SYT7; GAL; ASIC1; SYT1; CALM1; CACNA1A; RAB3A; SELM |
| GO:0002791 | regulation of peptide secretion | 2.27 | 2.22e-03 | 4.95e-02 | 15/113 | PRKCZ; WLS; NEUROD1; KLF7; CAPN10; BTN2A2; EZR; STX1A; SRI; SLC25A6; TMSB4X; CHD7; SYT7; GAPDH; CACNA1A |
| GO:0010975 | regulation of neuron projection development | 2.21 | 9.22e-05 | 7.52e-03 | 26/201 | ACAP3; PTPRF; CRABP2; AGT; MAP2; ITM2C; PTPRG; TNIK; ENC1; NR2F1; HDAC2; CAMK2B; FZD1; NEFL; DPYSL2; RAPGEF1; ILK; CFL1; SYT1; MYCBP2; EFNB3; RND2; NGFR; BAIAP2; PREX1; RTN4R |
| GO:0009636 | response to toxic substance | 2.12 | 7.60e-04 | 2.54e-02 | 21/169 | HPCA; JUN; CHRNA1; EEF1B2; GPX1; CASP3; GNB2L1; HDAC2; G6PD; NEFL; TNC; GSTP1; RPS3; FAM213A; ASIC1; HNRNPA1; ESD; CALM1; SNN; EEF2; PRDX2 |
| GO:0051098 | regulation of binding | 2.06 | 1.49e-03 | 4.00e-02 | 20/166 | STMN1; PTPRF; JUN; NUCKS1; SOX11; ID2; NEUROD1; MAP2; HMGB2; MDFI; HSP90AB1; HDAC2; SRI; TMSB4X; CTHRC1; CALM1; CCPG1; EIF4A3; AES; MFNG |
| GO:0050769 | positive regulation of neurogenesis | 2.05 | 1.61e-03 | 4.01e-02 | 20/167 | CRABP2; AGT; SOX11; ID2; NEUROD1; ENC1; HDAC2; CAMK2B; FZD1; NEFL; RAPGEF1; ILK; ZEB1; NAP1L1; SYT1; TCF12; RND2; NGFR; BAIAP2; PCP4 |
CHD2 : 21 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0007218 | neuropeptide signaling pathway | 4.51 | 7.06e-04 | 2.16e-02 | 6/10 | PCSK1N; GAL; SCG5; NMB; SSTR2; PTH2 |
| GO:0070972 | protein localization to endoplasmic reticulum | 4.36 | 0.00e+00 | 0.00e+00 | 69/119 | RPL22; RPL11; RPS8; RPL5; RPS27; RYR2; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; SPCS1; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; SEC63; RPS12; SEC61G; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; HSPA5; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| GO:0002181 | cytoplasmic translation | 3.40 | 1.93e-11 | 2.36e-09 | 33/73 | RPL11; RPL31; RPL32; RPL15; RPL24; RPL35A; RPL9; RPS23; RPL36A; RPL39; RPL30; EIF3E; EIF3H; RPL8; EIF3F; ZNF385A; RPL41; RPL6; RPLP0; DENR; RPS29; EIF3J; RPLP1; RPL26; RPL19; RPL38; RPL17; EEF2; RPS28; RPL18A; RPL18; RPL13A; EIF3L |
| GO:0006413 | translational initiation | 3.30 | 0.00e+00 | 0.00e+00 | 69/157 | RPL22; RPL11; RPS8; RPL5; RPS27; DDX1; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; DENR; RPL21; RPS29; EIF3J; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; EIF3L; RPL3 |
| GO:0090150 | establishment of protein localization to membrane | 3.10 | 0.00e+00 | 0.00e+00 | 84/204 | RPL22; RPL11; HPCA; RPS8; RPL5; RPS27; ATP1B1; CNST; RPS27A; REEP1; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL35A; CPLX1; NSG1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS10; SEC63; RPS12; FIS1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; RPL7A; RPL27A; RPS13; TIMM10; BAD; RPS3; RPS24; CACNB3; RPL41; RPL6; RPLP0; RPL21; RPS29; CALM1; HSP90AA1; SRP14; RPL4; RPLP1; RPS17; RAB26; RPS15A; RPL13; VAMP2; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RAB31; RPL17; ROMO1; RPS15; RPS28; RPL18A; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| GO:0010257 | NADH dehydrogenase complex assembly | 3.01 | 2.03e-07 | 2.13e-05 | 24/60 | NDUFS5; NDUFAF3; NDUFB4; NDUFA2; NDUFA5; NDUFB2; NDUFB11; NDUFA1; NDUFB9; NDUFS8; NDUFC2; NDUFB8; NDUFB10; NDUFAB1; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND1; MT-ND3; MT-ND4; MT-ND6 |
| GO:0001906 | cell killing | 2.95 | 5.30e-04 | 1.69e-02 | 11/28 | PRDX1; TUBB; RPL30; BAD; CADM1; GAPDH; B2M; EMP2; VAMP2; ROMO1; RPS19 |
| GO:0014074 | response to purine-containing compound | 2.89 | 7.00e-05 | 3.95e-03 | 15/39 | SEPN1; JUN; RYR2; HSPD1; IGFBP5; CDO1; HDAC2; EZR; AKAP9; HSPA5; RAPGEF1; LDHA; HSP90B1; NME1; BSG |
| GO:0006959 | humoral immune response | 2.82 | 2.50e-03 | 4.96e-02 | 9/24 | SPON2; RPL39; RPL30; SERPING1; GAPDH; C1QBP; PHB; ROMO1; RPS19 |
| GO:0050663 | cytokine secretion | 2.72 | 4.41e-04 | 1.47e-02 | 13/36 | PRKCZ; S100A13; SYT11; AGT; HSPD1; SELK; BTN2A2; EZR; TMSB4X; CADM1; GAPDH; HMGB1; MIF |
| GO:0050954 | sensory perception of mechanical stimulus | 2.70 | 2.87e-04 | 1.01e-02 | 14/39 | GPX1; CASP3; FAM65B; SOBP; CHD7; NDUFB9; NAV2; TIMM10; TIMM8B; CACNB3; SRRM4; SIX1; RPL38; RAB3A |
| GO:0046683 | response to organophosphorus | 2.65 | 9.26e-04 | 2.34e-02 | 12/34 | JUN; HSPD1; IGFBP5; CDO1; EZR; AKAP9; HSPA5; RAPGEF1; LDHA; HSP90B1; NME1; BSG |
| GO:0021953 | central nervous system neuron differentiation | 2.51 | 1.21e-04 | 5.94e-03 | 18/54 | DRAXIN; EPHB2; LMO4; NRP2; MAP2; ROBO1; UQCRQ; ID4; SOX4; MDGA1; FZD1; MTPN; NFIB; MYCBP2; HSP90AA1; CBLN1; DCC; ADARB1 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 2.42 | 3.65e-06 | 2.98e-04 | 28/87 | NDUFS5; NDUFAF3; COX17; NDUFB4; NDUFA2; UQCC2; NDUFA5; NDUFB2; NDUFB11; NDUFA1; UQCRB; NDUFB9; NDUFS8; NDUFC2; NDUFB8; NDUFB10; NDUFAB1; NDUFA11; PET100; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND1; MT-ND3; MT-ND4; MT-ND6 |
| GO:0006401 | RNA catabolic process | 2.39 | 3.36e-14 | 6.00e-12 | 75/236 | RPL22; RPL11; YBX1; RPS8; SERBP1; RPL5; RPS27; RPS27A; RPL31; SSB; RPL37A; RPL32; RPL15; RPSA; RPL14; PCBP4; RPL24; RPL35A; RPL9; RPL34; LSM6; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; RPL8; RPS6; RPL7A; RPL27A; RPS13; RNASEH2C; RPS3; HSPA8; RPS24; RPL41; CNOT2; RPL6; RPLP0; RPL21; HNRNPC; RPS29; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; CIRBP; RPS15; LSM7; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| GO:0006605 | protein targeting | 2.30 | 4.09e-14 | 6.00e-12 | 80/261 | RPL22; RPL11; HPCA; RPS8; RPL5; RPS27; RPS27A; RPL31; HSPD1; RPL37A; RPL32; RPL15; RPSA; RPL14; SPCS1; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; SEC63; RPS12; SEC61G; FIS1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; RPL7A; RPL27A; RPS13; TIMM10; RPS3; TIMM8B; HSPA8; RPS24; CACNB3; RPL41; RPL6; RPLP0; RPL21; DNAJC15; RPS29; HSP90AA1; SRP14; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; ROMO1; RPS15; RPS28; UBL5; RPL18A; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| GO:0031099 | regeneration | 2.24 | 8.15e-04 | 2.17e-02 | 17/57 | SEPN1; PTPRF; JUN; FKBP1B; GPX1; GAP43; NREP; MTPN; NEFL; TNC; GSTP1; PKM; ENO3; RPL19; PHB; RPS16; RTN4R |
| GO:0070661 | leukocyte proliferation | 2.17 | 2.36e-03 | 4.80e-02 | 15/52 | FKBP1B; HSPD1; IGFBP2; SATB1; SELK; CASP3; BTN2A2; RPS6; CD151; GSTP1; GAL; RPS3; CADM1; HMGB1; MIF |
| GO:0010959 | regulation of metal ion transport | 2.05 | 1.46e-04 | 6.68e-03 | 27/99 | SEPN1; NKAIN1; HPCA; NOS1AP; ATP1B1; AGT; RYR2; FKBP1B; GNAI2; SNCA; TSPAN13; CAMK2B; SRI; AKAP9; CHD7; GAL; FXYD6; KCNIP2; CACNB3; CD63; JPH4; CALM1; B2M; VAMP2; FKBP1A; NKAIN4; FXYD7 |
| GO:1902600 | proton transmembrane transport | 2.05 | 7.81e-04 | 2.17e-02 | 21/77 | COX5B; ATP5G3; COX17; ATP5I; ATP6V1G2; NDUFA4; ATP5J2; TMSB4X; COX7B; COX6C; ATP6V1G1; COX8A; ATP5B; COX5A; ATP5G1; ATP5H; ATP5A1; CYB5A; COX6B1; ATP5J; MT-ND4 |
| GO:0003012 | muscle system process | 2.01 | 9.05e-05 | 4.75e-03 | 30/112 | ENO1; NOS1AP; ATP1B1; RGS2; TNNI1; AGT; RYR2; FKBP1B; MYL1; IGFBP5; SORBS2; COL4A3BP; NEUROG1; ANXA6; HDAC2; CAMK2B; SRI; CALD1; MTPN; VCL; KCNIP2; MYL6B; MYL6; CALM1; ALDOA; SSTR2; MYL12A; MYL12B; EEF2; PDE9A |
DYRK1A : 2 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0046683 | response to organophosphorus | 7.55 | 0.000113 | 0.0415 | 6/34 | HSPD1; NDUFS4; EZR; HSPA5; LDHA; HSP90B1 |
| GO:0006457 | protein folding | 4.03 | 0.000016 | 0.0117 | 13/138 | PDIA6; FKBP1B; HSPD1; HSPE1; CANX; HSP90AB1; PRDX4; HSPA5; PFDN5; HSP90B1; PDIA3; PPIB; CALR |
PTEN : 22 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 4.52 | 0.00e+00 | 0.00e+00 | 84/119 | RPL22; RPL11; RPS8; RPL5; RPS27; RYR2; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; SEC63; RPS12; SEC61G; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; HSPA5; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; KDELR1; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0007218 | neuropeptide signaling pathway | 3.84 | 1.70e-03 | 3.37e-02 | 6/10 | PCSK1N; GAL; SCG5; NMB; SSTR2; PTH2 |
| GO:0014075 | response to amine | 3.84 | 1.70e-03 | 3.37e-02 | 6/10 | RGS2; HDAC2; RGS10; ASIC1; CALM1; NME1 |
| GO:0006413 | translational initiation | 3.75 | 0.00e+00 | 0.00e+00 | 92/157 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; DDX1; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; RPS18; RPS10; RPL10A; RPS12; HSPB1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; DENR; RPL21; RPS29; EIF3J; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; EIF2S2; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3; ATF4 |
| GO:0002181 | cytoplasmic translation | 3.51 | 7.99e-15 | 9.78e-13 | 40/73 | RPL11; RPL31; RPL32; RPL15; TMA7; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; RPL30; EIF3E; EIF3H; RPL8; EIF3F; EIF3M; RPS26; RPL41; RPL6; RPLP0; DENR; RPS29; EIF3J; RPLP1; RPL26; RPL19; RPL38; RPL17; RPS21; EEF2; RPL36; RPS28; RPL18A; RPL18; RPL13A; EIF3D; EIF3L |
| GO:0010644 | cell communication by electrical coupling | 3.45 | 1.59e-03 | 3.34e-02 | 7/13 | ATP1B1; RYR2; FKBP1B; DBN1; SRI; CALM1; HRC |
| GO:0015844 | monoamine transport | 3.39 | 3.92e-04 | 1.37e-02 | 9/17 | PARK7; SYT11; AGT; SNCA; ACTB; STX1A; GPM6B; SYT7; SNCG |
| GO:0051339 | regulation of lyase activity | 3.20 | 2.76e-03 | 4.82e-02 | 7/14 | PARK7; HPCA; GNAI2; CAP2; AKAP9; GABBR2; PHPT1 |
| GO:0090150 | establishment of protein localization to membrane | 3.14 | 0.00e+00 | 0.00e+00 | 100/204 | RPL22; RPL11; HPCA; RPS8; RPL5; RPS27; ATP1B1; CNST; RPS7; RPS27A; REEP1; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; NSG1; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPS10; RPL10A; CD24; SEC63; RPS12; FIS1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPL8; RPS6; SEC61B; RPL12; RPL7A; RPLP2; RPL27A; RPS13; TIMM10; BAD; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; CALM1; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; ARL6IP1; RPL13; YWHAE; VAMP2; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RAB31; RPL17; ROMO1; RPS21; RPS15; TIMM13; RPL36; RPS28; RPL18A; UBA52; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0010257 | NADH dehydrogenase complex assembly | 2.99 | 1.15e-08 | 1.20e-06 | 28/60 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFC1; NDUFS6; NDUFS4; NDUFA2; NDUFA5; NDUFA1; TMEM261; NDUFB6; NDUFB8; NDUFA12; NDUFB1; NDUFAB1; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND1; MT-ND2; MT-ND3; MT-ND4; MT-ND6 |
| GO:2001023 | regulation of response to drug | 2.65 | 7.63e-04 | 2.15e-02 | 12/29 | PARK7; SYT11; RGS2; AGT; NCOA1; SNCA; GNB2L1; TXN; SYT7; GAL; SNCG; ARL6IP1 |
