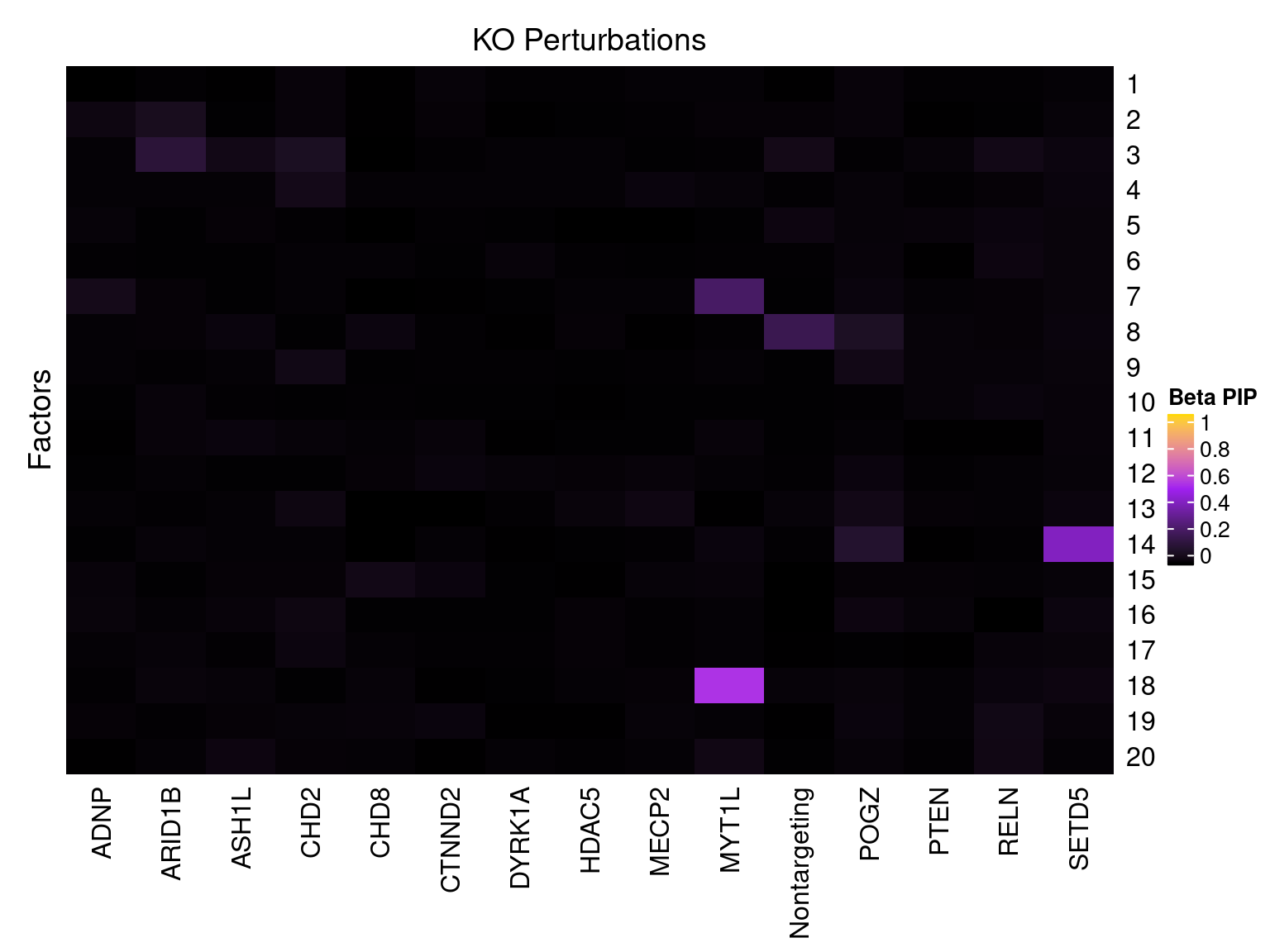
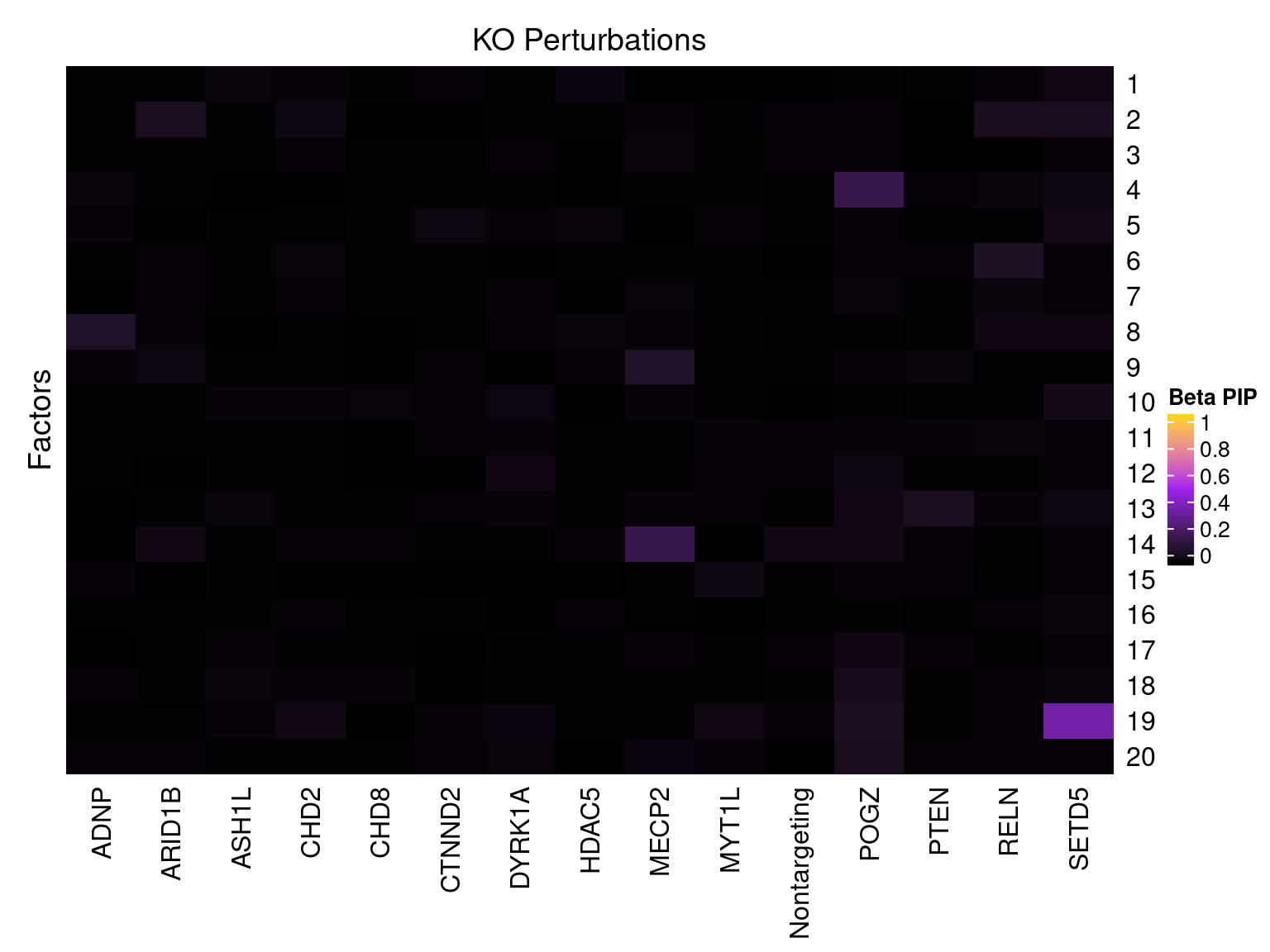
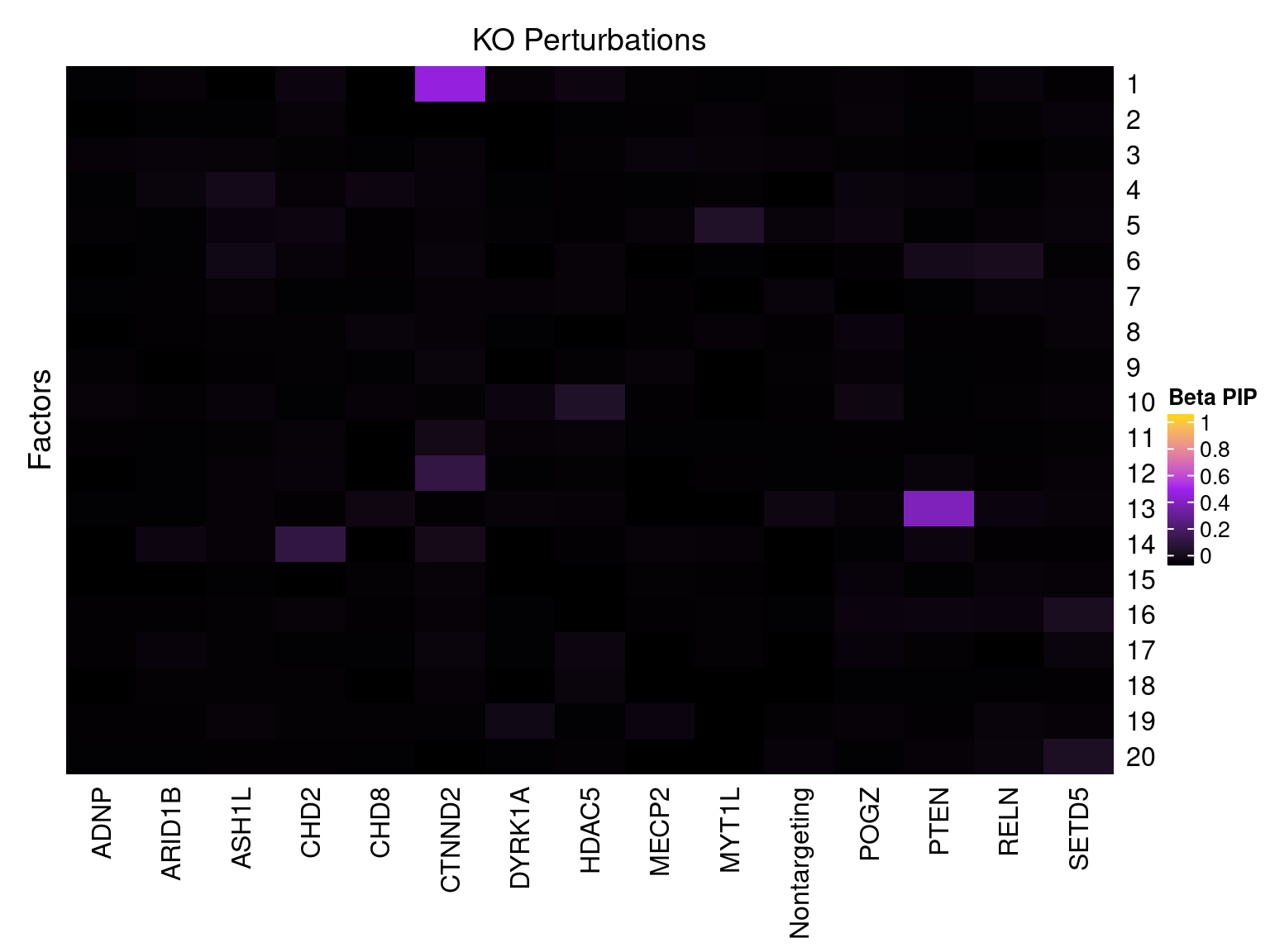
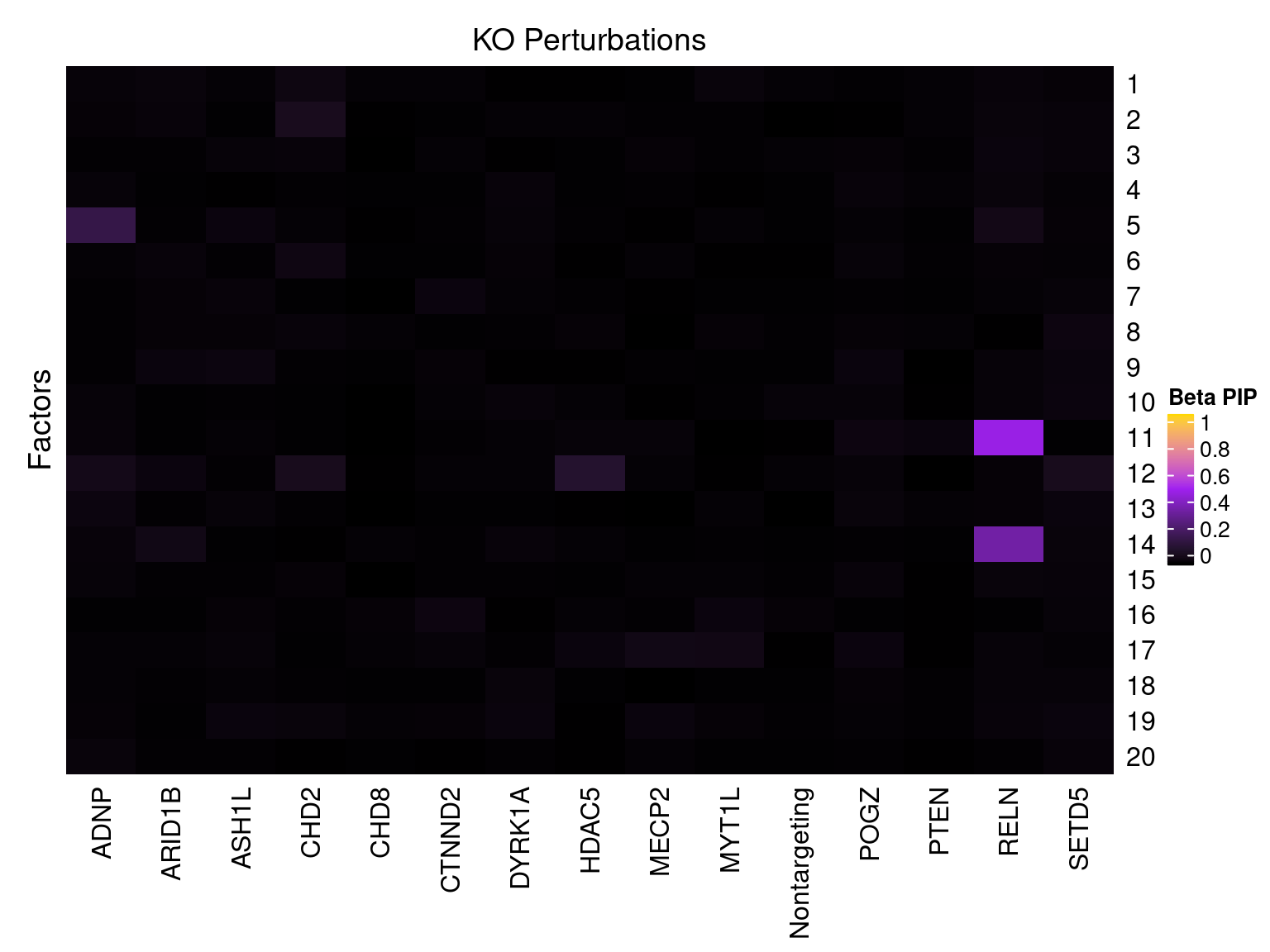
Single cell expression data
Source:
High-throughput single-cell functional elucidation of neurodevelopmental disease-associated genes reveals convergent mechanisms altering neuronal differentiation, GEO accession: GSE142078.
Perturbations:
CRISPR knock-down of 14 autism spectrum disorder (ASD)–associated genes (3 gRNAs per gene) + 5 non-targeting gRNAs.
Cells:
Lund human mesencephalic (LUHMES) neural progenitor cell line.
Cells from 3 batches were merged together into 1 analysis. All cells have only a single type of gRNA readout. Quality control resulted in 8708 cells.
Genes:
Only genes detected in > 10% of cells were kept, resulted in 6213 genes.
Normalization:
Seurat “LogNormalize”: log(count per 10K + 1).
Batch effect, unique UMI count, library size, and mitochondria percentage were all corrected for. The corrected and scaled expression data were used as input for subsequent factor analysis.
