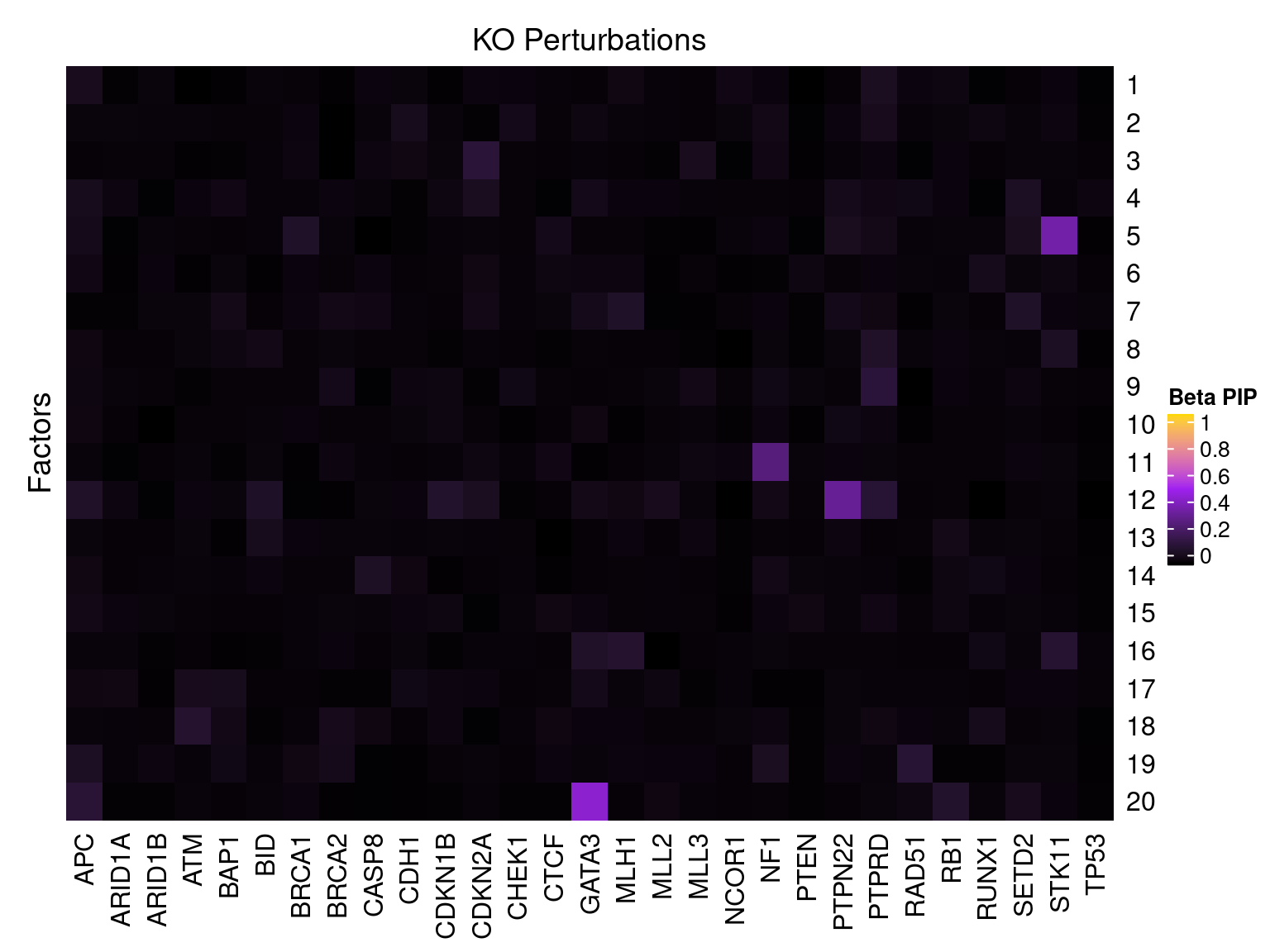
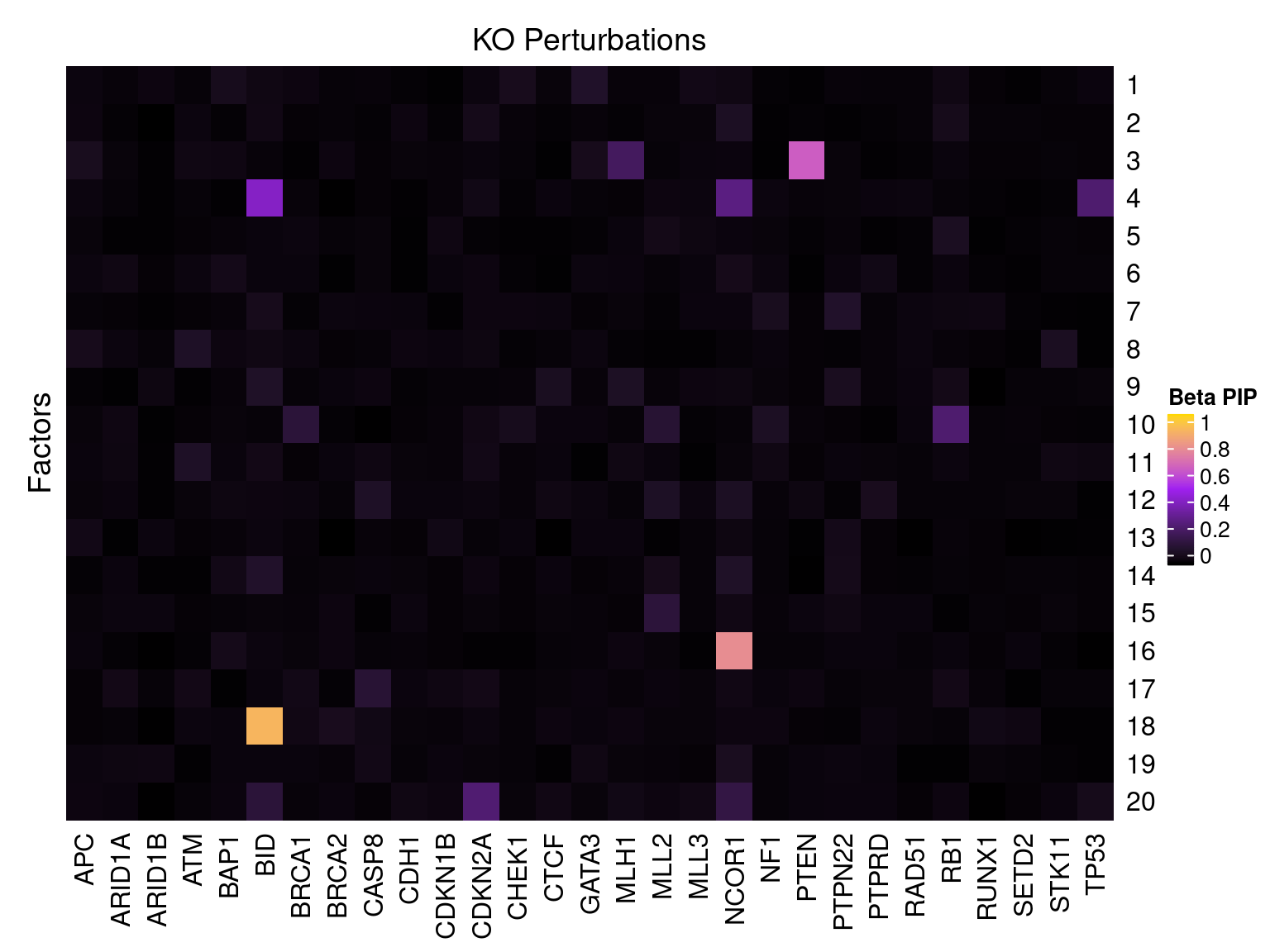
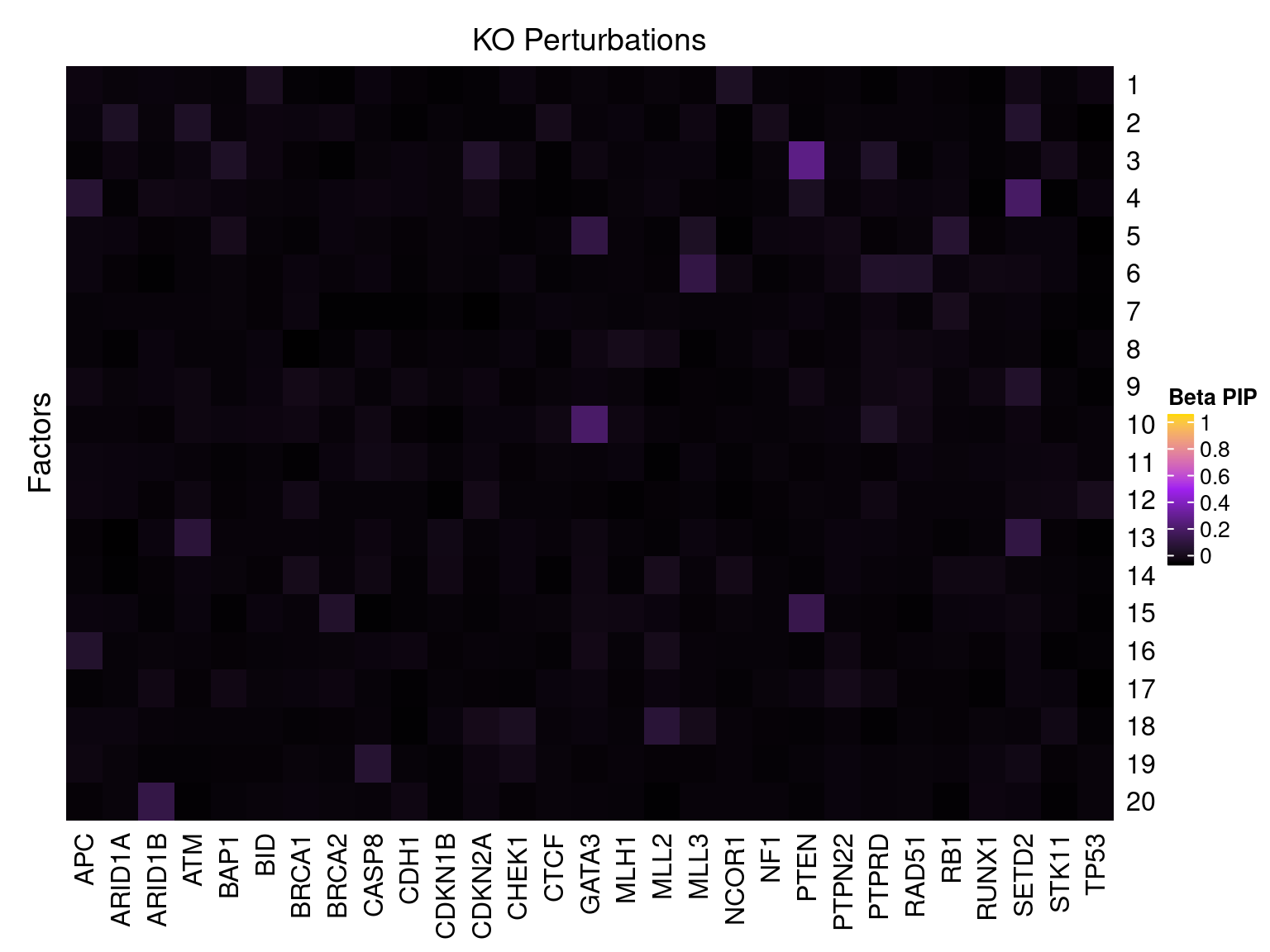
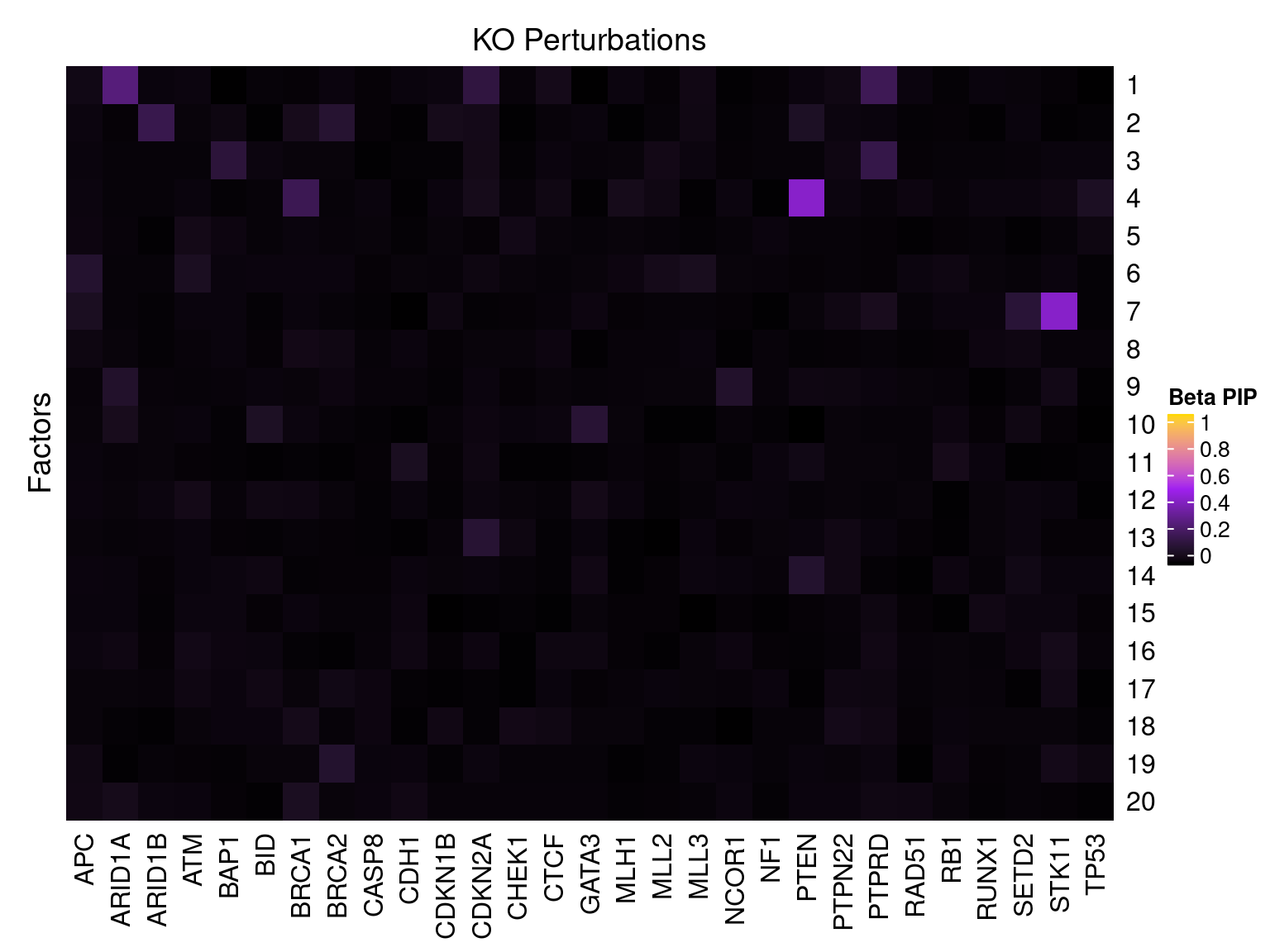
Single cell expression data
CROP-seq data are from this paper: On the design of CRISPR-based single cell molecular screens, GEO accession: GSE108699.
Perturbations:
Knock-outs of 29 tumor-suppressor genes (TP53, …), 1 non-targeting control.
Cells:
MCF10A cells (normal human breast epithelial cells) with exposure to a DNA damaging agent, doxorubicin.
Only cells with a single type of gRNA readout were kept, resulted in 4507 cells.
Genes:
Only genes detected in > 10% of cells were kept, resulted in 8046 genes.
Normalization:
Seurat “LogNormalize”: log(count per 10K + 1).
Library size was regressed out, so were the non-targeting control (NTC) condition per cell. The residuals were used as input.