| GO:1902600 | proton transmembrane transport | 2.58 | 1.34e-07 | 9.82e-06 | 31/77 | PARK7; COX7A2L; COX5B; ATP5G3; COX17; ATP5I; COX7C; ATP6V1G2; COX7A2; NDUFA4; ATP5J2; ATP6V1F; ATP6V0E2; TMSB4X; COX7B; COX6C; ATP6V1G1; COX8A; ATP5L; ATP5G2; COX6A1; ATP5G1; ATP5H; ATP5A1; CYB5A; ATP5E; NDUFS7; COX6B1; ATP5J; ATP5O; MT-ND4 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 2.58 | 2.10e-08 | 1.93e-06 | 35/87 | NDUFS5; COX20; NDUFB3; NDUFAF3; COX17; NDUFB4; NDUFC1; NDUFS6; NDUFS4; NDUFA2; NDUFA5; NDUFA1; TMEM261; NDUFB6; NDUFB8; COX14; NDUFA12; COX16; NDUFB1; NDUFAB1; COA3; NDUFS7; NDUFA11; PET100; NDUFA7; NDUFB7; NDUFA13; NDUFA3; UQCR10; NDUFA6; MT-ND1; MT-ND2; MT-ND3; MT-ND4; MT-ND6 |
| GO:0006401 | RNA catabolic process | 2.55 | 0.00e+00 | 0.00e+00 | 94/236 | RPL22; RPL11; RPS8; SERBP1; RPL5; RPS27; RPS7; RPS27A; RPL31; SSB; RPL37A; RPL32; LSM3; RPL15; RPSA; RPL14; PCBP4; RPL29; RPL24; RPL35A; RPL9; RPL34; LSM6; RPS3A; RPL37; RPS23; RPS14; NPM1; RPS18; RPS10; RPL10A; RPS12; LSM5; HSPB1; LINC01420; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; YWHAZ; EIF3E; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; CNOT2; RPL6; RPLP0; RPL21; RPS29; SLIRP; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; RPS21; RPS15; RPL36; RPS28; C19orf43; RPL18A; LSM4; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006605 | protein targeting | 2.50 | 0.00e+00 | 0.00e+00 | 102/261 | RPL22; RPL11; ATPIF1; HPCA; RPS8; RPL5; RPS27; TOMM20; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; SEC63; RPS12; AKAP12; TOMM7; SEC61G; FIS1; SLC25A6; TIMM17B; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; YWHAZ; RPL8; RPS6; SEC61B; RPL12; RPL7A; RPLP2; RPL27A; RPS13; IMMP1L; TIMM10; RPS3; TIMM8B; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; DNAJC15; AKAP6; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; ARL6IP1; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; ROMO1; RPS21; RPS15; TIMM13; RPL36; RPS28; UBL5; RPL18A; UBA52; HOMER3; NDUFA13; PDCD5; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0099132 | ATP hydrolysis coupled cation transmembrane transport | 2.40 | 2.11e-03 | 4.08e-02 | 12/32 | ATP2B4; ATP5G3; ATP6V1F; ATP6V0E2; ATP5L; ATP5G2; ATP2B1; ATP5G1; ATP5H; ATP5A1; ATP5E; ATP5O |
| GO:0014074 | response to purine-containing compound | 2.30 | 1.51e-03 | 3.25e-02 | 14/39 | SEPN1; RYR2; IGFBP5; NDUFS4; HDAC2; EZR; AKAP9; HSPA5; RAPGEF1; HSP90B1; PEBP1; AKAP6; NME1; BSG |
| GO:0032409 | regulation of transporter activity | 2.29 | 5.80e-06 | 3.04e-04 | 29/81 | PARK7; SEPN1; HPCA; NOS1AP; ATP1B1; RYR2; FKBP1B; COX17; ATP1B3; SNCA; MEF2C; GLRX; ACTB; SRI; AKAP9; TMSB4X; GAL; FXYD6; KIF5B; KCNIP2; REM2; JPH4; AKAP6; CALM1; YWHAE; VAMP2; FKBP1A; FXYD7; HRC |
| GO:0010959 | regulation of metal ion transport | 2.26 | 8.88e-07 | 5.43e-05 | 35/99 | SEPN1; NKAIN1; HPCA; NOS1AP; ATP1B1; ATP2B4; AGT; RYR2; FKBP1B; GNAI2; ATP1B3; SNCA; GLRX; TSPAN13; SRI; AKAP9; G6PD; CHD7; GAL; FXYD6; KIF5B; KCNIP2; CD63; REM2; JPH4; AKAP6; CALM1; YWHAE; VAMP2; FKBP1A; NKAIN4; HOMER3; FXYD7; HRC; ATF4 |
| GO:0034765 | regulation of ion transmembrane transport | 2.13 | 1.19e-06 | 6.73e-05 | 39/117 | PARK7; SEPN1; HPCA; NOS1AP; ATP1B1; RGS2; ATP2B4; AGT; RYR2; FKBP1B; SCN3A; SCN9A; COX17; ATP1B3; SNCA; MEF2C; GLRX; TSPAN13; SRI; AKAP9; TMSB4X; G6PD; CHD7; GAL; FXYD6; KIF5B; KCNIP2; CD63; REM2; JPH4; AKAP6; CALM1; ARL6IP1; YWHAE; VAMP2; FKBP1A; CACNA1A; FXYD7; HRC |
| GO:0034764 | positive regulation of transmembrane transport | 2.04 | 7.18e-04 | 2.11e-02 | 21/66 | PARK7; NOS1AP; ATP1B1; AGT; RYR2; CAPN10; COX17; ATP1B3; SNCA; GLRX; SRI; AKAP9; TMSB4X; G6PD; GAL; KIF5B; KCNIP2; OSBPL8; AKAP6; CALM1; ARL6IP1 |
| GO:0021953 | central nervous system neuron differentiation | 2.02 | 2.57e-03 | 4.71e-02 | 17/54 | DRAXIN; LMO4; NRP2; MAP2; UQCRQ; ID4; SOX4; MDGA1; HSP90AB1; FZD1; MTPN; NFIB; C12orf57; MYCBP2; CBLN1; DCC; ADARB1 |
SETD5 : 24 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0070972 | protein localization to endoplasmic reticulum | 9.76 | 0.00e+00 | 0.00e+00 | 81/119 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; SEC63; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; HSPA5; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; KDELR1; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006413 | translational initiation | 8.04 | 0.00e+00 | 0.00e+00 | 88/157 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; EIF3K; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| GO:0002181 | cytoplasmic translation | 7.46 | 0.00e+00 | 0.00e+00 | 38/73 | RPL11; RPL31; RPL32; RPL15; RPL29; RPL24; RPL35A; RPL9; RPS23; RPL10A; RPL36A; RPL39; EIF4EBP1; RPL30; EIF3E; EIF3H; RPL8; EIF3F; EIF3M; RPL41; RPL6; RPLP0; RPS29; RPLP1; RPL26; RPL19; RPL38; RPL17; RPS21; EEF2; RPL36; RPS28; RPL18A; EIF3K; RPL18; RPL13A; EIF3D; EIF3L |
| GO:0090150 | establishment of protein localization to membrane | 5.90 | 0.00e+00 | 0.00e+00 | 84/204 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; GNB2L1; RPS18; RPS10; RPL10A; SEC63; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; TIMM10; BAD; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; VAMP2; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; TIMM13; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006401 | RNA catabolic process | 5.53 | 0.00e+00 | 0.00e+00 | 91/236 | RPL22; RPL11; YBX1; RPS8; SERBP1; RPL5; RPS27; RPS7; RPS27A; RPL31; SSB; RPL37A; RPL32; RPL15; RPSA; RPL14; PCBP4; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; EIF3E; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; HNRNPC; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; DDX5; RPL38; RPL17; RPS21; CIRBP; RPS15; LSM7; RPL36; RPS28; C19orf43; RPL18A; LSM4; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0006605 | protein targeting | 4.72 | 0.00e+00 | 0.00e+00 | 86/261 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; TOMM20; RPS7; RPS27A; RPL31; HSPD1; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; SEC63; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; TIMM10; RPS3; TIMM8B; RPS25; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; TIMM13; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| GO:0046683 | response to organophosphorus | 4.22 | 6.90e-05 | 3.38e-03 | 10/34 | JUN; HSPD1; NDUFS4; EZR; HSPA5; LDHA; HSP90B1; NME1; BSG; SOD1 |
| GO:0006959 | humoral immune response | 4.18 | 9.26e-04 | 2.83e-02 | 7/24 | HMGN2; RBPJ; RPL39; RPL30; GAPDH; C1QBP; RPS19 |
| GO:0001906 | cell killing | 4.10 | 4.65e-04 | 1.48e-02 | 8/28 | HMGN2; PRDX1; HSP90AB1; RPL30; BAD; GAPDH; VAMP2; RPS19 |
| GO:0014074 | response to purine-containing compound | 3.68 | 2.44e-04 | 8.96e-03 | 10/39 | JUN; HSPD1; NDUFS4; EZR; HSPA5; LDHA; HSP90B1; NME1; BSG; SOD1 |
| GO:0032355 | response to estradiol | 3.23 | 1.39e-03 | 4.04e-02 | 9/40 | AGT; IGFBP2; H2AFZ; CASP3; BAD; GSTP1; SSTR2; EEF2; CALR |
| GO:0006090 | pyruvate metabolic process | 3.22 | 4.24e-04 | 1.43e-02 | 11/49 | ENO1; SLC16A1; VDAC1; LDHA; PGAM1; GAPDH; TPI1; LDHB; PKM; ALDOA; BSG |
| GO:0035821 | modification of morphology or physiology of other organism | 2.99 | 1.43e-03 | 4.04e-02 | 10/48 | HMGN2; JUN; NUCKS1; SLC25A6; RPL30; BAD; CFL1; GAPDH; PPIB; RPS19 |
| GO:1902600 | proton transmembrane transport | 2.98 | 5.95e-05 | 3.12e-03 | 16/77 | PARK7; COX7A2L; ATP5G3; COX7C; ATP6V1G2; NDUFA4; COX7B; COX8A; COX6A1; COX5A; ATP5G1; ATP5A1; ATP5D; COX6B1; MT-ND4; MT-CYB |
| GO:0071826 | ribonucleoprotein complex subunit organization | 2.79 | 8.49e-09 | 7.79e-07 | 36/185 | RPL11; RPL5; RPS27; RPSA; RPL24; RPS23; RPS14; NPM1; RPS10; HSP90AB1; RBMX; RPL10; EIF3E; EIF3H; RPL12; EIF3F; EIF3M; SNRPF; RPL6; RPLP0; C1QBP; RPL23A; RPL38; SNRPD1; CIRBP; RPS15; RPS28; LSM4; EIF3K; RPS19; GLTSCR2; RPL13A; RPS5; EIF3D; EIF3L; RPL3 |
| GO:0016072 | rRNA metabolic process | 2.68 | 1.58e-07 | 1.29e-05 | 32/171 | RPL11; RPS8; RPL5; RPS27; RPS7; NCL; RPSA; RPL14; RPL35A; RPS14; NHP2; RPL10A; RPL7; RPS6; RPL35; RPL7A; DDX21; RPS24; PA2G4; GTF3A; RPS17; RPS2; RPL26; RPL27; RPS21; RPS15; RPS28; C19orf43; RPS16; RPS19; GLTSCR2; RPS9 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 2.52 | 1.10e-11 | 1.15e-09 | 58/330 | RPL11; RPS8; RPL5; RPS27; RPS7; RPSA; RPL14; RPL24; RPL35A; RPS23; RPS14; NPM1; NHP2; RPS10; RPL10A; HSP90AB1; RBMX; RPL10; RPL7; EIF3E; EIF3H; RPS6; RPL35; RPL12; RPL7A; EIF3F; EIF3M; DDX21; RPS24; PA2G4; SNRPF; RPL6; RPLP0; RAN; GTF3A; RPS17; RPS2; C1QBP; RPL26; RPL23A; RPL27; RPL38; SNRPD1; RPS21; CIRBP; RPS15; RPS28; LSM4; EIF3K; RPS16; RPS19; GLTSCR2; RPL13A; RPS9; RPS5; EIF3D; EIF3L; RPL3 |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.50 | 5.44e-07 | 3.99e-05 | 33/189 | PARK7; ENO1; UQCRH; COX7A2L; ATP5G3; IMPDH2; NDUFS4; COX7C; NDUFA4; CYCS; NDUFB2; NDUFA1; LDHA; BAD; PGAM1; GAPDH; TPI1; COX6A1; RAN; PKM; COX5A; NME4; NDUFB10; ALDOA; DCTPP1; ATP5G1; NME1; ATP5A1; ATP5D; MT-ND2; MT-ND3; MT-ND4; MT-CYB |
| GO:0046677 | response to antibiotic | 2.49 | 7.45e-05 | 3.42e-03 | 21/121 | PARK7; JUN; FKBP1B; HSPD1; EEF1B2; CASP3; GNB2L1; NEFL; TNC; HSPA5; LDHA; BAD; GSTP1; RPS3; HNRNPA1; RPL23; PYCR1; EEF2; SOD1; MT-ND4; MT-CYB |
| GO:0009123 | nucleoside monophosphate metabolic process | 2.28 | 9.40e-06 | 5.75e-04 | 31/195 | PARK7; ENO1; UQCRH; COX7A2L; ATP5G3; IMPDH2; PAICS; NDUFS4; COX7C; HDDC2; NDUFA4; CYCS; NDUFB2; NDUFA1; LDHA; BAD; PGAM1; GAPDH; TPI1; COX6A1; PKM; COX5A; NDUFB10; ALDOA; ATP5G1; ATP5A1; ATP5D; MT-ND2; MT-ND3; MT-ND4; MT-CYB |
| GO:0009636 | response to toxic substance | 2.21 | 8.96e-05 | 3.65e-03 | 26/169 | PARK7; PRDX1; JUN; FKBP1B; SCN9A; HSPD1; EEF1B2; CASP3; GNB2L1; PRDX4; NEFL; TNC; LDHA; BAD; GSTP1; RPS3; RGS10; HNRNPA1; ESD; PYCR1; EEF2; PRDX2; UBA52; SOD1; MT-ND4; MT-CYB |
| GO:0010608 | posttranscriptional regulation of gene expression | 2.11 | 1.95e-06 | 1.30e-04 | 42/286 | YBX1; SERBP1; RPL5; SRP9; H3F3A; RPS27A; NCL; PCBP4; EIF4A2; RPS14; NPM1; GNB2L1; SOX4; HNRNPA2B1; RBM3; RPS4X; RPL10; EIF4EBP1; POLR2K; PABPC1; EIF3E; EIF3H; RPS3; GAPDH; PA2G4; UBC; RAN; HNRNPC; C1QBP; EIF5A; RPL26; DDX5; RPL38; CIRBP; EEF2; CALR; UBA52; POLR2I; EIF3K; RPL13A; RPS9; EIF3D |
| GO:0034248 | regulation of cellular amide metabolic process | 2.02 | 8.05e-05 | 3.48e-03 | 32/227 | RPL5; SRP9; NCL; EIF4A2; CASP3; RPS14; NPM1; GNB2L1; SOX4; EEF1A1; RBM3; RPS4X; RPL10; EIF4EBP1; PABPC1; EIF3E; EIF3H; RPS3; PGAM1; GAPDH; PA2G4; C1QBP; EIF5A; RPL26; RPL38; CIRBP; EEF2; CALR; EIF3K; RPL13A; RPS9; EIF3D |
| GO:0009259 | ribonucleotide metabolic process | 2.01 | 3.25e-05 | 1.83e-03 | 36/257 | PARK7; ENO1; UQCRH; COX7A2L; ATP5G3; IMPDH2; PAICS; NDUFS4; COX7C; HINT1; ELOVL4; NDUFA4; CYCS; NDUFB2; NDUFA1; LDHA; BAD; PGAM1; SCD; GAPDH; TPI1; COX6A1; RAN; PKM; COX5A; NME4; NDUFB10; ALDOA; ATP5G1; NME1; ATP5A1; ATP5D; MT-ND2; MT-ND3; MT-ND4; MT-CYB |
| Marker | ADNP | ARID1B | ASH1L | CHD2 | DYRK1A | PTEN | SETD5 |
| Signif_GO_terms | 25 | 20 | 33 | 21 | 2 | 22 | 24 |
6.2 Reactome Pathway Over-Representation Analysis
Gene sets: The Reactome pathway database.