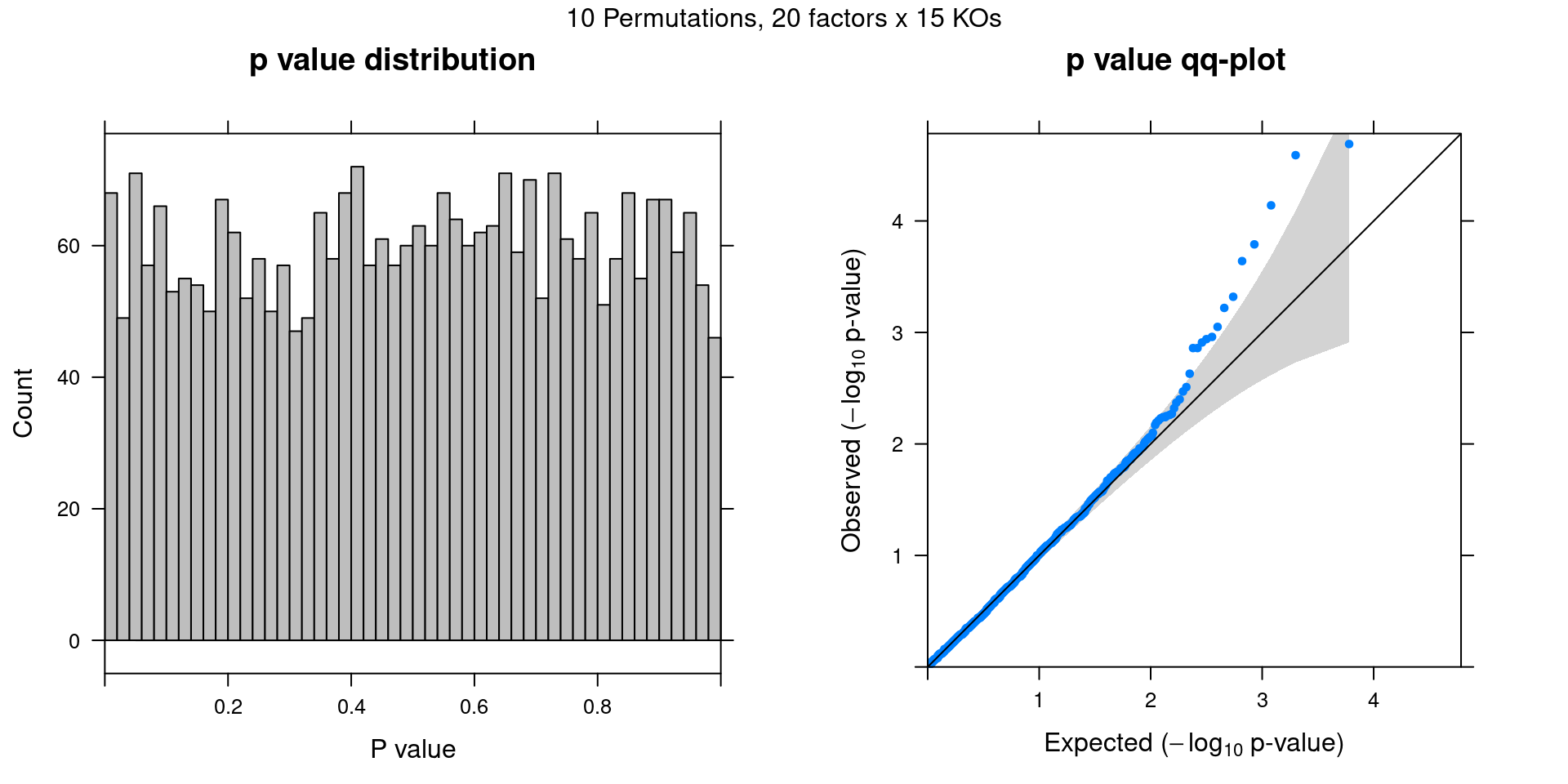
Permutation
Cell labels in the expression data were permuted randomly so that they are no longer correlated with the knock-down conditions. Then GSFA was performed still using all conditions as guides. Factor-guide association as well as the LFSR of each gene were evaluated as usual.
In total, 10 random permutation rounds like this were conducted.
