Guided Factor Analysis on TCGA BRCA Gene Expression Data
-- Normalization: TMM, Guides: LOF and Missense Mutations Separated
Yifan Zhou (zhouyf@uchicago.edu)
2022-03-23
1 Data Description
References: Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics.
Data source:
FireBrowse TCGA BRCA Archives.
Samples:
TCGA breast invasive carcinoma (BRCA) tumor samples in file "illuminahiseq_rnaseq-gene_expression".
To avoid possible confounding due to gender and race, we confined our study samples to be only from female Caucasian subjects.
In addition, only samples with somatic mutation annotation were kept, resulted in 573 samples.
Genes:
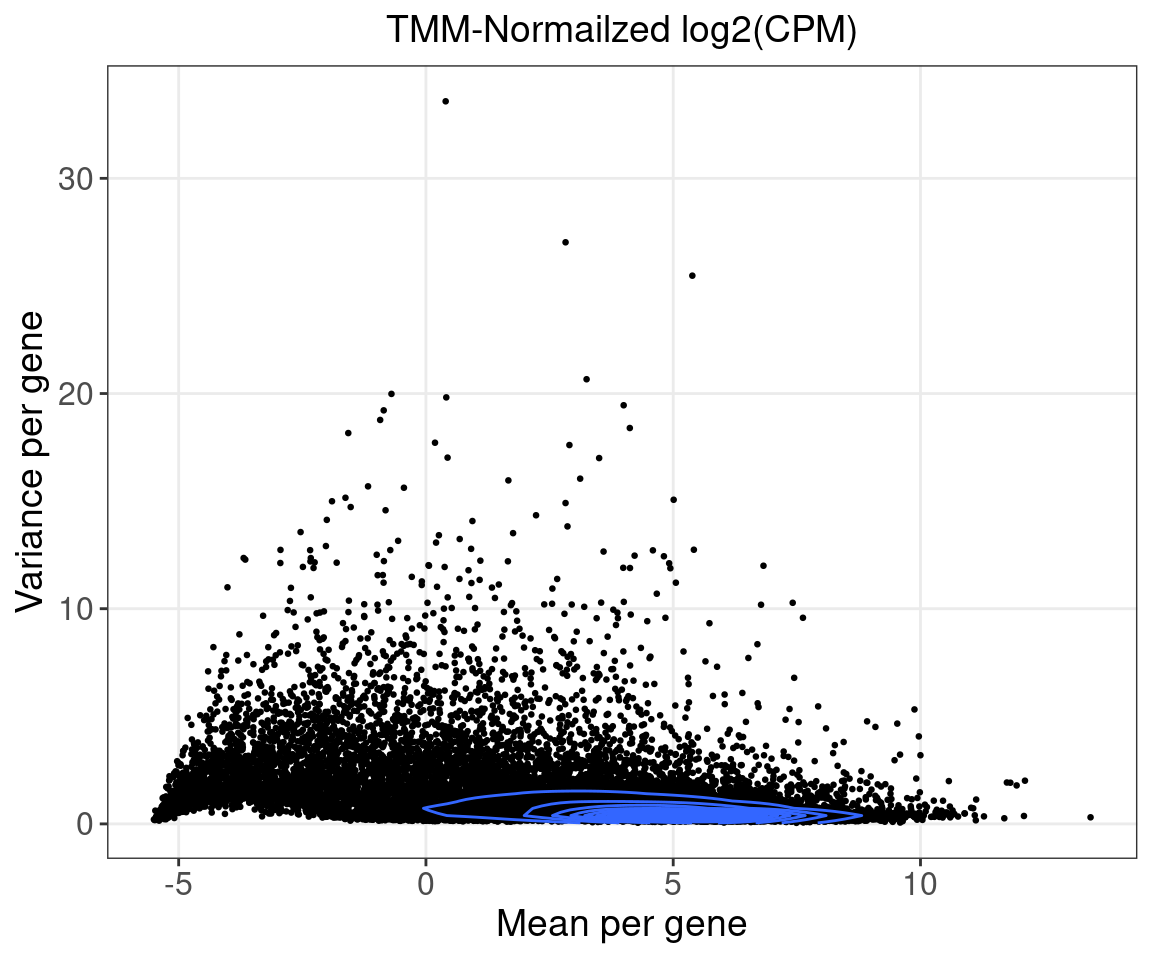
Only the top 10k variable genes in samples of consideration were kept. (Variance was calculated based on the TMM-normalized log2(CPM) data.)