ADNP : 137 significant Reactome pathways| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-156902 | Peptide chain elongation | 4.71 | 0.00e+00 | 0.00e+00 | 56/84 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; EEF1A1; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 4.68 | 0.00e+00 | 0.00e+00 | 57/86 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; EEF1B2; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; EEF1A1; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-192823 | Viral mRNA Translation | 4.68 | 0.00e+00 | 0.00e+00 | 55/83 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-2408557 | Selenocysteine synthesis | 4.57 | 0.00e+00 | 0.00e+00 | 55/85 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-445355 | Smooth Muscle Contraction | 4.57 | 2.28e-06 | 5.45e-05 | 11/17 | TPM3; ANXA6; CALD1; TPM2; VCL; MYL6B; MYL6; MYL12A; MYL12B; TPM4; CALM3 |
| R-HSA-72764 | Eukaryotic Translation Termination | 4.52 | 0.00e+00 | 0.00e+00 | 55/86 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 4.41 | 0.00e+00 | 0.00e+00 | 55/88 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-389960 | Formation of tubulin folding intermediates by CCT/TriC | 4.41 | 1.07e-05 | 2.42e-04 | 10/16 | CCT7; TUBA4A; CCT5; TUBB2A; TCP1; TUBB4B; TUBA1B; TUBA1A; TUBB3; TUBB4A |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 4.14 | 0.00e+00 | 0.00e+00 | 61/104 | RPL22; DDOST; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; SSR4; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; SPCS2; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; SRP14; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; SEC11C; RPN2; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 4.09 | 0.00e+00 | 0.00e+00 | 55/95 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-8955332 | Carboxyterminal post-translational modifications of tubulin | 4.04 | 2.09e-04 | 3.27e-03 | 8/14 | TUBA4A; TUBB2A; TUBB4B; TUBA1B; TUBA1A; TUBB3; TPGS2; TUBB4A |
| R-HSA-2408522 | Selenoamino acid metabolism | 3.92 | 0.00e+00 | 0.00e+00 | 55/99 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-190828 | Gap junction trafficking | 3.92 | 4.45e-05 | 8.34e-04 | 10/18 | TUBA4A; TUBB2A; CLTA; DNM1; TUBB4B; TUBA1B; TUBA1A; TUBB3; ACTG1; TUBB4A |
| R-HSA-5626467 | RHO GTPases activate IQGAPs | 3.92 | 4.45e-05 | 8.34e-04 | 10/18 | CDC42; TUBA4A; TUBB2A; TUBB4B; TUBA1B; TUBA1A; TUBB3; ACTG1; TUBB4A; CALM3 |
| R-HSA-373076 | Class A/1 (Rhodopsin-like receptors) | 3.85 | 1.92e-03 | 1.41e-02 | 6/11 | AGT; SST; GAL; PSAP; NMB; SSTR2 |
| R-HSA-983170 | Antigen Presentation: Folding, assembly and peptide loading of class I MHC | 3.85 | 1.92e-03 | 1.41e-02 | 6/11 | HLA-A; BCAP31; HSPA5; PDIA3; B2M; CALR |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 3.83 | 0.00e+00 | 0.00e+00 | 57/105 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; MAGOHB; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; EIF4A3; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 3.83 | 0.00e+00 | 0.00e+00 | 57/105 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; MAGOHB; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; EIF4A3; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-389977 | Post-chaperonin tubulin folding pathway | 3.80 | 8.65e-04 | 8.37e-03 | 7/13 | TUBA4A; TUBB2A; TUBB4B; TUBA1B; TUBA1A; TUBB3; TUBB4A |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 3.77 | 0.00e+00 | 0.00e+00 | 56/105 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; EIF2S2; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 3.73 | 0.00e+00 | 0.00e+00 | 56/106 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; EIF2S2; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-157858 | Gap junction trafficking and regulation | 3.72 | 8.23e-05 | 1.51e-03 | 10/19 | TUBA4A; TUBB2A; CLTA; DNM1; TUBB4B; TUBA1B; TUBA1A; TUBB3; ACTG1; TUBB4A |
| R-HSA-72613 | Eukaryotic Translation Initiation | 3.56 | 0.00e+00 | 0.00e+00 | 57/113 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; EIF4EBP1; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; EIF2S2; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 3.56 | 0.00e+00 | 0.00e+00 | 57/113 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; EIF4EBP1; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; EIF2S2; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-156590 | Glutathione conjugation | 3.53 | 3.38e-03 | 2.08e-02 | 6/12 | GSTM3; MGST3; MGST2; GGCT; GSTP1; ESD |
| R-HSA-888590 | GABA synthesis, release, reuptake and degradation | 3.53 | 7.60e-03 | 3.96e-02 | 5/10 | CPLX1; STX1A; HSPA8; SNAP25; RAB3A |
| R-HSA-5663084 | Diseases of carbohydrate metabolism | 3.53 | 7.60e-03 | 3.96e-02 | 5/10 | RPS27A; TALDO1; UBC; G6PC3; DCXR |
| R-HSA-390522 | Striated Muscle Contraction | 3.53 | 7.60e-03 | 3.96e-02 | 5/10 | TPM3; TPM2; ACTC1; TNNC2; TPM4 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 3.50 | 0.00e+00 | 0.00e+00 | 56/113 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; HSP90AA1; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 3.46 | 0.00e+00 | 0.00e+00 | 73/149 | RPL22; RPL11; RPS8; RPL5; PSMA5; PSMD4; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; PSMD6; ROBO1; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; PSMB1; PSMC2; RPS4X; RPL36A; RPL10; RPL7; TCEB1; RPL30; RPS6; RPL35; RPL7A; PSMD13; RPL27A; RPS13; RPS24; MAGOHB; RPS26; RPL41; RPL6; RPLP0; UBC; RPL21; PSME2; RPS29; PSMA3; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; RPL26; RPL23A; PSMB3; RPL19; RPL27; PSMC5; EIF4A3; PSMA7; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-500792 | GPCR ligand binding | 3.40 | 2.36e-05 | 4.96e-04 | 13/27 | GNG5; AGT; GNG4; SST; GRM4; FZD1; GABBR2; GAL; PSAP; NMB; SSTR2; GNG8; PTH2 |
| R-HSA-168255 | Influenza Life Cycle | 3.39 | 0.00e+00 | 0.00e+00 | 58/121 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; CLTA; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; HSP90AA1; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; CALR; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 3.38 | 1.80e-08 | 5.17e-07 | 23/48 | RPS8; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPS6; RPS13; RPS24; RPS26; RPS29; RPS17; RPS2; RPS15A; EIF2S2; RPS15; RPS28; RPS16; RPS19; RPS5 |
| R-HSA-389958 | Cooperation of Prefoldin and TriC/CCT in actin and tubulin folding | 3.38 | 1.11e-04 | 1.99e-03 | 11/23 | CCT7; TUBA4A; CCT5; TUBB2A; TCP1; TUBB4B; TUBA1B; TUBA1A; PFDN5; TUBB3; TUBB4A |
| R-HSA-70263 | Gluconeogenesis | 3.35 | 5.23e-04 | 6.17e-03 | 9/19 | ENO1; MDH1; MDH2; PGK1; PGAM1; TPI1; ALDOA; ENO3; G6PC3 |
| R-HSA-397014 | Muscle contraction | 3.34 | 2.96e-09 | 9.11e-08 | 26/55 | TPM3; ATP1B1; RYR2; SCN3A; ATP1B3; CAMK2D; ANXA6; CAMK2B; SRI; AKAP9; CALD1; TPM2; FXYD6; VCL; KCNIP2; CACNB3; MYL6B; MYL6; ACTC1; MYL12A; MYL12B; TNNC2; TPM4; FXYD1; FXYD7; CALM3 |
| R-HSA-168254 | Influenza Infection | 3.31 | 0.00e+00 | 0.00e+00 | 60/128 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; SLC25A6; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; CLTA; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; HSP90AA1; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; KPNA2; RPS15; RPS28; CALR; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-389957 | Prefoldin mediated transfer of substrate to CCT/TriC | 3.21 | 3.86e-04 | 5.19e-03 | 10/22 | CCT7; TUBA4A; CCT5; TUBB2A; TCP1; TUBB4B; TUBA1A; PFDN5; TUBB3; TUBB4A |
| R-HSA-5578775 | Ion homeostasis | 3.07 | 5.97e-04 | 6.51e-03 | 10/23 | ATP1B1; RYR2; ATP1B3; CAMK2D; CAMK2B; SRI; FXYD6; FXYD1; FXYD7; CALM3 |
| R-HSA-376176 | Signaling by ROBO receptors | 3.06 | 0.00e+00 | 0.00e+00 | 77/178 | RPL22; CDC42; RPL11; RPS8; RPL5; PSMA5; PSMD4; RPS27A; RPL31; RPL37A; SRGAP3; RPL32; RPL15; RPSA; RPL14; PSMD6; ROBO1; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; CAP2; RPS10; RPS12; PSMB1; PSMC2; RPS4X; RPL36A; RPL10; RPL7; TCEB1; RPL30; RPS6; RPL35; RPL7A; PSMD13; RPL27A; RPS13; RPS24; MAGOHB; RPS26; RPL41; SRGAP1; RPL6; RPLP0; UBC; RPL21; PSME2; RPS29; PSMA3; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; RPL26; RPL23A; PSMB3; RPL19; RPL27; PSMC5; EIF4A3; PSMA7; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 3.03 | 1.30e-07 | 3.49e-06 | 24/56 | RPS8; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; EIF4EBP1; RPS6; RPS13; RPS24; RPS26; RPS29; RPS17; RPS2; RPS15A; EIF2S2; RPS15; RPS28; RPS16; RPS19; RPS5 |
| R-HSA-8852276 | The role of GTSE1 in G2/M progression after G2 checkpoint | 2.95 | 4.10e-07 | 1.01e-05 | 23/55 | PSMA5; PSMD4; RPS27A; TUBA4A; PSMD6; TUBB2A; PSMB1; PSMC2; TUBB4B; PSMD13; TUBA1B; TUBA1A; UBC; PSME2; PSMA3; HSP90AA1; PSMA4; PSMD7; TUBB3; PSMB3; PSMC5; PSMA7; TUBB4A |
| R-HSA-72649 | Translation initiation complex formation | 2.95 | 4.10e-07 | 1.01e-05 | 23/55 | RPS8; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPS6; RPS13; RPS24; RPS26; RPS29; RPS17; RPS2; RPS15A; EIF2S2; RPS15; RPS28; RPS16; RPS19; RPS5 |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 2.95 | 4.10e-07 | 1.01e-05 | 23/55 | RPS8; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPS6; RPS13; RPS24; RPS26; RPS29; RPS17; RPS2; RPS15A; EIF2S2; RPS15; RPS28; RPS16; RPS19; RPS5 |
| R-HSA-437239 | Recycling pathway of L1 | 2.92 | 2.97e-04 | 4.26e-03 | 12/29 | TUBA4A; TUBB2A; EZR; DPYSL2; CLTA; DNM1; TUBB4B; TUBA1B; TUBA1A; TUBB3; ACTG1; TUBB4A |
| R-HSA-977225 | Amyloid fiber formation | 2.88 | 6.23e-04 | 6.71e-03 | 11/27 | H3F3A; RPS27A; SNCA; HIST1H4C; HIST1H2AC; UBE2L6; UBC; ITM2B; B2M; CST3; PSENEN |
| R-HSA-5576891 | Cardiac conduction | 2.83 | 1.44e-04 | 2.43e-03 | 14/35 | ATP1B1; RYR2; SCN3A; ATP1B3; CAMK2D; CAMK2B; SRI; AKAP9; FXYD6; KCNIP2; CACNB3; FXYD1; FXYD7; CALM3 |
| R-HSA-114608 | Platelet degranulation | 2.79 | 3.34e-05 | 6.54e-04 | 17/43 | TAGLN2; TUBA4A; MANF; WDR1; SPARC; PPIA; MAGED2; HSPA5; APLP2; PSAP; VCL; CD63; SCG3; ALDOA; LGALS3BP; CALM3; SOD1 |
| R-HSA-76005 | Response to elevated platelet cytosolic Ca2+ | 2.79 | 3.34e-05 | 6.54e-04 | 17/43 | TAGLN2; TUBA4A; MANF; WDR1; SPARC; PPIA; MAGED2; HSPA5; APLP2; PSAP; VCL; CD63; SCG3; ALDOA; LGALS3BP; CALM3; SOD1 |
| R-HSA-983189 | Kinesins | 2.76 | 2.73e-03 | 1.81e-02 | 9/23 | TUBA4A; TUBB2A; TUBB4B; KIF5B; TUBA1B; TUBA1A; TUBB3; KIF19; TUBB4A |
| R-HSA-163210 | Formation of ATP by chemiosmotic coupling | 2.75 | 8.46e-03 | 4.33e-02 | 7/18 | ATP5F1; ATP5G3; ATP5C1; ATP5G2; ATP5B; ATP5H; ATP5J |
| R-HSA-1236974 | ER-Phagosome pathway | 2.68 | 2.24e-05 | 4.83e-04 | 19/50 | PSMA5; PSMD4; RPS27A; PSMD6; HLA-A; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PDIA3; B2M; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7; CALR |
| R-HSA-936837 | Ion transport by P-type ATPases | 2.65 | 3.83e-03 | 2.27e-02 | 9/24 | ATP1B1; ATP1B3; CAMK2D; CAMK2B; SRI; FXYD6; FXYD1; FXYD7; CALM3 |
| R-HSA-1236975 | Antigen processing-Cross presentation | 2.63 | 3.11e-05 | 6.38e-04 | 19/51 | PSMA5; PSMD4; RPS27A; PSMD6; HLA-A; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PDIA3; B2M; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7; CALR |
| R-HSA-180534 | Vpu mediated degradation of CD4 | 2.63 | 1.35e-04 | 2.32e-03 | 16/43 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69229 | Ubiquitin-dependent degradation of Cyclin D1 | 2.57 | 1.84e-04 | 2.94e-03 | 16/44 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; CDK4; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-75815 | Ubiquitin-dependent degradation of Cyclin D | 2.57 | 1.84e-04 | 2.94e-03 | 16/44 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; CDK4; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-174113 | SCF-beta-TrCP mediated degradation of Emi1 | 2.57 | 1.84e-04 | 2.94e-03 | 16/44 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-187577 | SCF(Skp2)-mediated degradation of p27/p21 | 2.56 | 1.25e-04 | 2.20e-03 | 17/47 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; CDK4; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 2.53 | 2.22e-16 | 8.31e-15 | 75/209 | RPL22; RPL11; RPS8; RPL5; PSMA5; PHGDH; PSMD4; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; PDHB; PSMD6; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; PSMB1; ASNS; PSMC2; PDHA1; HSD17B10; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; PSMD13; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; PXMP2; RPL21; PSME2; RPS29; PSMA3; CKB; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; RPL26; RPL23A; PSMB3; RPL19; RPL27; PSMC5; PSMA7; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 2.52 | 5.24e-12 | 1.80e-10 | 55/154 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-8854050 | FBXL7 down-regulates AURKA during mitotic entry and in early mitosis | 2.51 | 2.49e-04 | 3.77e-03 | 16/45 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-373760 | L1CAM interactions | 2.49 | 8.41e-06 | 1.96e-04 | 24/68 | CNTN2; SCN3A; TUBA4A; ALCAM; GAP43; ANK2; TUBB2A; CSNK2B; EZR; DCX; DPYSL2; SDCBP; CLTA; DNM1; VAV2; TUBB4B; NCAM1; HSPA8; ANK3; TUBA1B; TUBA1A; TUBB3; ACTG1; TUBB4A |
| R-HSA-211733 | Regulation of activated PAK-2p34 by proteasome mediated degradation | 2.46 | 4.92e-04 | 5.89e-03 | 15/43 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69601 | Ubiquitin Mediated Degradation of Phosphorylated Cdc25A | 2.46 | 4.92e-04 | 5.89e-03 | 15/43 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69610 | p53-Independent DNA Damage Response | 2.46 | 4.92e-04 | 5.89e-03 | 15/43 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69613 | p53-Independent G1/S DNA damage checkpoint | 2.46 | 4.92e-04 | 5.89e-03 | 15/43 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5610787 | Hedgehog 'off' state | 2.46 | 1.12e-05 | 2.48e-04 | 24/69 | PSMA5; PSMD4; RPS27A; TUBA4A; PSMD6; IFT57; SKP1; TUBB2A; PSMB1; PSMC2; TUBB4B; PSMD13; TUBA1B; TUBA1A; UBC; PSME2; PSMA3; PSMA4; PSMD7; TUBB3; PSMB3; PSMC5; PSMA7; TUBB4A |