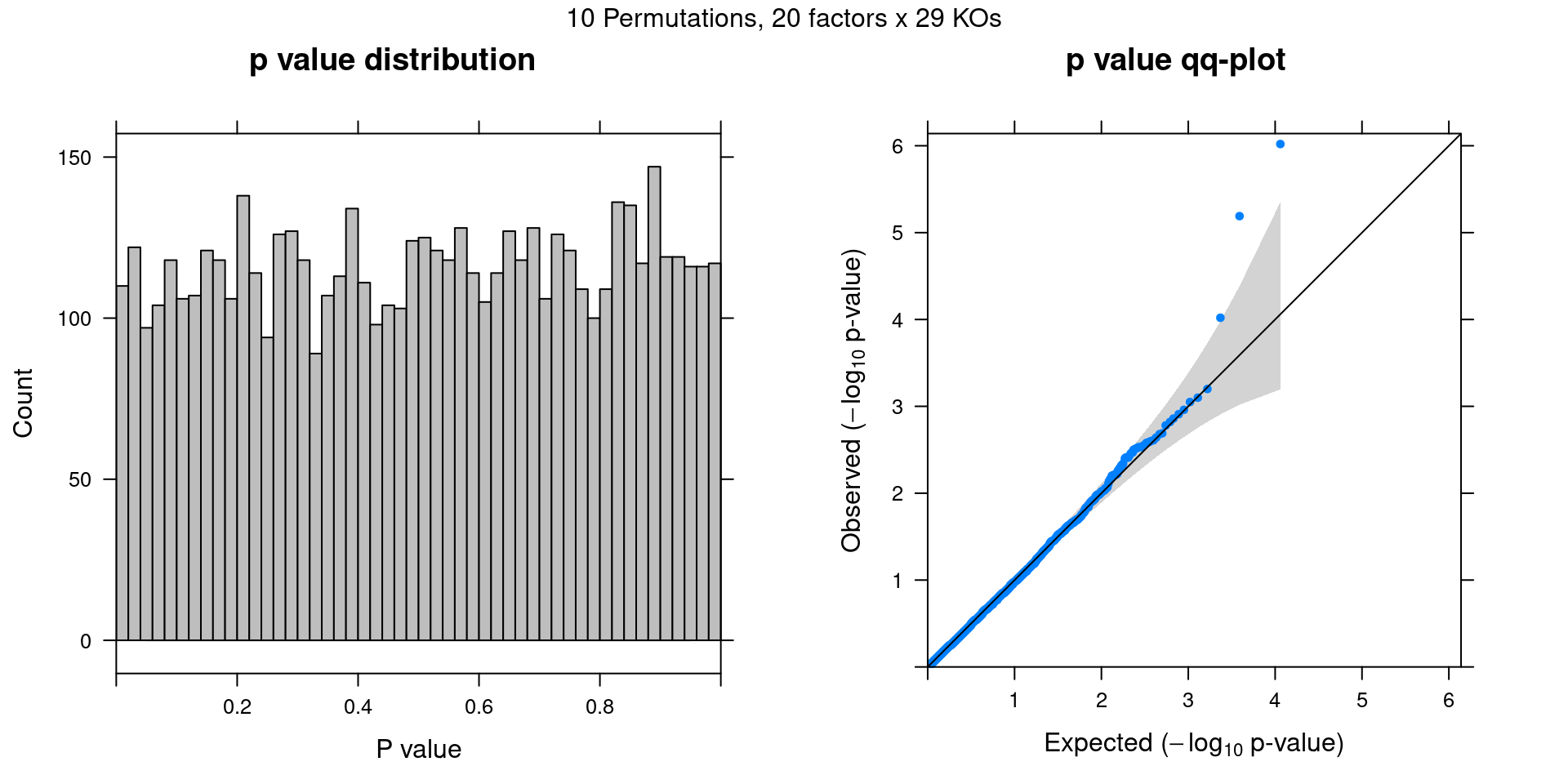
Permutation
Cell labels in the expression data were permuted randomly so that they are no longer correlated with the KO conditions. Then GSFA was performed still using all conditions other than NTC as guides.