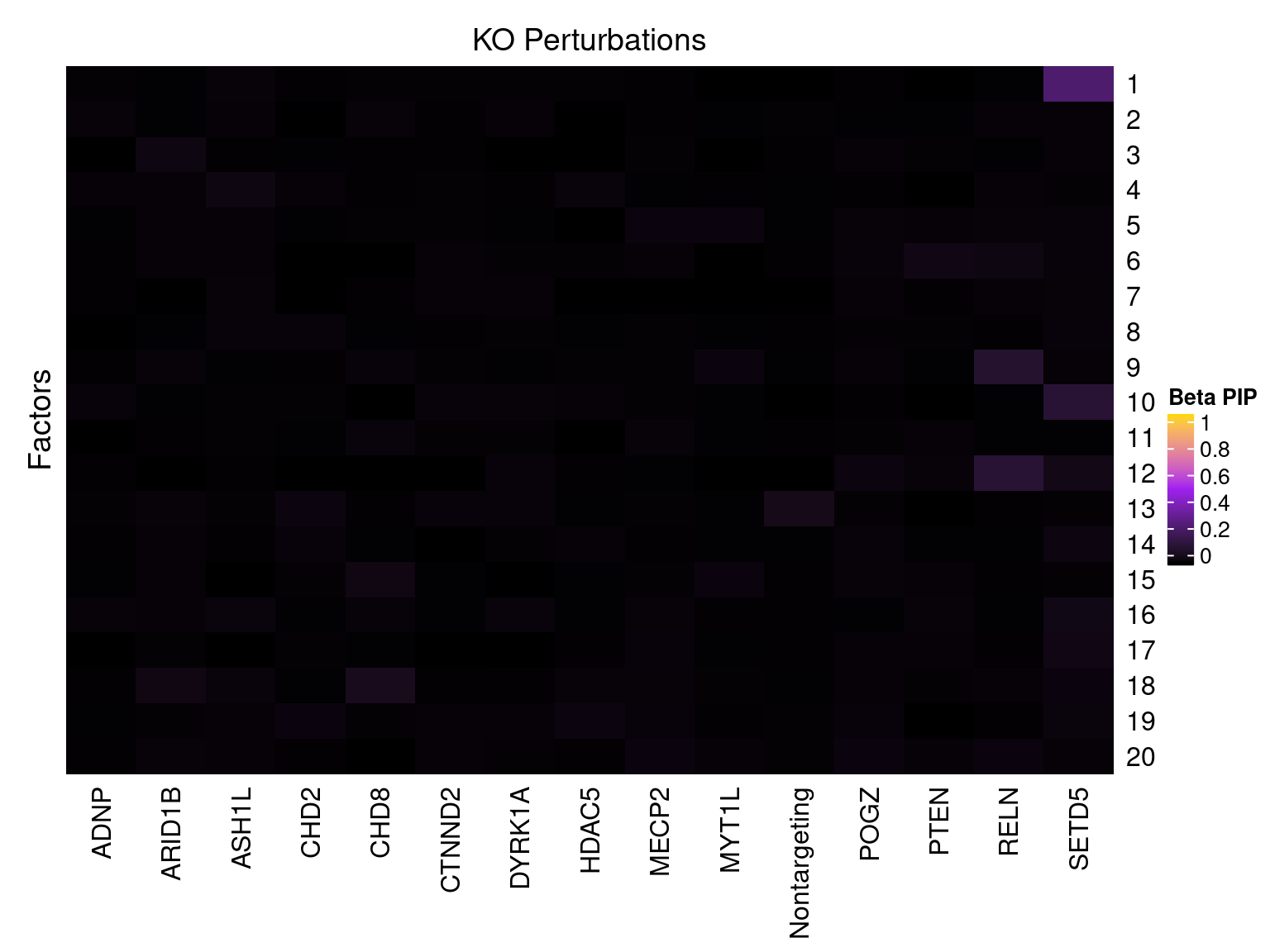
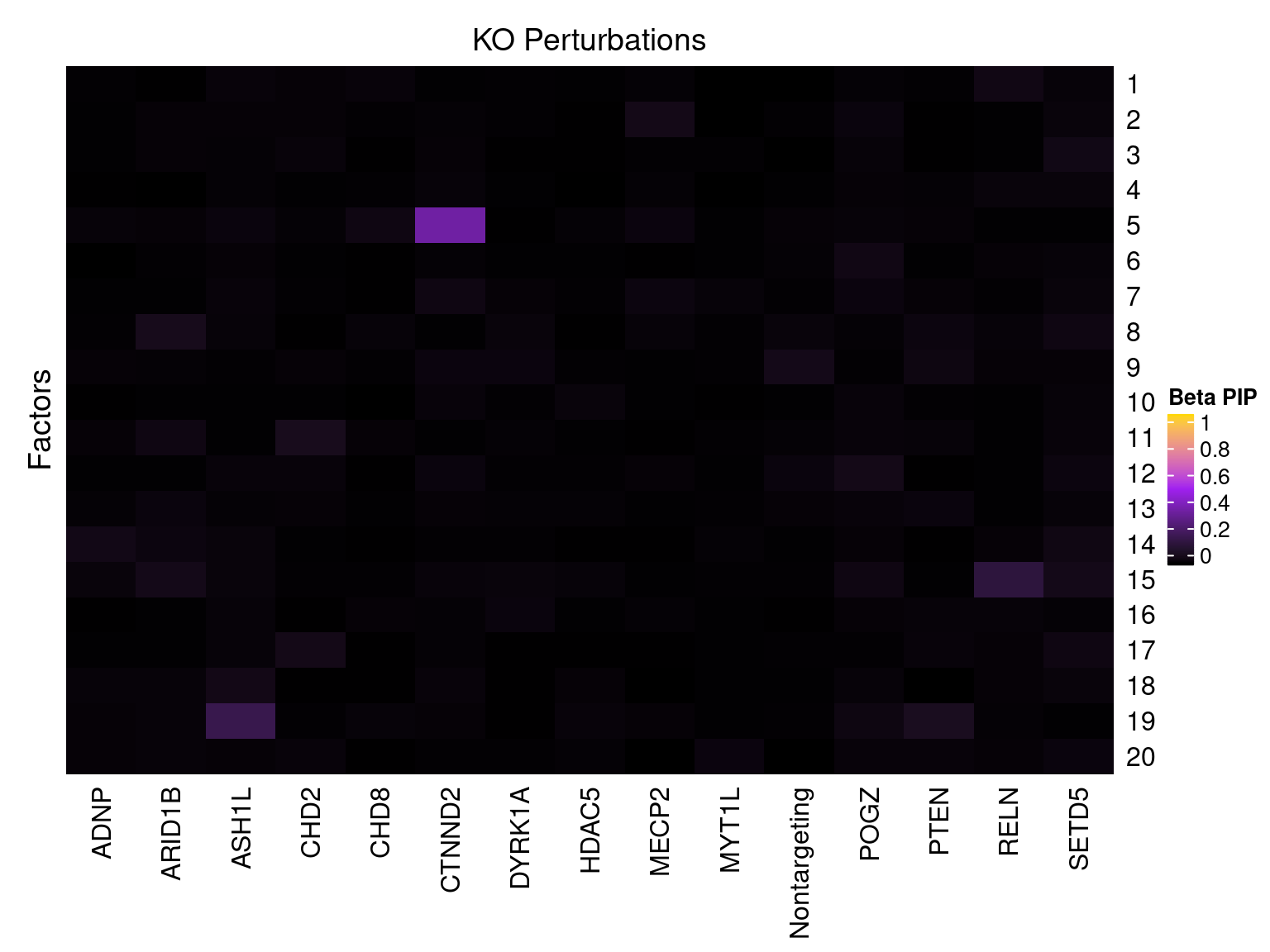
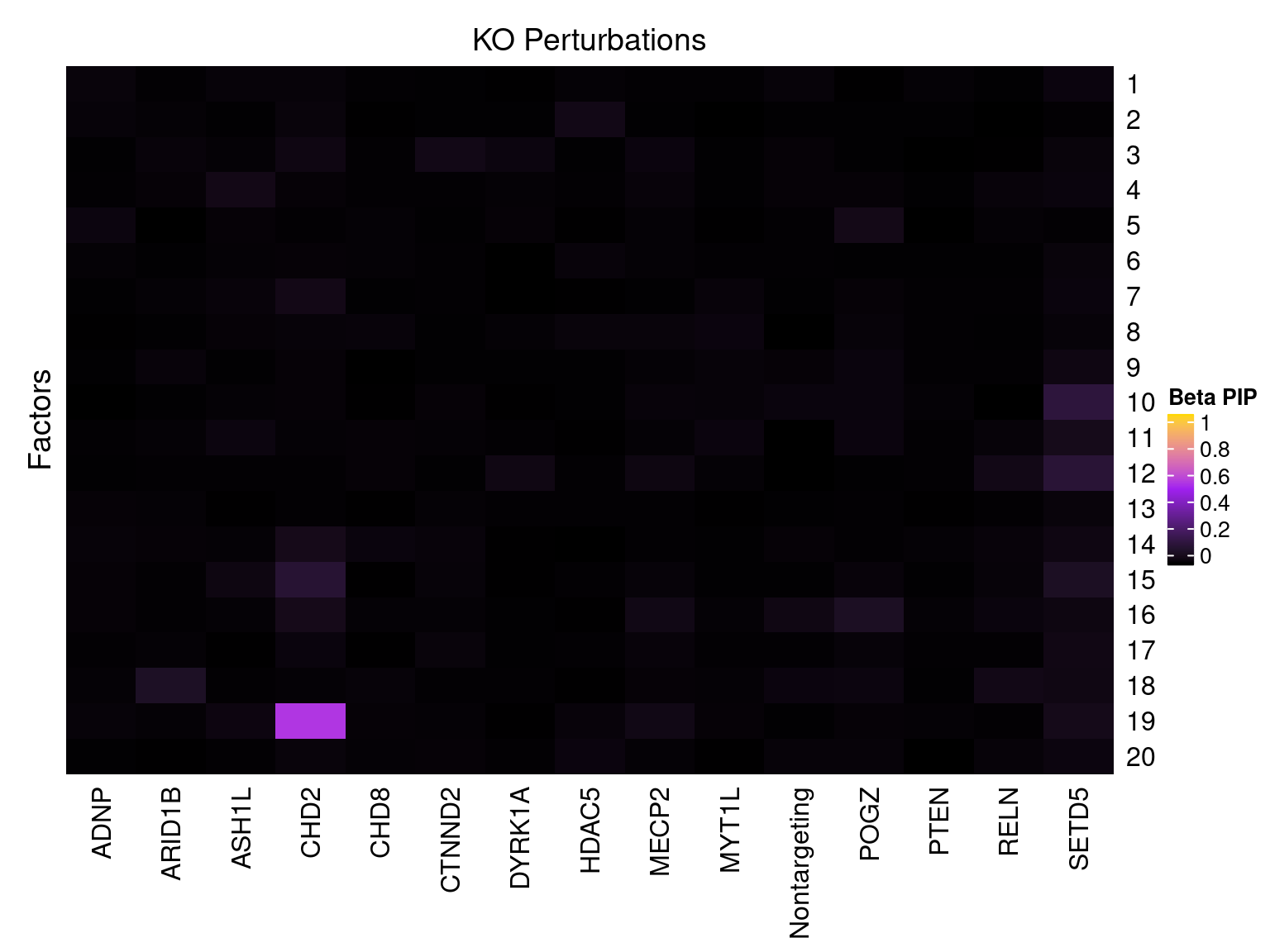
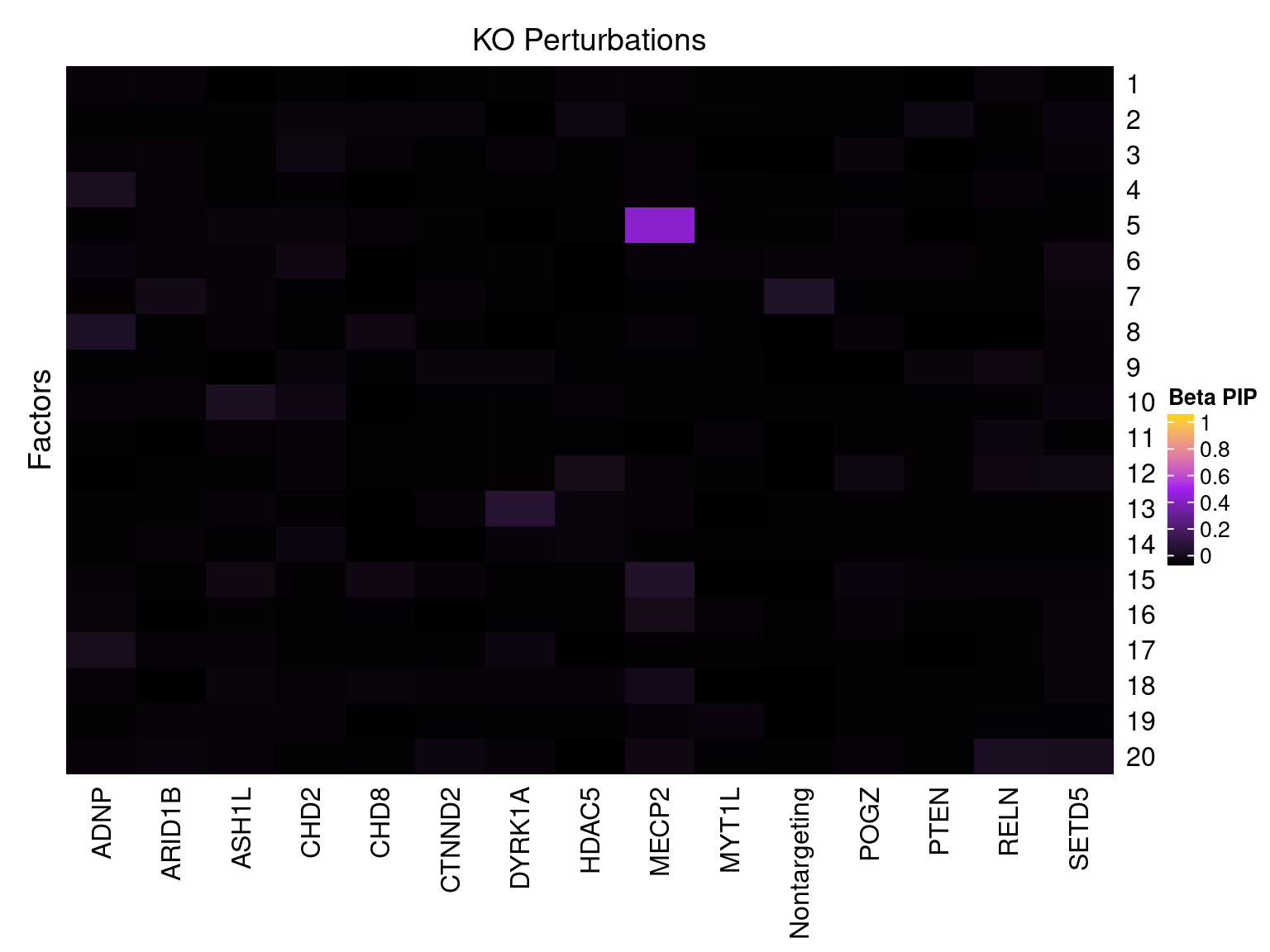
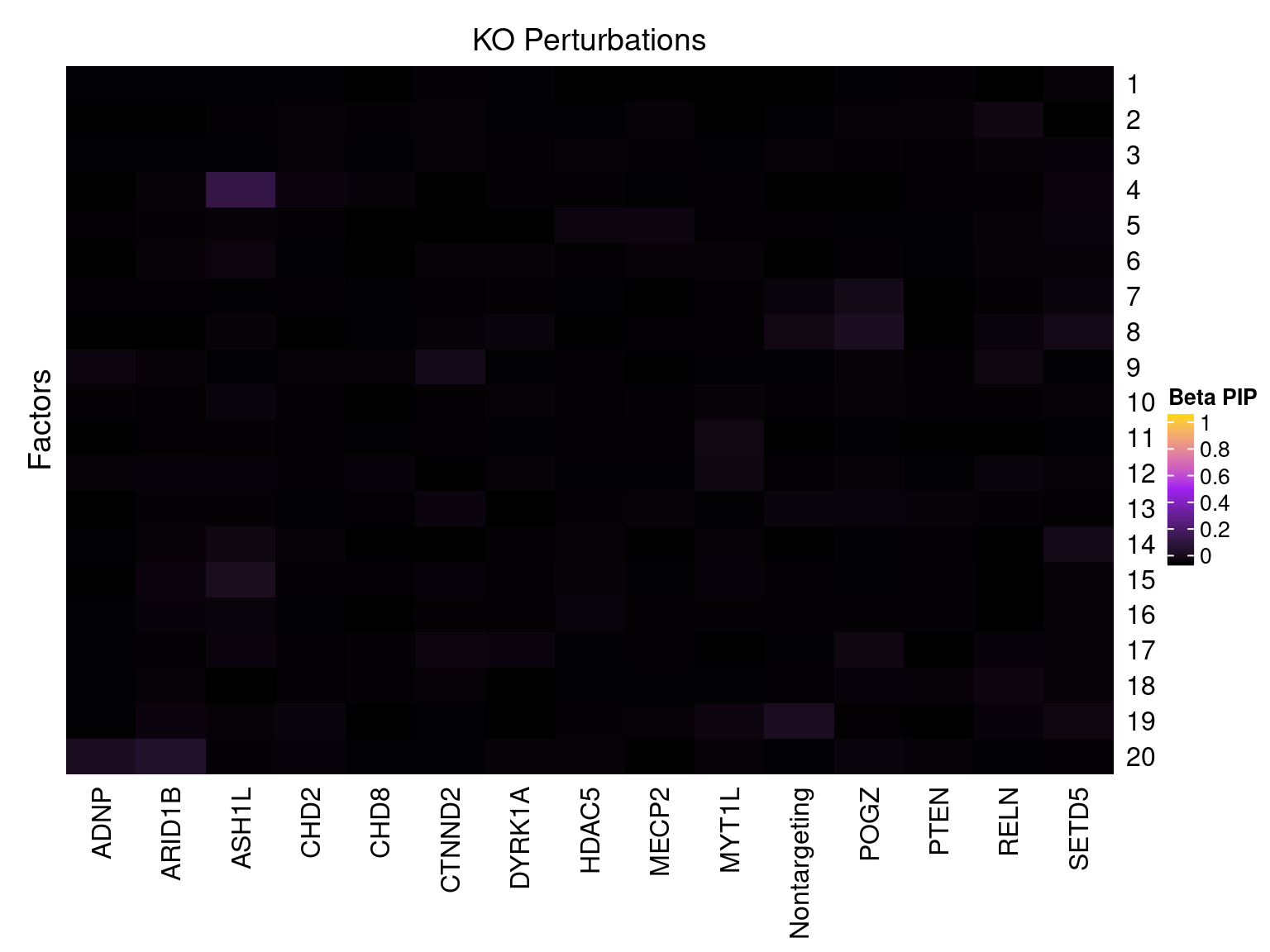
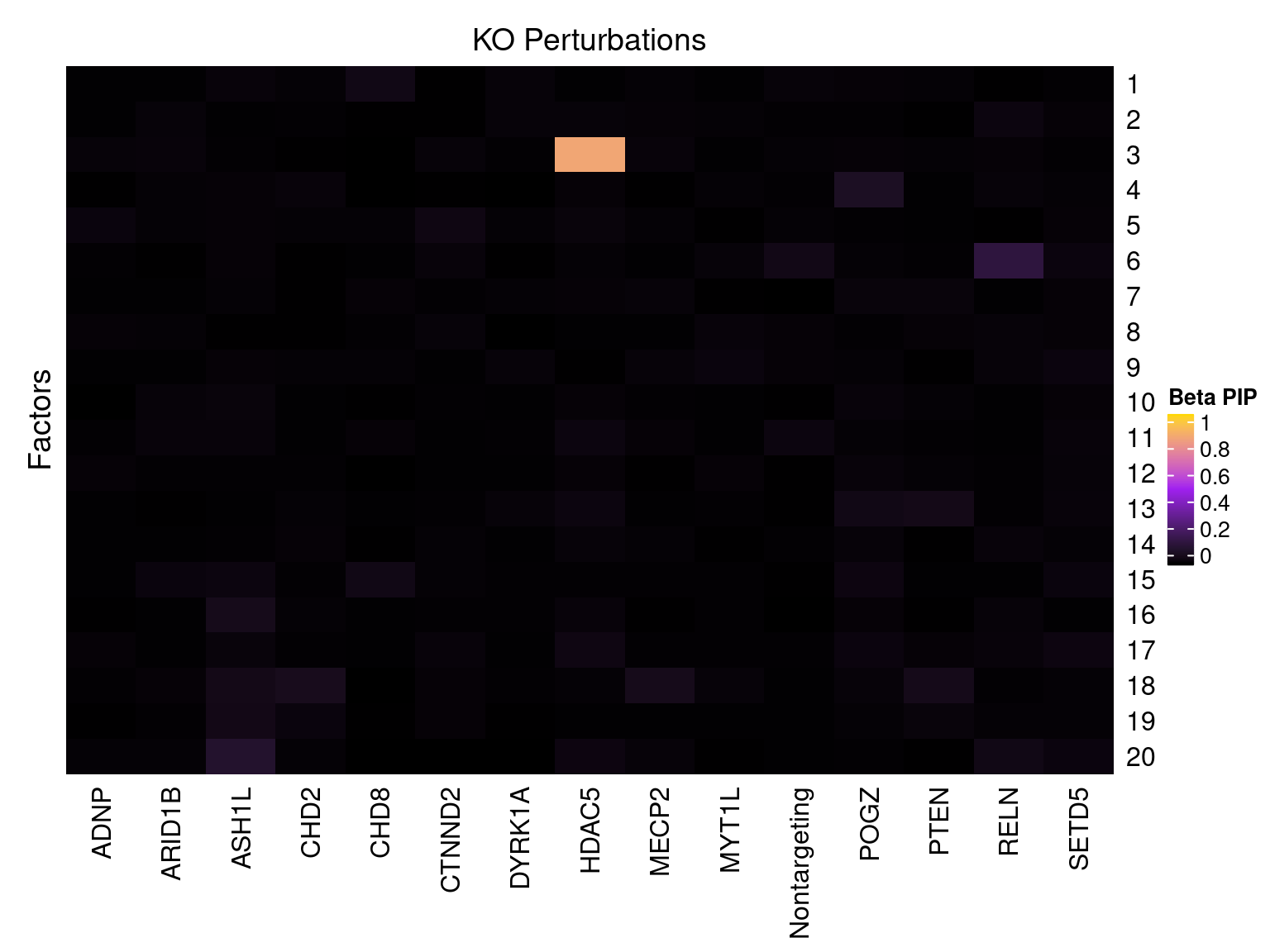
Factor ~ KO Posterior Association
LFSR
Permutation 1, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 2, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 3, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 4, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 5, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 6, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 7, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 8, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 9, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 10, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
ADNP
|
ARID1B
|
ASH1L
|
CHD2
|
CHD8
|
CTNND2
|
DYRK1A
|
HDAC5
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
MECP2
|
MYT1L
|
Nontargeting
|
POGZ
|
PTEN
|
RELN
|
SETD5
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|