| R-HSA-180585 | Vif-mediated degradation of APOBEC3G | 2.46 | 3.33e-04 | 4.70e-03 | 16/46 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; TCEB1; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 2.43 | 3.02e-11 | 9.63e-10 | 55/160 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-422475 | Axon guidance | 2.41 | 0.00e+00 | 0.00e+00 | 112/328 | RPL22; CDC42; RPL11; RPS8; RPL5; PSMA5; PSMD4; SHC1; EFNA3; CNTN2; RPS27A; RPL31; SCN3A; RPL37A; ARPC2; TUBA4A; SRGAP3; RPL32; RPL15; RPSA; RPL14; PSMD6; ROBO1; ALCAM; GAP43; RPL35A; RPL9; RPL34; ANK2; RPS3A; RPS23; RPS14; TUBB2A; CAP2; CSNK2B; RPS10; RPS12; EZR; PSMB1; ARPC1A; PSMC2; RPS4X; RPL36A; DCX; RPL10; DPYSL2; SDCBP; RPL7; TCEB1; RPL30; RPS6; CLTA; RPL35; DNM1; RPL7A; VAV2; TUBB4B; PSMD13; RPL27A; RPS13; NCAM1; HSPA8; ANK3; RPS24; MAGOHB; CACNB3; TUBA1B; TUBA1A; RPS26; RPL41; MYL6; SRGAP1; RPL6; RPLP0; UBC; RPL21; PSME2; RPS29; PSMA3; HSP90AA1; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; TUBB3; EFNB3; RPL26; RPL23A; PSMB3; RPL19; RPL27; PSMC5; EIF4A3; ACTG1; MYL12A; MYL12B; PSMA7; RPS15; TUBB4A; RPS28; RPL18A; PSENEN; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72312 | rRNA processing | 2.41 | 2.66e-11 | 8.82e-10 | 56/164 | RPL22; RPL11; RPS8; RPL5; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL35A; RPL9; RPL34; RPS3A; RPS23; RPS14; RPS10; RPS12; HSD17B10; RPS4X; RPL36A; RPL10; RPL7; RPL30; RPS6; RPL35; RPL7A; RPL27A; RPS13; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL19; RPL27; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-169911 | Regulation of Apoptosis | 2.41 | 6.52e-04 | 6.93e-03 | 15/44 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-450408 | AUF1 (hnRNP D0) binds and destabilizes mRNA | 2.40 | 4.41e-04 | 5.58e-03 | 16/47 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; HSPA8; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5607761 | Dectin-1 mediated noncanonical NF-kB signaling | 2.40 | 4.41e-04 | 5.58e-03 | 16/47 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5676590 | NIK-->noncanonical NF-kB signaling | 2.40 | 4.41e-04 | 5.58e-03 | 16/47 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-9604323 | Negative regulation of NOTCH4 signaling | 2.40 | 4.41e-04 | 5.58e-03 | 16/47 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-68949 | Orc1 removal from chromatin | 2.35 | 5.76e-04 | 6.36e-03 | 16/48 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-1169091 | Activation of NF-kappaB in B cells | 2.35 | 5.76e-04 | 6.36e-03 | 16/48 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-4608870 | Asymmetric localization of PCP proteins | 2.35 | 5.76e-04 | 6.36e-03 | 16/48 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; FZD1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5610780 | Degradation of GLI1 by the proteasome | 2.35 | 5.76e-04 | 6.36e-03 | 16/48 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5668541 | TNFR2 non-canonical NF-kB pathway | 2.35 | 5.76e-04 | 6.36e-03 | 16/48 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-349425 | Autodegradation of the E3 ubiquitin ligase COP1 | 2.35 | 8.53e-04 | 8.35e-03 | 15/45 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-4641258 | Degradation of DVL | 2.35 | 8.53e-04 | 8.35e-03 | 15/45 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-68827 | CDT1 association with the CDC6:ORC:origin complex | 2.35 | 8.53e-04 | 8.35e-03 | 15/45 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-8941858 | Regulation of RUNX3 expression and activity | 2.35 | 8.53e-04 | 8.35e-03 | 15/45 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-1236978 | Cross-presentation of soluble exogenous antigens (endosomes) | 2.35 | 1.88e-03 | 1.41e-02 | 13/39 | PSMA5; PSMD4; PSMD6; PSMB1; PSMC2; PSMD13; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5610783 | Degradation of GLI2 by the proteasome | 2.31 | 7.46e-04 | 7.65e-03 | 16/49 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5610785 | GLI3 is processed to GLI3R by the proteasome | 2.31 | 7.46e-04 | 7.65e-03 | 16/49 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-4641257 | Degradation of AXIN | 2.30 | 1.10e-03 | 1.05e-02 | 15/46 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-3371497 | HSP90 chaperone cycle for steroid hormone receptors (SHR) | 2.29 | 3.61e-03 | 2.16e-02 | 12/37 | TUBA4A; TUBB2A; DNAJA1; TUBB4B; HSPA8; TUBA1B; TUBA1A; DYNLL1; ACTR10; HSP90AA1; TUBB3; TUBB4A |
| R-HSA-5362768 | Hh mutants that don't undergo autocatalytic processing are degraded by ERAD | 2.25 | 1.42e-03 | 1.24e-02 | 15/47 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5387390 | Hh mutants abrogate ligand secretion | 2.25 | 1.42e-03 | 1.24e-02 | 15/47 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-68867 | Assembly of the pre-replicative complex | 2.25 | 1.42e-03 | 1.24e-02 | 15/47 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-8939236 | RUNX1 regulates transcription of genes involved in differentiation of HSCs | 2.24 | 3.79e-04 | 5.18e-03 | 19/60 | PSMA5; PSMD4; H3F3A; RPS27A; PSMD6; HIST1H4C; HIST1H2AC; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; TCF12; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-8939902 | Regulation of RUNX2 expression and activity | 2.22 | 1.22e-03 | 1.13e-02 | 16/51 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69541 | Stabilization of p53 | 2.21 | 1.80e-03 | 1.41e-02 | 15/48 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-1445148 | Translocation of SLC2A4 (GLUT4) to the plasma membrane | 2.20 | 2.66e-03 | 1.79e-02 | 14/45 | RAB13; YWHAQ; TUBA4A; TUBB2A; TUBB4B; TUBA1B; TUBA1A; RAB11A; TUBB3; YWHAE; ACTG1; YWHAB; TUBB4A; CALM3 |
| R-HSA-1168372 | Downstream signaling events of B Cell Receptor (BCR) | 2.19 | 7.06e-04 | 7.41e-03 | 18/58 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PPIA; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7; CALM3 |
| R-HSA-5663205 | Infectious disease | 2.18 | 6.49e-14 | 2.33e-12 | 85/275 | RPL22; RPL11; RPS8; RPL5; PSMA5; PSMD4; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; PSMD6; RPL35A; RPL9; RPL34; RPS3A; RPS23; SKP1; RPS14; HMGA1; RPS10; RPS12; PSMB1; PPIA; STX1A; PSMC2; SLC25A6; RPS4X; RPL36A; RPL10; RPL7; TCEB1; RPL30; RPS6; CHMP5; CLTA; RPL35; RPL7A; PSMD13; RPL27A; RPS13; TSG101; RPS24; RPS26; RPL41; RPL6; RPLP0; UBC; RPL21; PSME2; RPS29; PSMA3; HSP90AA1; B2M; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; RPL26; RPL23A; PSMB3; RPL19; RPL27; PSMC5; KPNA2; SNAP25; PSMA7; RPS15; RPS28; CALR; RPL18A; RPS16; RPS19; CALM3; RPL18; RPL13A; RPS5; CHMP2A; RPL3 |
| R-HSA-69563 | p53-Dependent G1 DNA Damage Response | 2.17 | 1.53e-03 | 1.26e-02 | 16/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ZNF385A; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69580 | p53-Dependent G1/S DNA damage checkpoint | 2.17 | 1.53e-03 | 1.26e-02 | 16/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ZNF385A; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69615 | G1/S DNA Damage Checkpoints | 2.17 | 1.53e-03 | 1.26e-02 | 16/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ZNF385A; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-1234176 | Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha | 2.17 | 1.53e-03 | 1.26e-02 | 16/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; TCEB1; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-174084 | Autodegradation of Cdh1 by Cdh1:APC/C | 2.17 | 1.53e-03 | 1.26e-02 | 16/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5678895 | Defective CFTR causes cystic fibrosis | 2.16 | 2.26e-03 | 1.62e-02 | 15/49 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-983712 | Ion channel transport | 2.16 | 8.85e-04 | 8.47e-03 | 18/59 | ATP6V0B; ATP1B1; RYR2; RPS27A; ATP1B3; CAMK2D; CAMK2B; SRI; ATP6AP1; ATP6V1G1; FXYD6; ASIC1; UBC; ATP6V0D1; TTYH2; FXYD1; FXYD7; CALM3 |
| R-HSA-176408 | Regulation of APC/C activators between G1/S and early anaphase | 2.14 | 1.30e-03 | 1.18e-02 | 17/56 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-350562 | Regulation of ornithine decarboxylase (ODC) | 2.14 | 4.93e-03 | 2.76e-02 | 13/43 | PSMA5; PSMD4; PSMD6; PSMB1; PSMC2; PSMD13; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-2871837 | FCERI mediated NF-kB activation | 2.13 | 1.92e-03 | 1.41e-02 | 16/53 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69017 | CDK-mediated phosphorylation and removal of Cdc6 | 2.13 | 1.92e-03 | 1.41e-02 | 16/53 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-202424 | Downstream TCR signaling | 2.13 | 1.92e-03 | 1.41e-02 | 16/53 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-390466 | Chaperonin-mediated protein folding | 2.13 | 1.92e-03 | 1.41e-02 | 16/53 | GNG5; GNG4; CCT7; TUBA4A; GNAI2; CCT5; TUBB2A; CSNK2B; TCP1; TUBB4B; TUBA1B; TUBA1A; PFDN5; TUBB3; TUBB4A; GNG8 |
| R-HSA-5358346 | Hedgehog ligand biogenesis | 2.12 | 2.82e-03 | 1.85e-02 | 15/50 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69202 | Cyclin E associated events during G1/S transition | 2.11 | 1.62e-03 | 1.28e-02 | 17/57 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; CDK4; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69656 | Cyclin A:Cdk2-associated events at S phase entry | 2.11 | 1.62e-03 | 1.28e-02 | 17/57 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; CDK4; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-174143 | APC/C-mediated degradation of cell cycle proteins | 2.11 | 1.62e-03 | 1.28e-02 | 17/57 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-453276 | Regulation of mitotic cell cycle | 2.11 | 1.62e-03 | 1.28e-02 | 17/57 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-174154 | APC/C:Cdc20 mediated degradation of Securin | 2.09 | 2.38e-03 | 1.64e-02 | 16/54 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-174178 | APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 | 2.09 | 2.38e-03 | 1.64e-02 | 16/54 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-174184 | Cdc20:Phospho-APC/C mediated degradation of Cyclin A | 2.09 | 2.38e-03 | 1.64e-02 | 16/54 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-179419 | APC:Cdc20 mediated degradation of cell cycle proteins prior to satisfation of the cell cycle checkpoint | 2.09 | 2.38e-03 | 1.64e-02 | 16/54 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5619084 | ABC transporter disorders | 2.08 | 3.49e-03 | 2.13e-02 | 15/51 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-9013694 | Signaling by NOTCH4 | 2.07 | 2.00e-03 | 1.46e-02 | 17/58 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7; PSENEN |
| R-HSA-5358351 | Signaling by Hedgehog | 2.07 | 2.60e-04 | 3.86e-03 | 24/82 | PSMA5; PSMD4; RPS27A; TUBA4A; PSMD6; IFT57; SKP1; TUBB2A; PSMB1; PSMC2; TUBB4B; PSMD13; TUBA1B; TUBA1A; UBC; PSME2; PSMA3; PSMA4; PSMD7; TUBB3; PSMB3; PSMC5; PSMA7; TUBB4A |
| R-HSA-8948751 | Regulation of PTEN stability and activity | 2.05 | 2.92e-03 | 1.87e-02 | 16/55 | PSMA5; PSMD4; RPS27A; PSMD6; CSNK2B; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-176409 | APC/C:Cdc20 mediated degradation of mitotic proteins | 2.05 | 2.92e-03 | 1.87e-02 | 16/55 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-176814 | Activation of APC/C and APC/C:Cdc20 mediated degradation of mitotic proteins | 2.05 | 2.92e-03 | 1.87e-02 | 16/55 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5658442 | Regulation of RAS by GAPs | 2.04 | 4.28e-03 | 2.44e-02 | 15/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69002 | DNA Replication Pre-Initiation | 2.04 | 4.28e-03 | 2.44e-02 | 15/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-5632684 | Hedgehog 'on' state | 2.04 | 4.28e-03 | 2.44e-02 | 15/52 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-69052 | Switching of origins to a post-replicative state | 2.04 | 2.45e-03 | 1.66e-02 | 17/59 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; ANAPC5; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-9020702 | Interleukin-1 signaling | 2.04 | 2.45e-03 | 1.66e-02 | 17/59 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PELI2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7 |
| R-HSA-76002 | Platelet activation, signaling and aggregation | 2.03 | 2.67e-04 | 3.89e-03 | 25/87 | PRKCZ; CDC42; GNG5; SHC1; TAGLN2; GNG4; TUBA4A; GNAI2; MANF; WDR1; SPARC; PPIA; MAGED2; HSPA5; VAV2; APLP2; PSAP; VCL; CD63; SCG3; ALDOA; LGALS3BP; CALM3; GNG8; SOD1 |
| R-HSA-382556 | ABC-family proteins mediated transport | 2.02 | 3.57e-03 | 2.15e-02 | 16/56 | PSMA5; PSMD4; RPS27A; PSMD6; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; EIF2S2; PSMA7 |
| R-HSA-5607764 | CLEC7A (Dectin-1) signaling | 2.00 | 2.98e-03 | 1.87e-02 | 17/60 | PSMA5; PSMD4; RPS27A; PSMD6; SKP1; PSMB1; PSMC2; PSMD13; UBC; PSME2; PSMA3; PSMA4; PSMD7; PSMB3; PSMC5; PSMA7; CALM3 |
| R-HSA-1266738 | Developmental Biology | 2.00 | 0.00e+00 | 0.00e+00 | 126/445 | RPL22; CDC42; RPL11; RPS8; JUN; RPL5; PSMA5; PSMD4; SHC1; EFNA3; CNTN2; H3F3A; NCOA1; RPS27A; RPL31; PKP4; SCN3A; NEUROD1; RPL37A; ARPC2; TUBA4A; SRGAP3; RPL32; RPL15; RPSA; RPL14; PSMD6; ROBO1; ALCAM; GAP43; RPL35A; RPL9; RPL34; ANK2; RPS3A; RPS23; RPS14; EBF1; TUBB2A; CAP2; HIST1H4C; HIST1H2AC; CSNK2B; RPS10; RPS12; EZR; PSMB1; STX1A; ARPC1A; PSMC2; RPS4X; RPL36A; DCX; RPL10; DPYSL2; SDCBP; RPL7; TCEB1; RPL30; RPS6; CLTA; RPL35; DNM1; RPL7A; VAV2; TUBB4B; PSMD13; RPL27A; RPS13; NCAM1; HSPA8; ANK3; RPS24; MAGOHB; CACNB3; TUBA1B; TUBA1A; RPS26; RPL41; MYL6; CDK4; SRGAP1; RPL6; RPLP0; UBC; RPL21; PSME2; RPS29; PSMA3; HSP90AA1; TCF12; RPL4; RPLP1; PSMA4; RPS17; RPS2; RPS15A; PSMD7; RPL13; TUBB3; EFNB3; RPL26; RPL23A; PSMB3; RPL19; RPL27; PSMC5; EIF4A3; ACTG1; MYL12A; MYL12B; TCF4; PSMA7; RPS15; TUBB4A; RPS28; RPL18A; LGI4; PSENEN; CAPNS1; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