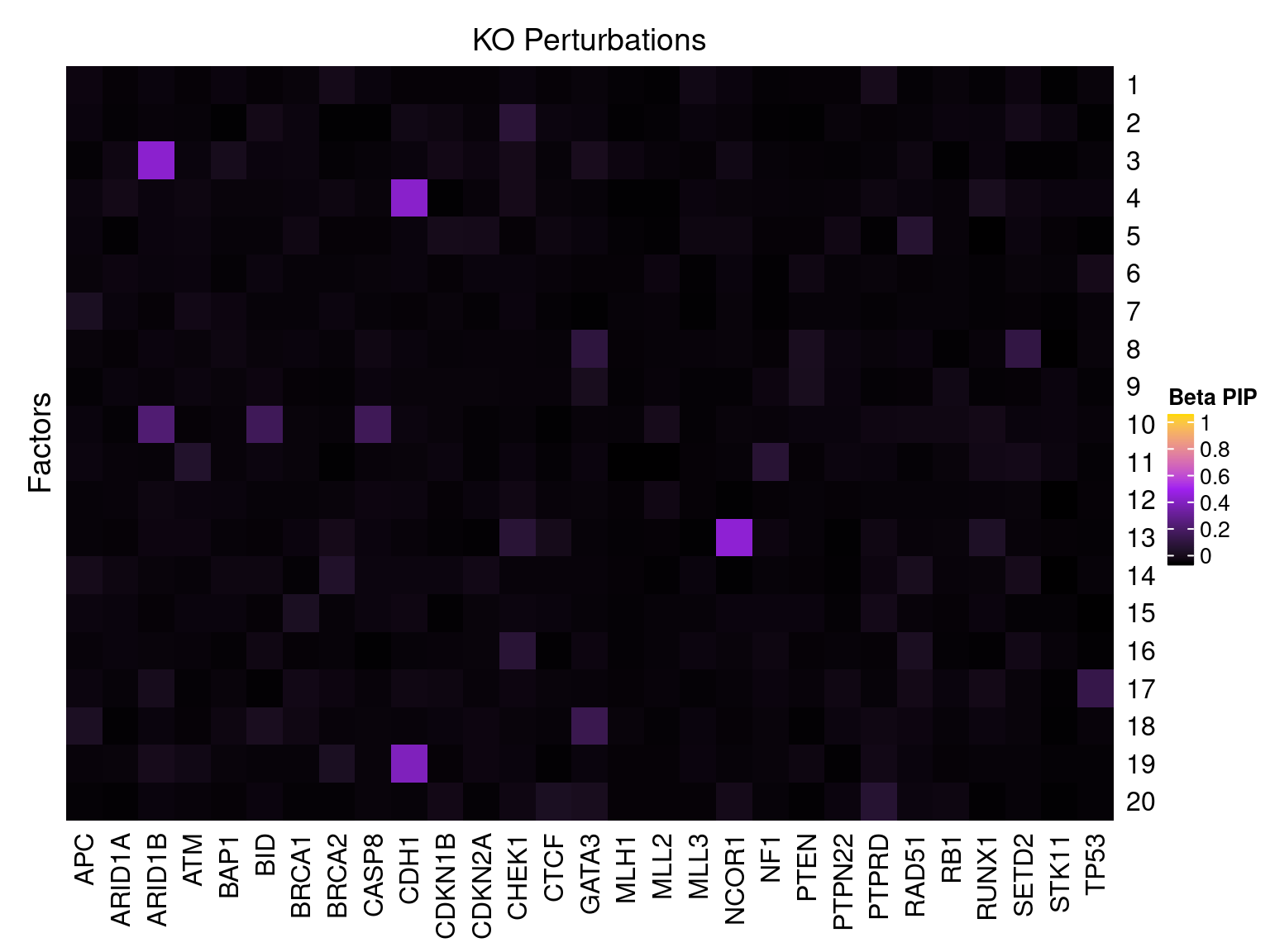
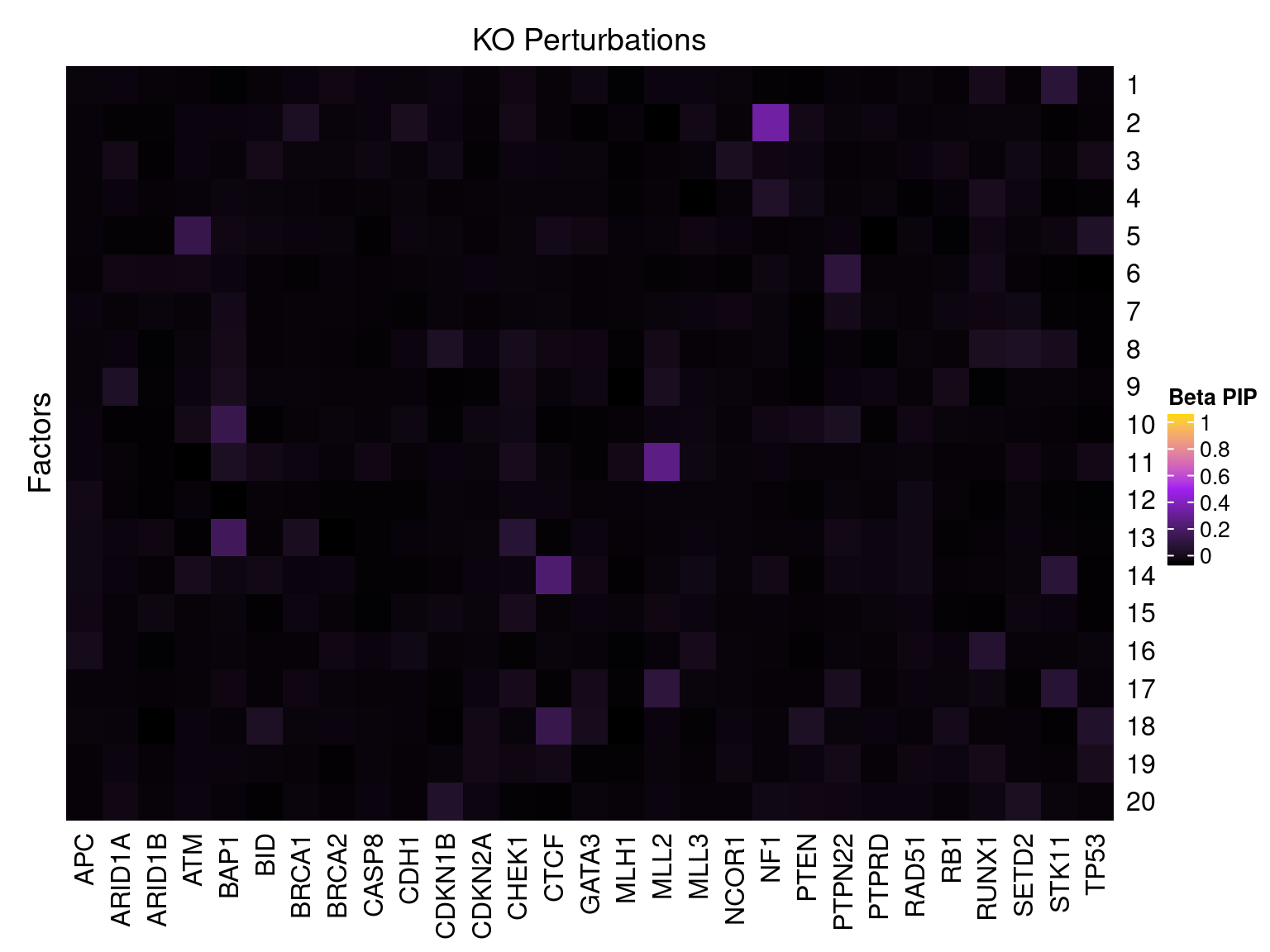
Factor ~ KO Posterior Association
LFSR
Permutation 1, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 2, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 3, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 4, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
371
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 5, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 6, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 7, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 8, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 9, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|
Permutation 10, # of genes that passed LFSR cutoff of 0.05:
|
KO
|
APC
|
ARID1A
|
ARID1B
|
ATM
|
BAP1
|
BID
|
BRCA1
|
BRCA2
|
CASP8
|
CDH1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
CDKN1B
|
CDKN2A
|
CHEK1
|
CTCF
|
GATA3
|
MLH1
|
MLL2
|
MLL3
|
NCOR1
|
NF1
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
|
KO
|
PTEN
|
PTPN22
|
PTPRD
|
RAD51
|
RB1
|
RUNX1
|
SETD2
|
STK11
|
TP53
|
|
|
Num_genes
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
0
|
NA
|