ARID1B : 59 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-192823 | Viral mRNA Translation | 8.36 | 0.00e+00 | 0.00e+00 | 49/83 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-156902 | Peptide chain elongation | 8.26 | 0.00e+00 | 0.00e+00 | 49/84 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-2408557 | Selenocysteine synthesis | 8.16 | 0.00e+00 | 0.00e+00 | 49/85 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 8.07 | 0.00e+00 | 0.00e+00 | 49/86 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-72764 | Eukaryotic Translation Termination | 8.07 | 0.00e+00 | 0.00e+00 | 49/86 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 7.90 | 0.00e+00 | 0.00e+00 | 53/95 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; EIF3L; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 7.88 | 0.00e+00 | 0.00e+00 | 49/88 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 7.35 | 0.00e+00 | 0.00e+00 | 54/104 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; SPCS1; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SEC61G; RPS4X; RPL36A; RPL39; SSR4; RPS20; RPL7; RPL30; RPS6; SEC61B; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 7.15 | 0.00e+00 | 0.00e+00 | 53/105 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; EIF3L; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 7.08 | 0.00e+00 | 0.00e+00 | 53/106 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; EIF3L; RPL3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 7.08 | 5.55e-16 | 1.59e-14 | 24/48 | RPS8; RPS27; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS26; RPS17; RPS2; RPS15A; RPS19; EIF3L |
| R-HSA-163210 | Formation of ATP by chemiosmotic coupling | 7.08 | 1.05e-06 | 2.32e-05 | 9/18 | ATP5G3; ATP5I; ATP5J2; ATP5L; ATP5G2; ATP5G1; ATP5H; ATP5E; ATP5J |
| R-HSA-2408522 | Selenoamino acid metabolism | 7.01 | 0.00e+00 | 0.00e+00 | 49/99 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 6.76 | 0.00e+00 | 0.00e+00 | 54/113 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; EIF3L; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 6.76 | 0.00e+00 | 0.00e+00 | 54/113 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; EIF3L; RPL3 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 6.76 | 0.00e+00 | 0.00e+00 | 54/113 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; POLR2J; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; POLR2K; RPS6; RPL7A; POLR2L; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; POLR2I; RPS19; RPL18; RPL13A; POLR2F; RPL3 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 6.61 | 0.00e+00 | 0.00e+00 | 49/105 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 6.61 | 0.00e+00 | 0.00e+00 | 49/105 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 6.32 | 4.11e-15 | 1.14e-13 | 25/56 | RPS8; RPS27; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; EIF4EBP1; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS26; RPS17; RPS2; RPS15A; RPS19; EIF3L |
| R-HSA-168255 | Influenza Life Cycle | 6.32 | 0.00e+00 | 0.00e+00 | 54/121 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; POLR2J; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; POLR2K; RPS6; RPL7A; POLR2L; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; POLR2I; RPS19; RPL18; RPL13A; POLR2F; RPL3 |
| R-HSA-3299685 | Detoxification of Reactive Oxygen Species | 6.29 | 1.30e-05 | 2.74e-04 | 8/18 | PRDX1; ATOX1; CYCS; TXN; PRDX5; GSTP1; PRDX2; SOD1 |
| R-HSA-6799198 | Complex I biogenesis | 6.26 | 7.04e-14 | 1.78e-12 | 23/52 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFA2; NDUFB2; NDUFB11; NDUFA1; NDUFB9; NDUFB6; NDUFS8; NDUFC2; NDUFB8; NDUFB1; NDUFB10; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND3 |
| R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 6.21 | 0.00e+00 | 0.00e+00 | 50/114 | NDUFS5; UQCRH; COX20; COX5B; ATP5G3; NDUFB3; NDUFAF3; NDUFB4; ATP5I; COX7C; UQCRQ; NDUFA2; NDUFA4; CYCS; ATP5J2; NDUFB2; NDUFB11; COX7B; NDUFA1; COX6C; NDUFB9; NDUFB6; COX8A; NDUFS8; NDUFC2; ATP5L; NDUFB8; COX14; ATP5G2; COX16; NDUFB1; COX5A; NDUFB10; COX4I1; ATP5G1; ATP5H; ATP5E; NDUFS7; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; NDUFA3; UQCR10; NDUFA6; ATP5J; MT-CO1; MT-ND3 |
| R-HSA-72649 | Translation initiation complex formation | 6.18 | 2.83e-14 | 7.39e-13 | 24/55 | RPS8; RPS27; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS26; RPS17; RPS2; RPS15A; RPS19; EIF3L |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 6.18 | 2.83e-14 | 7.39e-13 | 24/55 | RPS8; RPS27; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS26; RPS17; RPS2; RPS15A; RPS19; EIF3L |
| R-HSA-611105 | Respiratory electron transport | 6.11 | 0.00e+00 | 0.00e+00 | 41/95 | NDUFS5; UQCRH; COX20; COX5B; NDUFB3; NDUFAF3; NDUFB4; COX7C; UQCRQ; NDUFA2; NDUFA4; CYCS; NDUFB2; NDUFB11; COX7B; NDUFA1; COX6C; NDUFB9; NDUFB6; COX8A; NDUFS8; NDUFC2; NDUFB8; COX14; COX16; NDUFB1; COX5A; NDUFB10; COX4I1; NDUFS7; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; NDUFA3; UQCR10; NDUFA6; MT-CO1; MT-ND3 |
| R-HSA-168254 | Influenza Infection | 5.97 | 0.00e+00 | 0.00e+00 | 54/128 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; POLR2J; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; POLR2K; RPS6; RPL7A; POLR2L; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; POLR2I; RPS19; RPL18; RPL13A; POLR2F; RPL3 |
| R-HSA-5601884 | PIWI-interacting RNA (piRNA) biogenesis | 5.44 | 1.36e-03 | 2.49e-02 | 5/13 | POLR2J; POLR2K; POLR2L; POLR2I; POLR2F |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 5.04 | 0.00e+00 | 0.00e+00 | 53/149 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SHFM1; RPS4X; RPL36A; RPL39; RPS20; RPL7; TCEB1; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3; RBX1 |
| R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | 4.95 | 0.00e+00 | 0.00e+00 | 50/143 | NDUFS5; UQCRH; COX20; COX5B; ATP5G3; NDUFB3; NDUFAF3; NDUFB4; ATP5I; COX7C; UQCRQ; NDUFA2; NDUFA4; CYCS; ATP5J2; NDUFB2; NDUFB11; COX7B; NDUFA1; COX6C; NDUFB9; NDUFB6; COX8A; NDUFS8; NDUFC2; ATP5L; NDUFB8; COX14; ATP5G2; COX16; NDUFB1; COX5A; NDUFB10; COX4I1; ATP5G1; ATP5H; ATP5E; NDUFS7; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; NDUFA3; UQCR10; NDUFA6; ATP5J; MT-CO1; MT-ND3 |
| R-HSA-5628897 | TP53 Regulates Metabolic Genes | 4.72 | 2.13e-10 | 4.82e-09 | 22/66 | PRDX1; LAMTOR5; LAMTOR2; COX20; COX5B; COX7C; NDUFA4; CYCS; LAMTOR4; COX7B; COX6C; YWHAZ; TXN; COX8A; PRDX5; COX14; COX16; COX5A; COX4I1; PRDX2; COX6B1; MT-CO1 |
| R-HSA-8949613 | Cristae formation | 4.72 | 2.20e-05 | 4.51e-04 | 10/30 | MINOS1; ATP5G3; ATP5I; ATP5J2; ATP5L; ATP5G2; ATP5G1; ATP5H; ATP5E; ATP5J |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 4.69 | 0.00e+00 | 0.00e+00 | 53/160 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; NCL; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; NOP10; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3; SNU13 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 4.69 | 0.00e+00 | 0.00e+00 | 51/154 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; NCL; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3; SNU13 |
| R-HSA-72312 | rRNA processing | 4.66 | 0.00e+00 | 0.00e+00 | 54/164 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; NCL; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS18; RPL10A; RPS12; HSD17B10; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; NOP10; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3; SNU13 |
| R-HSA-376176 | Signaling by ROBO receptors | 4.29 | 0.00e+00 | 0.00e+00 | 54/178 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SHFM1; RPS4X; RPL36A; RPL39; RPS20; RPL7; TCEB1; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; PFN1; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; RPL3; RBX1 |
| R-HSA-167160 | RNA Pol II CTD phosphorylation and interaction with CE during HIV infection | 4.25 | 1.95e-03 | 3.17e-02 | 6/20 | GTF2H5; POLR2J; POLR2K; POLR2L; POLR2I; POLR2F |
| R-HSA-77075 | RNA Pol II CTD phosphorylation and interaction with CE | 4.25 | 1.95e-03 | 3.17e-02 | 6/20 | GTF2H5; POLR2J; POLR2K; POLR2L; POLR2I; POLR2F |
| R-HSA-73777 | RNA Polymerase I Chain Elongation | 4.04 | 4.97e-04 | 9.52e-03 | 8/28 | H3F3A; H2AFZ; HIST1H4J; GTF2H5; CBX3; POLR2K; POLR2L; POLR2F |
| R-HSA-72165 | mRNA Splicing - Minor Pathway | 3.89 | 6.50e-06 | 1.40e-04 | 14/51 | SNRPE; SF3B6; SNRPG; SF3B5; POLR2J; POLR2K; POLR2L; SF3B2; SNRPF; SNRPD1; POLR2I; SNRPD2; POLR2F; SNU13 |
| R-HSA-72086 | mRNA Capping | 3.86 | 3.33e-03 | 4.86e-02 | 6/22 | GTF2H5; POLR2J; POLR2K; POLR2L; POLR2I; POLR2F |
| R-HSA-167238 | Pausing and recovery of Tat-mediated HIV elongation | 3.67 | 2.10e-03 | 3.29e-02 | 7/27 | POLR2J; TCEB1; POLR2K; POLR2L; TCEB2; POLR2I; POLR2F |
| R-HSA-167243 | Tat-mediated HIV elongation arrest and recovery | 3.67 | 2.10e-03 | 3.29e-02 | 7/27 | POLR2J; TCEB1; POLR2K; POLR2L; TCEB2; POLR2I; POLR2F |
| R-HSA-72766 | Translation | 3.64 | 0.00e+00 | 0.00e+00 | 66/257 | MRPL20; RPL11; RPS8; RPL5; MRPS21; RPS27; MRPL33; RPS27A; RPL37A; RPL32; RPL15; RPSA; SPCS1; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SEC61G; RPS4X; RPL36A; RPL39; SSR4; EIF4EBP1; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPS6; SEC61B; RPL7A; MRPL41; EIF3F; RPL27A; RPS13; RPS3; CHCHD1; RPS24; RPS26; RPL6; RPLP0; RPL21; MRPL52; RPL36AL; SRP14; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; RPS19; RPL18; RPL13A; EIF3L; RPL3; MRPS6 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 3.52 | 0.00e+00 | 0.00e+00 | 52/209 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; RPS12; SHFM1; HSD17B10; RPS4X; RPL36A; RPL39; RPS20; RPL7; RPL30; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; OAZ1; RPL18A; RPS19; RPL18; RPL13A; RPL3 |
| R-HSA-5663205 | Infectious disease | 3.50 | 0.00e+00 | 0.00e+00 | 68/275 | RPL11; RPS8; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; SKP1; RPS14; RPS18; RPL10A; RPS12; GTF2H5; PPIA; SHFM1; POLR2J; RPS4X; RPL36A; RPL39; RPS20; RPL7; TCEB1; RPL30; POLR2K; RPS6; RPL7A; POLR2L; RPL27A; RPS13; BANF1; RPS3; RPS24; RPS26; RPL6; RPLP0; RPL21; RPL36AL; CALM1; GTF2A2; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL18A; POLR2I; RPS19; CALM3; AP2S1; RPL18; RPL13A; CHMP2A; RANBP1; POLR2F; RPL3; RBX1 |
| R-HSA-167169 | HIV Transcription Elongation | 3.43 | 1.63e-03 | 2.81e-02 | 8/33 | GTF2H5; POLR2J; TCEB1; POLR2K; POLR2L; TCEB2; POLR2I; POLR2F |
| R-HSA-167200 | Formation of HIV-1 elongation complex containing HIV-1 Tat | 3.43 | 1.63e-03 | 2.81e-02 | 8/33 | GTF2H5; POLR2J; TCEB1; POLR2K; POLR2L; TCEB2; POLR2I; POLR2F |
| R-HSA-167246 | Tat-mediated elongation of the HIV-1 transcript | 3.43 | 1.63e-03 | 2.81e-02 | 8/33 | GTF2H5; POLR2J; TCEB1; POLR2K; POLR2L; TCEB2; POLR2I; POLR2F |
| R-HSA-167287 | HIV elongation arrest and recovery | 3.42 | 3.27e-03 | 4.86e-02 | 7/29 | POLR2J; TCEB1; POLR2K; POLR2L; TCEB2; POLR2I; POLR2F |
| R-HSA-167290 | Pausing and recovery of HIV elongation | 3.42 | 3.27e-03 | 4.86e-02 | 7/29 | POLR2J; TCEB1; POLR2K; POLR2L; TCEB2; POLR2I; POLR2F |
| R-HSA-6782210 | Gap-filling DNA repair synthesis and ligation in TC-NER | 3.27 | 1.23e-03 | 2.30e-02 | 9/39 | RPS27A; GTF2H5; RPA3; POLR2J; POLR2K; POLR2L; POLR2I; POLR2F; RBX1 |
| R-HSA-1592230 | Mitochondrial biogenesis | 3.25 | 5.95e-05 | 1.16e-03 | 14/61 | MINOS1; ATP5G3; ATP5I; CYCS; ATP5J2; SSBP1; ATP5L; ATP5G2; CALM1; ATP5G1; ATP5H; ATP5E; CALM3; ATP5J |
| R-HSA-167152 | Formation of HIV elongation complex in the absence of HIV Tat | 3.24 | 2.44e-03 | 3.76e-02 | 8/35 | GTF2H5; POLR2J; TCEB1; POLR2K; POLR2L; TCEB2; POLR2I; POLR2F |
| R-HSA-6782135 | Dual incision in TC-NER | 3.11 | 1.79e-03 | 3.02e-02 | 9/41 | RPS27A; GTF2H5; RPA3; POLR2J; POLR2K; POLR2L; POLR2I; POLR2F; RBX1 |
| R-HSA-422475 | Axon guidance | 2.63 | 1.28e-13 | 3.16e-12 | 61/328 | RPL11; RPS8; RPL5; RPS27; ARPC5; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; RPS14; RPS18; RPL10A; CD24; RPS12; SHFM1; RPS4X; RPL36A; RPL39; RPS20; RPL7; TCEB1; RPL30; RPS6; RPL7A; RPL27A; RPS13; CFL1; RPS3; RPS24; RPS26; MYL6; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; PFN1; RPL26; RPL23A; RPL23; RPL19; MYL12B; RPL18A; PSENEN; RPS19; AP2S1; RPL18; RPL13A; RPL3; RBX1 |
| R-HSA-1643685 | Disease | 2.23 | 2.17e-12 | 5.19e-11 | 73/464 | RPL11; RPS8; PRDX1; RPL5; RPS27; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; RPS3A; RPL37; RPS23; SKP1; RPS14; RPS18; RPL10A; RPS12; GTF2H5; PPIA; SHFM1; POLR2J; RPS4X; RPL36A; RPL39; RPS20; RPL7; TCEB1; RPL30; POLR2K; RPS6; RPL7A; POLR2L; RPL27A; RPS13; BANF1; RPS3; RPS24; RPS26; RPL6; PEBP1; RPLP0; RPL21; RPL36AL; CALM1; GTF2A2; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; DCXR; PRDX2; RPL18A; PSENEN; POLR2I; RPS19; CALM3; AP2S1; RPL18; RPL13A; CHMP2A; RANBP1; POLR2F; RPL3; RBX1 |
| R-HSA-1266738 | Developmental Biology | 2.19 | 2.26e-11 | 5.25e-10 | 69/445 | RPL11; RPS8; RPL5; RPS27; ARPC5; H3F3A; RPS27A; RPL37A; RPL32; RPL15; RPSA; RPL22L1; RPL9; H2AFZ; RPS3A; RPL37; RPS23; RPS14; HIST1H4J; RPS18; RPL10A; CD24; RPS12; SHFM1; POLR2J; RPS4X; RPL36A; RPL39; RPS20; RPL7; TCEB1; RPL30; POLR2K; RPS6; RPL7A; POLR2L; RPL27A; RPS13; CFL1; RPS3; RPS24; RPS26; MYL6; RPL6; RPLP0; RPL21; RPL36AL; RPL4; RPS17; RPS2; TCEB2; RPS15A; RPL13; PFN1; RPL26; RPL23A; RPL23; RPL19; MYL12B; RPL18A; PSENEN; POLR2I; RPS19; AP2S1; RPL18; RPL13A; POLR2F; RPL3; RBX1 |
| R-HSA-3700989 | Transcriptional Regulation by TP53 | 2.11 | 2.88e-05 | 5.76e-04 | 32/215 | PRDX1; LAMTOR5; LAMTOR2; COX20; RPS27A; COX5B; COX7C; GTF2H5; RPA3; NDUFA4; CYCS; LAMTOR4; POLR2J; COX7B; TCEB1; COX6C; POLR2K; YWHAZ; TXN; POLR2L; COX8A; PRDX5; COX14; COX16; COX5A; TCEB2; COX4I1; PRDX2; COX6B1; POLR2I; POLR2F; MT-CO1 |
ASH1L : 13 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-8876725 | Protein methylation | 7.52 | 2.85e-04 | 0.02454 | 5/12 | EEF1A1; CALM1; EEF2; ETFB |
| R-HSA-5339562 | Uptake and actions of bacterial toxins | 6.37 | 1.98e-04 | 0.02387 | 6/17 | HSP90AB1; STX1A; SYT1; CALM1; EEF2 |
| R-HSA-500792 | GPCR ligand binding | 6.02 | 8.01e-06 | 0.00345 | 9/27 | GNG5; AGT; GNG4; SST; GRM4; FZD1; GAL; SSTR2; PTH2 |
| R-HSA-5578775 | Ion homeostasis | 5.50 | 1.65e-04 | 0.02364 | 7/23 | ATP1B1; RYR2; CAMK2B; SRI; CALM1; FXYD7 |
| R-HSA-2672351 | Stimuli-sensing channels | 5.27 | 2.22e-04 | 0.02387 | 7/24 | RYR2; ASIC4; SRI; ASIC1; CALM1; TTYH2 |
| R-HSA-5576891 | Cardiac conduction | 4.64 | 8.19e-05 | 0.01410 | 9/35 | ATP1B1; RYR2; SCN3A; CAMK2B; SRI; AKAP9; CALM1; FXYD7 |
| R-HSA-112314 | Neurotransmitter receptors and postsynaptic signal transmission | 4.05 | 5.17e-05 | 0.01112 | 11/49 | GNG5; GNG4; CHRNA1; GNAI2; AP2M1; GRIA2; CAMK2B; NEFL; CALM1; KCNJ6 |
| R-HSA-397014 | Muscle contraction | 3.28 | 7.06e-04 | 0.04808 | 10/55 | ATP1B1; RYR2; SCN3A; CAMK2B; SRI; AKAP9; CALD1; CALM1; FXYD7 |
| R-HSA-112315 | Transmission across Chemical Synapses | 3.26 | 3.55e-05 | 0.01020 | 15/83 | GNG5; GNG4; CHRNA1; GNAI2; AP2M1; GRIA2; CAMK2B; STX1A; NEFL; SYT1; CALM1; CACNA1A; RAB3A; KCNJ6 |
| R-HSA-112316 | Neuronal System | 3.21 | 4.06e-06 | 0.00345 | 19/107 | PTPRF; GNG5; GNG4; NRXN1; CHRNA1; GNAI2; AP2M1; GRIA2; CAMK2B; STX1A; NEFL; SYT7; SYT1; CALM1; CACNA1A; RAB3A; HOMER3; KCNJ6 |
| R-HSA-418594 | G alpha (i) signalling events | 3.06 | 7.26e-04 | 0.04808 | 11/65 | GNG5; RGS16; AGT; GNG4; GNAI2; SST; GRM4; GAL; CALM1; SSTR2 |
| R-HSA-372790 | Signaling by GPCR | 2.56 | 2.59e-04 | 0.02454 | 17/120 | GNG5; RHOC; RGS16; AGT; GNG4; REEP1; GNAI2; ABHD6; SST; GRM4; FZD1; GAL; CALM1; SSTR2; PREX1; PTH2 |
| R-HSA-388396 | GPCR downstream signalling | 2.49 | 5.37e-04 | 0.04201 | 16/116 | GNG5; RHOC; RGS16; AGT; GNG4; REEP1; GNAI2; ABHD6; SST; GRM4; GAL; CALM1; SSTR2; PREX1; PTH2 |
CHD2 : 48 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-156902 | Peptide chain elongation | 5.82 | 0.00e+00 | 0.00e+00 | 66/84 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; EEF2; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 5.77 | 0.00e+00 | 0.00e+00 | 67/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; EEF1B2; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; EEF2; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-192823 | Viral mRNA Translation | 5.71 | 0.00e+00 | 0.00e+00 | 64/83 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72764 | Eukaryotic Translation Termination | 5.60 | 0.00e+00 | 0.00e+00 | 65/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; TRMT112; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-2408557 | Selenocysteine synthesis | 5.57 | 0.00e+00 | 0.00e+00 | 64/85 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 5.38 | 0.00e+00 | 0.00e+00 | 64/88 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 5.38 | 0.00e+00 | 0.00e+00 | 69/95 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; EIF3L; RPL3 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 4.98 | 0.00e+00 | 0.00e+00 | 70/104 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; SPCS1; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; SEC61G; RPS4X; RPL36A; RPL39; SSR4; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; SRP14; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPN2; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 4.87 | 0.00e+00 | 0.00e+00 | 69/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; EIF3L; RPL3 |
| R-HSA-2408522 | Selenoamino acid metabolism | 4.86 | 0.00e+00 | 0.00e+00 | 65/99 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; PAPSS1; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 4.82 | 0.00e+00 | 0.00e+00 | 69/106 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; EIF3L; RPL3 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 4.58 | 0.00e+00 | 0.00e+00 | 65/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 4.58 | 0.00e+00 | 0.00e+00 | 65/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 4.52 | 0.00e+00 | 0.00e+00 | 69/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; EIF3L; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 4.52 | 0.00e+00 | 0.00e+00 | 69/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL7A; EIF3F; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; EIF3L; RPL3 |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 4.46 | 0.00e+00 | 0.00e+00 | 68/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL7A; POLR2L; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; HSP90AA1; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; POLR2F; RPL3 |
| R-HSA-445355 | Smooth Muscle Contraction | 4.36 | 1.48e-05 | 3.10e-04 | 10/17 | CALM2; ANXA6; CALD1; VCL; MYL6B; MYL6; CALM1; MYL12A; MYL12B; CALM3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 4.32 | 2.67e-13 | 7.93e-12 | 28/48 | RPS8; RPS27; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS29; EIF3J; RPS17; RPS15A; RPS15; RPS28; RPS16; RPS19; RPS5; EIF3L |
| R-HSA-8876725 | Protein methylation | 4.32 | 3.37e-04 | 6.59e-03 | 7/12 | CALM2; EEF1A1; HSPA8; CALM1; EEF2; CALM3; ETFB |
| R-HSA-5576892 | Phase 0 - rapid depolarisation | 4.32 | 3.37e-04 | 6.59e-03 | 7/12 | CALM2; SCN3A; SCN9A; CAMK2B; CACNB3; CALM1; CALM3 |
| R-HSA-168255 | Influenza Life Cycle | 4.28 | 0.00e+00 | 0.00e+00 | 70/121 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; CANX; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL7A; POLR2L; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; HSP90AA1; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; CALR; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; POLR2F; RPL3 |
| R-HSA-168254 | Influenza Infection | 4.05 | 0.00e+00 | 0.00e+00 | 70/128 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; CANX; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL7A; POLR2L; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; HSP90AA1; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; CALR; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; POLR2F; RPL3 |
| R-HSA-983170 | Antigen Presentation: Folding, assembly and peptide loading of class I MHC | 4.04 | 1.49e-03 | 2.62e-02 | 6/11 | CANX; HLA-A; HSPA5; PDIA3; B2M; CALR |
| R-HSA-72649 | Translation initiation complex formation | 3.77 | 2.42e-11 | 6.50e-10 | 28/55 | RPS8; RPS27; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS29; EIF3J; RPS17; RPS15A; RPS15; RPS28; RPS16; RPS19; RPS5; EIF3L |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 3.77 | 2.42e-11 | 6.50e-10 | 28/55 | RPS8; RPS27; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS29; EIF3J; RPS17; RPS15A; RPS15; RPS28; RPS16; RPS19; RPS5; EIF3L |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 3.70 | 4.23e-11 | 1.10e-09 | 28/56 | RPS8; RPS27; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS10; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; RPS3; RPS24; RPS29; EIF3J; RPS17; RPS15A; RPS15; RPS28; RPS16; RPS19; RPS5; EIF3L |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 3.58 | 0.00e+00 | 0.00e+00 | 72/149 | RPL22; RPL11; RPS8; RPL5; RPS27; LHX9; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; ROBO1; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; SHFM1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; TCEB2; RPS15A; RPL13; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; PSMA7; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-500792 | GPCR ligand binding | 3.56 | 1.40e-05 | 3.01e-04 | 13/27 | GNG5; AGT; GNG4; SST; GRM4; FZD1; GABBR2; GAL; NMB; SSTR2; GNAS; GNG8; PTH2 |
| R-HSA-5578775 | Ion homeostasis | 3.54 | 7.12e-05 | 1.46e-03 | 11/23 | ATP1B1; RYR2; FKBP1B; CALM2; CAMK2B; SRI; FXYD6; ATP2B1; CALM1; FXYD7; CALM3 |
| R-HSA-6799198 | Complex I biogenesis | 3.42 | 8.54e-09 | 1.94e-07 | 24/52 | NDUFS5; NDUFAF3; NDUFB4; NDUFA2; NDUFA5; NDUFB2; NDUFB11; NDUFA1; NDUFB9; NDUFS8; NDUFC2; NDUFB8; NDUFB10; NDUFAB1; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND1; MT-ND3; MT-ND4; MT-ND6 |
| R-HSA-5576891 | Cardiac conduction | 3.38 | 3.29e-06 | 7.27e-05 | 16/35 | ATP1B1; RYR2; FKBP1B; CALM2; SCN3A; SCN9A; CAMK2B; SRI; AKAP9; FXYD6; KCNIP2; CACNB3; ATP2B1; CALM1; FXYD7; CALM3 |
| R-HSA-397014 | Muscle contraction | 3.37 | 6.13e-09 | 1.43e-07 | 25/55 | ATP1B1; TNNI1; RYR2; FKBP1B; CALM2; SCN3A; SCN9A; MYL1; ANXA6; CAMK2B; SRI; AKAP9; CALD1; FXYD6; VCL; KCNIP2; CACNB3; MYL6B; MYL6; ATP2B1; CALM1; MYL12A; MYL12B; FXYD7; CALM3 |
| R-HSA-163210 | Formation of ATP by chemiosmotic coupling | 3.29 | 1.32e-03 | 2.37e-02 | 8/18 | ATP5G3; ATP5I; ATP5J2; ATP5B; ATP5G1; ATP5H; ATP5A1; ATP5J |
| R-HSA-376176 | Signaling by ROBO receptors | 3.16 | 0.00e+00 | 0.00e+00 | 76/178 | RPL22; RPL11; RPS8; RPL5; RPS27; LHX9; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; ROBO1; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; CAP2; RPS10; RPS12; SHFM1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; RPL41; SRGAP1; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; TCEB2; RPS15A; RPL13; PFN1; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; DCC; PSMA7; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 3.15 | 0.00e+00 | 0.00e+00 | 68/160 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; TRMT112; RPS3; DDX21; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; NOP10; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 3.12 | 0.00e+00 | 0.00e+00 | 65/154 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; DDX21; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-72312 | rRNA processing | 3.11 | 0.00e+00 | 0.00e+00 | 69/164 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS10; RPS12; HSD17B10; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; TRMT112; RPS3; DDX21; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; NOP10; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS15; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 2.86 | 8.16e-12 | 2.34e-10 | 44/114 | NDUFS5; COX5B; ATP5G3; NDUFAF3; NDUFB4; ATP5I; UQCRQ; NDUFA2; NDUFA4; CYCS; ATP5J2; NDUFA5; NDUFB2; NDUFB11; COX7B; NDUFA1; UQCRB; COX6C; NDUFB9; COX8A; NDUFS8; NDUFC2; NDUFB8; ATP5B; COX5A; NDUFB10; NDUFAB1; ATP5G1; ATP5H; ATP5A1; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; ETFB; NDUFA3; NDUFA6; ATP5J; MT-ND1; MT-ND3; MT-ND4; MT-ND6 |
| R-HSA-611105 | Respiratory electron transport | 2.81 | 1.32e-09 | 3.24e-08 | 36/95 | NDUFS5; COX5B; NDUFAF3; NDUFB4; UQCRQ; NDUFA2; NDUFA4; CYCS; NDUFA5; NDUFB2; NDUFB11; COX7B; NDUFA1; UQCRB; COX6C; NDUFB9; COX8A; NDUFS8; NDUFC2; NDUFB8; COX5A; NDUFB10; NDUFAB1; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; ETFB; NDUFA3; NDUFA6; MT-ND1; MT-ND3; MT-ND4; MT-ND6 |
| R-HSA-936837 | Ion transport by P-type ATPases | 2.78 | 2.76e-03 | 4.66e-02 | 9/24 | ATP1B1; CALM2; CAMK2B; SRI; FXYD6; ATP2B1; CALM1; FXYD7; CALM3 |
| R-HSA-418346 | Platelet homeostasis | 2.71 | 1.18e-03 | 2.16e-02 | 11/30 | GNG5; GNG4; CALM2; PRKG2; SRI; ATP2B1; CALM1; GNAS; CALM3; GNG8; PDE9A |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 2.69 | 0.00e+00 | 0.00e+00 | 76/209 | RPL22; RPL11; RPS8; RPL5; PHGDH; RPS27; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; RPL35A; RPL9; PAPSS1; RPL34; RPS3A; RPL37; RPS23; CDO1; RPS14; RPS10; RPS12; SHFM1; SAT1; HSD17B10; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL7A; RPL27A; RPS13; RPS3; RPS24; BCAT1; RPL41; RPL6; RPLP0; PXMP2; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS15A; NDUFAB1; RPL13; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; RPL38; RPL17; PSMA7; RPS15; OAZ1; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
| R-HSA-5663205 | Infectious disease | 2.42 | 0.00e+00 | 0.00e+00 | 90/275 | RPL22; RPL11; RPS8; RPL5; RPS27; CALM2; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL24; RPL22L1; AP2M1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; SKP1; RPS14; CANX; HMGA1; RPS10; RPS12; GTF2H5; STX1A; SHFM1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; POLR2K; RPL8; RPS6; RPL7A; POLR2L; RPL27A; RPS13; BANF1; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; CALM1; HSP90AA1; B2M; GTF2A2; RPL4; RPLP1; RPS17; TCEB2; RPS15A; RPL13; VAMP2; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; RPL38; RPL17; PSMA7; RPS15; EEF2; RPS28; CALR; RPL18A; RPS16; RPS19; CALM3; RPL18; RPL13A; RPS5; CHMP2A; RANBP1; POLR2F; RPL3 |
| R-HSA-72766 | Translation | 2.42 | 1.11e-16 | 3.54e-15 | 84/257 | MRPL20; RPL22; RPL11; RPS8; RPL5; MRPS21; RPS27; MRPL33; RPS27A; RPL31; EEF1B2; RPL37A; RPL32; RPL15; RPSA; RPL14; SPCS1; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS10; MRPL14; EEF1A1; RPS12; SEC61G; RPS4X; RPL36A; RPL39; SSR4; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; SEC61B; RPL7A; EIF3F; RPL27A; RPS13; TRMT112; RPS3; RPS24; RPL41; RPL6; RPLP0; RPL21; MRPL52; RPS29; RPL36AL; SRP14; EIF3J; RPL4; RPLP1; RPS17; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPN2; RPS15; EEF2; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; EIF3L; RPL3 |
| R-HSA-114608 | Platelet degranulation | 2.41 | 1.02e-03 | 1.92e-02 | 14/43 | TAGLN2; CALM2; MANF; TMSB4X; HSPA5; SERPING1; APLP2; VCL; CD63; CALM1; ALDOA; PFN1; LGALS3BP; CALM3 |
| R-HSA-76005 | Response to elevated platelet cytosolic Ca2+ | 2.41 | 1.02e-03 | 1.92e-02 | 14/43 | TAGLN2; CALM2; MANF; TMSB4X; HSPA5; SERPING1; APLP2; VCL; CD63; CALM1; ALDOA; PFN1; LGALS3BP; CALM3 |
| R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | 2.38 | 3.40e-09 | 8.13e-08 | 46/143 | NDUFS5; COX5B; ATP5G3; NDUFAF3; NDUFB4; ATP5I; UQCRQ; NDUFA2; NDUFA4; CYCS; ATP5J2; NDUFA5; NDUFB2; NDUFB11; COX7B; NDUFA1; UQCRB; COX6C; NDUFB9; LDHA; COX8A; NDUFS8; NDUFC2; NDUFB8; ATP5B; COX5A; NDUFB10; NDUFAB1; ATP5G1; ATP5H; ATP5A1; BSG; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; ETFB; NDUFA3; NDUFA6; ATP5J; MT-ND1; MT-ND3; MT-ND4; MT-ND6 |
| R-HSA-422475 | Axon guidance | 2.28 | 0.00e+00 | 0.00e+00 | 101/328 | RPL22; EPHB2; RPL11; RPS8; RPL5; RPS27; SHC1; LHX9; RPS27A; RPL31; SCN3A; SCN9A; NRP2; RPL37A; RPL32; RPL15; RPSA; RPL14; ROBO1; RPL24; ALCAM; GAP43; RPL22L1; AP2M1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RGMB; DPYSL3; RPS14; PDLIM7; TUBB2B; CAP2; RPS10; RPS12; EZR; SHFM1; RPS4X; RPL36A; DCX; RPL39; RPL10; DPYSL2; ANK1; RPS20; RPL7; TCEB1; RPL30; RPL8; RPS6; SPTAN1; RPL7A; RPL27A; RPS13; RPS3; HSPA8; RPS24; CACNB3; RPL41; MYL6; SRGAP1; RPL6; RPLP0; RPL21; RPS29; RPL36AL; HSP90AA1; RPL4; RPLP1; RPS17; TCEB2; RPS15A; RPL13; PFN1; EFNB3; RPL26; RPL23A; PSMB3; RPL23; RPL19; RPL27; RPL38; EIF4A3; MYL12A; MYL12B; RPL17; DCC; PSMA7; RPS15; TUBB4A; RPS28; RPL18A; RPS16; RPS19; RPL18; RPL13A; RPS5; RPL3 |
PTEN : 46 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-156902 | Peptide chain elongation | 5.94 | 0.00e+00 | 0.00e+00 | 80/84 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-192823 | Viral mRNA Translation | 5.87 | 0.00e+00 | 0.00e+00 | 78/83 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 5.81 | 0.00e+00 | 0.00e+00 | 80/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72764 | Eukaryotic Translation Termination | 5.73 | 0.00e+00 | 0.00e+00 | 79/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; TRMT112; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-2408557 | Selenocysteine synthesis | 5.73 | 0.00e+00 | 0.00e+00 | 78/85 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 5.60 | 0.00e+00 | 0.00e+00 | 79/88 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 5.58 | 0.00e+00 | 0.00e+00 | 85/95 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 5.23 | 0.00e+00 | 0.00e+00 | 88/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; EIF2S2; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 5.12 | 0.00e+00 | 0.00e+00 | 87/106 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; EIF2S2; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 5.07 | 0.00e+00 | 0.00e+00 | 39/48 | RPS8; RPS27; RPS7; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPS29; EIF3J; RPS17; RPS2; RPS15A; EIF2S2; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5; EIF3D; EIF3L |
| R-HSA-2408522 | Selenoamino acid metabolism | 4.98 | 0.00e+00 | 0.00e+00 | 79/99 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; QARS; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 4.92 | 0.00e+00 | 0.00e+00 | 82/104 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SEC61G; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; SEC61B; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; SRP14; RPL4; RPLP1; RPS17; SEC11A; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 4.86 | 0.00e+00 | 0.00e+00 | 88/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; EIF2S2; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 4.86 | 0.00e+00 | 0.00e+00 | 88/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; EIF3J; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; EIF2S2; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 4.76 | 0.00e+00 | 0.00e+00 | 80/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 4.76 | 0.00e+00 | 0.00e+00 | 80/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72649 | Translation initiation complex formation | 4.65 | 0.00e+00 | 0.00e+00 | 41/55 | RPS8; RPS27; RPS7; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; PABPC1; EIF3E; EIF3H; RPS6; EIF3F; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPS29; EIF3J; RPS17; RPS2; RPS15A; EIF2S2; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5; EIF3D; EIF3L |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 4.57 | 0.00e+00 | 0.00e+00 | 41/56 | RPS8; RPS27; RPS7; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; PABPC1; EIF3E; EIF3H; RPS6; EIF3F; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPS29; EIF3J; RPS17; RPS2; RPS15A; EIF2S2; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5; EIF3D; EIF3L |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 4.54 | 0.00e+00 | 0.00e+00 | 40/55 | RPS8; RPS27; RPS7; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS26; RPS29; EIF3J; RPS17; RPS2; RPS15A; EIF2S2; RPS21; RPS15; RPS28; RPS16; RPS19; RPS11; RPS9; RPS5; EIF3D; EIF3L |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 4.53 | 0.00e+00 | 0.00e+00 | 82/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; POLR2J; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL12; RPL7A; RPLP2; POLR2L; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; POLR2F; RPL3 |
| R-HSA-168255 | Influenza Life Cycle | 4.33 | 0.00e+00 | 0.00e+00 | 84/121 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; CANX; RPS18; RPS10; RPL10A; RPS12; POLR2J; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL12; RPL7A; RPLP2; POLR2L; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; CALR; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; POLR2F; RPL3 |
| R-HSA-168254 | Influenza Infection | 4.19 | 0.00e+00 | 0.00e+00 | 86/128 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; CANX; RPS18; RPS10; RPL10A; RPS12; POLR2J; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL12; RPL7A; RPLP2; POLR2L; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; KPNA2; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; CALR; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; POLR2F; RPL3 |
| R-HSA-163210 | Formation of ATP by chemiosmotic coupling | 3.81 | 1.76e-05 | 3.99e-04 | 11/18 | ATP5G3; ATP5I; ATP5J2; ATP5L; ATP5G2; ATP5G1; ATP5H; ATP5A1; ATP5E; ATP5J; ATP5O |
| R-HSA-8876725 | Protein methylation | 3.64 | 9.90e-04 | 1.89e-02 | 7/12 | CALM2; EEF1A1; CALM1; RPS2; EEF2; ETFB |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 3.60 | 0.00e+00 | 0.00e+00 | 86/149 | RPL22; RPL11; RPS8; RPL5; RPS27; LHX9; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SHFM1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; PABPC1; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; TCEB2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; PSMA7; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3; RBX1 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 3.28 | 0.00e+00 | 0.00e+00 | 81/154 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; NCL; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; DDX21; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3; SNU13 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 3.28 | 0.00e+00 | 0.00e+00 | 84/160 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; NCL; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; TRMT112; RPS3; RPS25; DDX21; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; NOP10; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3; SNU13 |
| R-HSA-5578775 | Ion homeostasis | 3.26 | 6.28e-05 | 1.32e-03 | 12/23 | ATP1B1; ATP2B4; RYR2; FKBP1B; CALM2; ATP1B3; SRI; FXYD6; ATP2B1; CALM1; FXYD7 |
| R-HSA-6799198 | Complex I biogenesis | 3.24 | 1.71e-09 | 3.98e-08 | 27/52 | NDUFS5; NDUFB3; NDUFAF3; NDUFB4; NDUFC1; NDUFS6; NDUFS4; NDUFA2; NDUFA5; NDUFA1; NDUFB6; NDUFB8; NDUFA12; NDUFB1; NDUFAB1; NDUFS7; NDUFA11; NDUFA7; NDUFB7; NDUFA13; NDUFA3; NDUFA6; MT-ND1; MT-ND2; MT-ND3; MT-ND4; MT-ND6 |
| R-HSA-72312 | rRNA processing | 3.23 | 0.00e+00 | 0.00e+00 | 85/164 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; NCL; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS18; RPS10; RPL10A; RPS12; HSD17B10; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; TRMT112; RPS3; RPS25; DDX21; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; NOP10; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3; SNU13 |
| R-HSA-376176 | Signaling by ROBO receptors | 3.16 | 0.00e+00 | 0.00e+00 | 90/178 | RPL22; RPL11; RPS8; RPL5; RPS27; LHX9; RPS7; RPS27A; RPL31; RPL37A; SRGAP3; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; CAP2; RPS18; RPS10; RPL10A; RPS12; SHFM1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; PABPC1; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPS26; RPL41; SRGAP1; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; TCEB2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; RPL17; DCC; PSMA7; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3; RBX1 |
| R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 3.01 | 2.22e-16 | 5.79e-15 | 55/114 | NDUFS5; COX20; COX7A2L; COX5B; ATP5G3; NDUFB3; NDUFAF3; NDUFB4; ATP5I; NDUFC1; NDUFS6; NDUFS4; COX7C; UQCRQ; NDUFA2; NDUFA4; CYCS; ATP5J2; NDUFA5; COX7B; NDUFA1; COX6C; NDUFB6; COX8A; ATP5L; NDUFB8; COX14; ATP5G2; NDUFA12; COX6A1; COX16; NDUFB1; NDUFAB1; ATP5G1; ATP5H; ATP5A1; ATP5E; NDUFS7; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; ETFB; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-ND1; MT-ND2; MT-ND3; MT-ND4; MT-ND6 |
| R-HSA-500792 | GPCR ligand binding | 3.01 | 9.09e-05 | 1.86e-03 | 13/27 | GNG5; AGT; SST; GRM4; FZD1; GABBR2; GNG3; GAL; NMB; SSTR2; GNAS; GNG8; PTH2 |
| R-HSA-611105 | Respiratory electron transport | 2.89 | 1.81e-12 | 4.45e-11 | 44/95 | NDUFS5; COX20; COX7A2L; COX5B; NDUFB3; NDUFAF3; NDUFB4; NDUFC1; NDUFS6; NDUFS4; COX7C; UQCRQ; NDUFA2; NDUFA4; CYCS; NDUFA5; COX7B; NDUFA1; COX6C; NDUFB6; COX8A; NDUFB8; COX14; NDUFA12; COX6A1; COX16; NDUFB1; NDUFAB1; NDUFS7; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; ETFB; NDUFA3; UQCR10; NDUFA6; MT-ND1; MT-ND2; MT-ND3; MT-ND4; MT-ND6 |
| R-HSA-5576891 | Cardiac conduction | 2.85 | 3.14e-05 | 6.93e-04 | 16/35 | ATP1B1; ATP2B4; RYR2; FKBP1B; CALM2; SCN3A; SCN9A; ATP1B3; SRI; AKAP9; FXYD6; KCNIP2; ATP2B1; CALM1; FXYD7 |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 2.63 | 0.00e+00 | 0.00e+00 | 88/209 | RPL22; RPL11; RPS8; RPL5; PHGDH; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; QARS; RPL29; RPL24; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SHFM1; SAT1; HSD17B10; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; BCAT1; RPS26; RPL41; RPL6; RPLP0; PXMP2; RPL21; RPS29; RPL36AL; CKB; RPL4; RPLP1; RPS17; RPS2; RPS15A; NDUFAB1; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; PSMA7; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-2672351 | Stimuli-sensing channels | 2.60 | 2.38e-03 | 4.19e-02 | 10/24 | RYR2; FKBP1B; CALM2; RPS27A; SRI; ASIC1; CALM1; TTYH2; UBA52 |
| R-HSA-936837 | Ion transport by P-type ATPases | 2.60 | 2.38e-03 | 4.19e-02 | 10/24 | ATP1B1; ATP2B4; CALM2; ATP1B3; SRI; FXYD6; ATP2B1; CALM1; FXYD7 |
| R-HSA-72766 | Translation | 2.57 | 0.00e+00 | 0.00e+00 | 106/257 | AURKAIP1; MRPL20; RPL22; RPL11; RPS8; RPL5; MRPS21; RPS27; RPS7; MRPL33; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; QARS; RPL29; RPL24; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; MRPL14; EEF1A1; RPS12; SEC61G; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; SEC61B; RPL12; RPL7A; MRPL41; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; TRMT112; RPS3; RPS25; CHCHD1; RPS24; MRPL51; RPS26; RPL41; TSFM; RPL6; RPLP0; RPL21; MRPL52; RPS29; RPL36AL; SRP14; EIF3J; RPL4; RPLP1; RPS17; SEC11A; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; EIF2S2; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | 2.53 | 5.53e-13 | 1.40e-11 | 58/143 | NDUFS5; SLC16A1; COX20; COX7A2L; COX5B; ATP5G3; NDUFB3; NDUFAF3; NDUFB4; ATP5I; NDUFC1; NDUFS6; NDUFS4; COX7C; UQCRQ; VDAC1; NDUFA2; NDUFA4; CYCS; ATP5J2; NDUFA5; COX7B; NDUFA1; COX6C; NDUFB6; COX8A; ATP5L; NDUFB8; COX14; ATP5G2; NDUFA12; COX6A1; COX16; NDUFB1; NDUFAB1; ATP5G1; ATP5H; ATP5A1; ATP5E; BSG; NDUFS7; UQCR11; NDUFA11; NDUFA7; NDUFB7; NDUFA13; COX6B1; ETFB; NDUFA3; UQCR10; NDUFA6; ATP5J; ATP5O; MT-ND1; MT-ND2; MT-ND3; MT-ND4; MT-ND6 |
| R-HSA-8949613 | Cristae formation | 2.50 | 1.36e-03 | 2.55e-02 | 12/30 | MINOS1; ATP5G3; ATP5I; ATP5J2; ATP5L; ATP5G2; ATP5G1; ATP5H; ATP5A1; ATP5E; ATP5J; ATP5O |
| R-HSA-5663205 | Infectious disease | 2.47 | 0.00e+00 | 0.00e+00 | 109/275 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; CALM2; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; AP2M1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; CANX; RPS18; HMGA1; RPS10; RPL10A; HSP90AB1; RPS12; GTF2H5; PPIA; STX1A; SHFM1; POLR2J; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; TCEB1; RPL30; POLR2K; RPL8; RPS6; RPL12; RPL7A; RPLP2; POLR2L; RPL27A; RPS13; BANF1; RPS3; RPS25; RPS24; RPS26; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL36AL; CALM1; GTF2A2; RPL4; RPLP1; RPS17; RPS2; TCEB2; RPS15A; RPL13; VAMP2; RPL26; RPL23A; RPL23; RPL19; RPL27; SUPT4H1; KPNA2; RPL38; RPL17; PSMA7; RPS21; RPS15; EEF2; RPL36; RPS28; CALR; RPL18A; UBA52; RPS16; RPS19; AP2S1; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; CHMP2A; RANBP1; POLR2F; RPL3; RBX1 |
| R-HSA-397014 | Muscle contraction | 2.38 | 5.32e-05 | 1.15e-03 | 21/55 | ATP1B1; ATP2B4; RYR2; FKBP1B; CALM2; SCN3A; SCN9A; MYL1; ATP1B3; SRI; AKAP9; CALD1; FXYD6; KCNIP2; MYL6B; ATP2B1; CALM1; ACTC1; MYL12A; FXYD7 |
| R-HSA-422475 | Axon guidance | 2.17 | 0.00e+00 | 0.00e+00 | 114/328 | RPL22; RPL11; RPS8; RPL5; RPS27; LHX9; RPS7; RPS27A; RPL31; SCN3A; SCN9A; NRP2; RPL37A; SRGAP3; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; ALCAM; GAP43; AP2M1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; SEMA6A; DPYSL3; RPS14; PDLIM7; CAP2; RPS18; RPS10; RPL10A; HSP90AB1; CD24; RPS12; EZR; ACTB; SHFM1; RPS4X; RPL36A; RPL39; RPL10; DPYSL2; ANK1; RPS20; RPL7; TCEB1; RPL30; PABPC1; RPL8; RPS6; RPL12; RPL7A; VAV2; RPLP2; RPL27A; RPS13; CFL1; RPS3; RPS25; RPS24; TUBA1B; RPS26; RPL41; SRGAP1; RPL6; RPLP0; RPL21; RPS29; RPL36AL; RPL4; RPLP1; RPS17; RPS2; TCEB2; RPS15A; RPL13; EFNB3; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; EIF4A3; ACTG1; MYL12A; RPL17; DCC; PSMA7; RPS21; RPS15; RPL36; TUBB4A; RPS28; RPL18A; UBA52; PSENEN; RPS16; RPS19; AP2S1; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3; RBX1 |
| R-HSA-983712 | Ion channel transport | 2.12 | 5.19e-04 | 1.02e-02 | 20/59 | ATP1B1; ATP2B4; RYR2; FKBP1B; CALM2; RPS27A; ATP1B3; ATP6V1G2; SRI; ATP6V1F; ATP6V0E2; ATP6V1G1; FXYD6; ASIC1; ATP2B1; CALM1; TTYH2; UBA52; FXYD7 |
| R-HSA-5628897 | TP53 Regulates Metabolic Genes | 2.08 | 3.61e-04 | 7.23e-03 | 22/66 | PRDX1; LAMTOR5; LAMTOR2; COX20; COX7A2L; COX5B; COX7C; NDUFA4; CYCS; LAMTOR4; COX7B; G6PD; COX6C; YWHAZ; TXN; COX8A; PRDX5; COX14; COX6A1; COX16; YWHAE; COX6B1 |
SETD5 : 41 significant Reactome pathways
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| R-HSA-156902 | Peptide chain elongation | 11.70 | 0.00e+00 | 0.000000 | 80/84 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-156842 | Eukaryotic Translation Elongation | 11.50 | 0.00e+00 | 0.000000 | 81/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; EEF1B2; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-192823 | Viral mRNA Translation | 11.50 | 0.00e+00 | 0.000000 | 78/83 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-2408557 | Selenocysteine synthesis | 11.20 | 0.00e+00 | 0.000000 | 78/85 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72764 | Eukaryotic Translation Termination | 11.10 | 0.00e+00 | 0.000000 | 78/86 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975956 | Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) | 11.00 | 0.00e+00 | 0.000000 | 79/88 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72689 | Formation of a pool of free 40S subunits | 11.00 | 0.00e+00 | 0.000000 | 85/95 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; EIF3K; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-156827 | L13a-mediated translational silencing of Ceruloplasmin expression | 10.10 | 0.00e+00 | 0.000000 | 87/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; EIF3K; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-72706 | GTP hydrolysis and joining of the 60S ribosomal subunit | 9.94 | 0.00e+00 | 0.000000 | 86/106 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; EIF3E; EIF3H; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; EIF3K; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-2408522 | Selenoamino acid metabolism | 9.77 | 0.00e+00 | 0.000000 | 79/99 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; QARS; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-1799339 | SRP-dependent cotranslational protein targeting to membrane | 9.54 | 0.00e+00 | 0.000000 | 81/104 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; SSR4; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPN2; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72613 | Eukaryotic Translation Initiation | 9.54 | 0.00e+00 | 0.000000 | 88/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; EIF3K; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-72737 | Cap-dependent Translation Initiation | 9.54 | 0.00e+00 | 0.000000 | 88/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; EIF3K; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-72695 | Formation of the ternary complex, and subsequently, the 43S complex | 9.44 | 0.00e+00 | 0.000000 | 37/48 | RPS8; RPS27; RPS7; RPS27A; RPSA; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; RPS21; RPS15; RPS28; EIF3K; RPS16; RPS19; RPS11; RPS9; RPS5; EIF3D; EIF3L |
| R-HSA-927802 | Nonsense-Mediated Decay (NMD) | 9.21 | 0.00e+00 | 0.000000 | 79/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-975957 | Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) | 9.21 | 0.00e+00 | 0.000000 | 79/105 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72662 | Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S | 8.75 | 0.00e+00 | 0.000000 | 40/56 | RPS8; RPS27; RPS7; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; EIF4EBP1; RPS20; PABPC1; EIF3E; EIF3H; RPS6; EIF3F; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; RPS21; RPS15; RPS28; EIF3K; RPS16; RPS19; RPS11; RPS9; RPS5; EIF3D; EIF3L |
| R-HSA-72649 | Translation initiation complex formation | 8.68 | 0.00e+00 | 0.000000 | 39/55 | RPS8; RPS27; RPS7; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; PABPC1; EIF3E; EIF3H; RPS6; EIF3F; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; RPS21; RPS15; RPS28; EIF3K; RPS16; RPS19; RPS11; RPS9; RPS5; EIF3D; EIF3L |
| R-HSA-168273 | Influenza Viral RNA Transcription and Replication | 8.67 | 0.00e+00 | 0.000000 | 80/113 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; POLR2I; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72702 | Ribosomal scanning and start codon recognition | 8.46 | 0.00e+00 | 0.000000 | 38/55 | RPS8; RPS27; RPS7; RPS27A; RPSA; EIF4A2; RPS3A; RPS23; RPS14; RPS18; RPS10; RPS12; RPS4X; RPS20; EIF3E; EIF3H; RPS6; EIF3F; RPS13; EIF3M; RPS3; RPS25; RPS24; RPS29; RPS17; RPS2; RPS15A; RPS21; RPS15; RPS28; EIF3K; RPS16; RPS19; RPS11; RPS9; RPS5; EIF3D; EIF3L |
| R-HSA-168255 | Influenza Life Cycle | 8.40 | 0.00e+00 | 0.000000 | 83/121 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; CANX; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RAN; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; CALR; RPL18A; UBA52; POLR2I; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-168254 | Influenza Infection | 8.04 | 0.00e+00 | 0.000000 | 84/128 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; CANX; RPS18; RPS10; RPL10A; RPS12; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RAN; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; CALR; RPL18A; UBA52; POLR2I; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-9010553 | Regulation of expression of SLITs and ROBOs | 6.57 | 0.00e+00 | 0.000000 | 80/149 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-6791226 | Major pathway of rRNA processing in the nucleolus and cytosol | 6.44 | 0.00e+00 | 0.000000 | 81/154 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; NCL; RPL32; RPL15; RPSA; RPL14; RPL29; GNL3; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; DDX21; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-8868773 | rRNA processing in the nucleus and cytosol | 6.28 | 0.00e+00 | 0.000000 | 82/160 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; NCL; RPL32; RPL15; RPSA; RPL14; RPL29; GNL3; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; DDX21; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72312 | rRNA processing | 6.12 | 0.00e+00 | 0.000000 | 82/164 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; NCL; RPL32; RPL15; RPSA; RPL14; RPL29; GNL3; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NHP2; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; DDX21; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-376176 | Signaling by ROBO receptors | 5.57 | 0.00e+00 | 0.000000 | 81/178 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; PFN1; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-983170 | Antigen Presentation: Folding, assembly and peptide loading of class I MHC | 5.57 | 1.07e-03 | 0.023716 | 5/11 | CANX; HLA-A; HSPA5; PDIA3; CALR |
| R-HSA-71291 | Metabolism of amino acids and derivatives | 4.86 | 0.00e+00 | 0.000000 | 83/209 | RPL22; RPL11; RPS8; RPL5; PHGDH; RPS27; RPS7; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; QARS; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; RPS12; SAT1; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; BCAT1; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; PYCR1; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-72766 | Translation | 4.62 | 0.00e+00 | 0.000000 | 97/257 | RPL22; RPL11; RPS8; RPL5; RPS27; SRP9; RPS7; RPS27A; RPL31; EEF1B2; RPL37A; RPL32; RPL15; RPSA; RPL14; QARS; RPL29; RPL24; MRPL3; RPL22L1; EIF4A2; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; RPS18; RPS10; RPL10A; EEF1A1; RPS12; RPS4X; RPL36A; RPL39; SSR4; RPL10; EIF4EBP1; RPS20; RPL7; RPL30; PABPC1; EIF3E; EIF3H; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; EIF3F; RPL27A; RPS13; EIF3M; RPS3; RPS25; RPS24; RPL41; RPL6; RPLP0; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPN2; RPS21; RPS15; EEF2; RPL36; RPS28; GADD45GIP1; RPL18A; UBA52; EIF3K; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; EIF3D; EIF3L; RPL3 |
| R-HSA-901042 | Calnexin/calreticulin cycle | 4.59 | 1.12e-03 | 0.024031 | 6/16 | RPS27A; CANX; UBC; PDIA3; CALR; UBA52 |
| R-HSA-5663205 | Infectious disease | 4.19 | 0.00e+00 | 0.000000 | 94/275 | RPL22; RPL11; RPS8; RPL5; RPS27; RPS7; CALM2; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; CANX; RPS18; HMGA1; RPS10; RPL10A; HSP90AB1; RPS12; PPIA; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; RPS3; RPS25; RPS24; RPL41; SYT1; RPL6; RPLP0; UBC; RAN; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; VAMP2; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; RPS21; RPS15; EEF2; RPL36; RPS28; CALR; RPL18A; UBA52; POLR2I; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RANBP1; RPL3 |
| R-HSA-422475 | Axon guidance | 3.40 | 0.00e+00 | 0.000000 | 91/328 | RPL22; RPL11; RPS8; RPL5; RPS27; ARPC5; RPS7; RPS27A; RPL31; SCN9A; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; GAP43; RPL22L1; RPL35A; RPL9; RPL34; RPS3A; RPL37; RPS23; DPYSL3; RPS14; RPS18; RPS10; RPL10A; HSP90AB1; RPS12; EZR; RPS4X; RPL36A; DCX; RPL39; RPL10; RPS20; RPL7; RPL30; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; CFL1; RPS3; RPS25; RPS24; TUBA1A; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; PFN1; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; MYL12A; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-381426 | Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) | 3.24 | 1.20e-03 | 0.025223 | 9/34 | PDIA6; IGFBP2; TNC; RCN1; APLP2; HSP90B1; SCG3; CHGB; CST3 |
| R-HSA-114608 | Platelet degranulation | 3.13 | 4.78e-04 | 0.010822 | 11/43 | TAGLN2; CALM2; MANF; PPIA; HSPA5; CFL1; APLP2; SCG3; ALDOA; PFN1; SOD1 |
| R-HSA-76005 | Response to elevated platelet cytosolic Ca2+ | 3.13 | 4.78e-04 | 0.010822 | 11/43 | TAGLN2; CALM2; MANF; PPIA; HSPA5; CFL1; APLP2; SCG3; ALDOA; PFN1; SOD1 |
| R-HSA-1643685 | Disease | 2.69 | 0.00e+00 | 0.000000 | 102/464 | RPL22; RPL11; RPS8; PRDX1; JUN; RPL5; SLC16A1; RPS27; RPS7; CALM2; RPS27A; RPL31; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; RPL22L1; RPL35A; RBPJ; RPL9; RPL34; RPS3A; RPL37; RPS23; RPS14; NPM1; CANX; RPS18; HMGA1; RPS10; RPL10A; HSP90AB1; RPS12; PPIA; SLC25A6; RPS4X; RPL36A; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; BAD; RPS3; RPS25; RPS24; RPL41; SYT1; RPL6; RPLP0; UBC; RAN; RPL21; GPC6; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; VAMP2; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; RPL17; FKBP1A; RPS21; RPS15; EEF2; RPL36; RPS28; PRDX2; CALR; RPL18A; UBA52; POLR2I; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RANBP1; RPL3 |
| R-HSA-1266738 | Developmental Biology | 2.67 | 0.00e+00 | 0.000000 | 97/445 | RPL22; RPL11; RPS8; JUN; RPL5; RPS27; ARPC5; H3F3A; RPS7; RPS27A; RPL31; SCN9A; RPL37A; RPL32; RPL15; RPSA; RPL14; RPL29; RPL24; GAP43; RPL22L1; RPL35A; RBPJ; RPL9; H2AFZ; RPL34; RPS3A; RPL37; RPS23; DPYSL3; RPS14; RPS18; RPS10; RPL10A; HSP90AB1; RPS12; EZR; RPS4X; RPL36A; DCX; RPL39; RPL10; RPS20; RPL7; RPL30; POLR2K; PABPC1; RPL8; RPS6; RPL35; RPL12; RPL7A; RPLP2; RPL27A; RPS13; CFL1; RPS3; RPS25; RPS24; TUBA1A; RPL41; RPL6; RPLP0; UBC; RPL21; RPS29; RPL4; RPLP1; RPS17; RPS2; RPS15A; RPL13; PFN1; RPL26; RPL23A; RPL23; RPL19; RPL27; RPL38; MYL12A; RPL17; RPS21; RPS15; RPL36; RPS28; RPL18A; UBA52; POLR2I; RPS16; RPS19; RPL18; RPL13A; RPS11; RPS9; RPL28; RPS5; RPL3 |
| R-HSA-163200 | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 2.47 | 3.09e-05 | 0.000760 | 23/114 | UQCRH; COX7A2L; ATP5G3; NDUFS4; COX7C; NDUFA4; CYCS; NDUFB2; NDUFB11; COX7B; NDUFA1; COX8A; COX6A1; COX5A; NDUFB10; ATP5G1; ATP5A1; ATP5D; COX6B1; MT-ND2; MT-ND3; MT-ND4; MT-CYB |
| R-HSA-611105 | Respiratory electron transport | 2.45 | 1.74e-04 | 0.004160 | 19/95 | UQCRH; COX7A2L; NDUFS4; COX7C; NDUFA4; CYCS; NDUFB2; NDUFB11; COX7B; NDUFA1; COX8A; COX6A1; COX5A; NDUFB10; COX6B1; MT-ND2; MT-ND3; MT-ND4; MT-CYB |
| R-HSA-1428517 | The citric acid (TCA) cycle and respiratory electron transport | 2.40 | 7.53e-06 | 0.000191 | 28/143 | UQCRH; SLC16A1; COX7A2L; ATP5G3; NDUFS4; COX7C; VDAC1; NDUFA4; CYCS; NDUFB2; NDUFB11; COX7B; NDUFA1; LDHA; COX8A; LDHB; COX6A1; COX5A; NDUFB10; ATP5G1; ATP5A1; BSG; ATP5D; COX6B1; MT-ND2; MT-ND3; MT-ND4; MT-CYB |
| Marker | ADNP | ARID1B | ASH1L | CHD2 | PTEN | SETD5 |
| Signif_GO_terms | 137 | 59 | 13 | 48 | 46 | 41 |