Normalization:
Raw counts normalized by TMM using edgeR, converted to log2(CPM), then corrected for subject age and tumor purity. The corrected and scaled expression data were used as input for subsequent factor analysis.
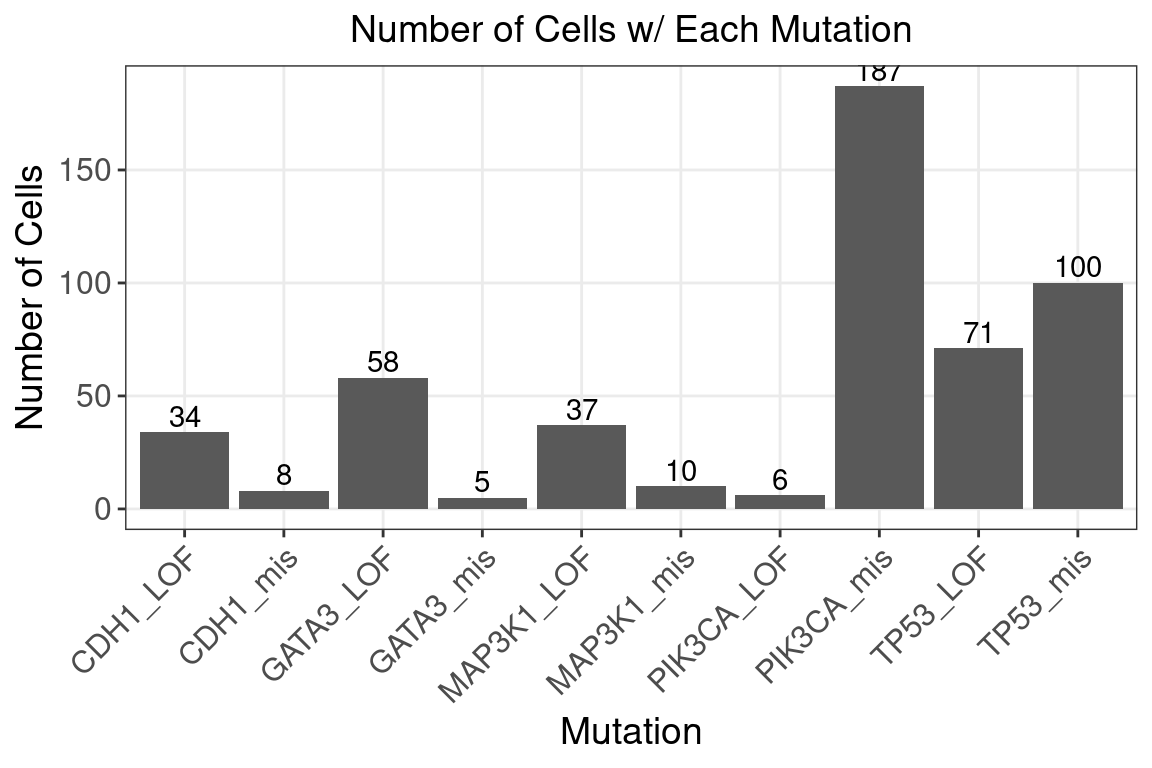
Perturbations:
Somatic mutation status of selected frequently mutated driver genes (TP53, CDH1, GATA3, MAP3K1, PIK3CA).
Mutations were classified into 2 types: missense mutations and loss of function (LOF) mutations.
2 GSFA results
Here, our "guide", \(G\) matrix, consists of the somatic mutation status (missense and LOF separated) of top 5 driver genes across samples.
We specified 20 factors in our model. In each case, Gibbs sampling was conducted for 3000 iterations, and the posterior mean estimates were averaged over the last 1000 iterations.
3 Factor ~ Perturbation Association
3.1 Perturbation effects on factors
Fisrt of all, we look at the estimated effects of gene perturbations on factors inferred by GSFA.
Factors from bulk data were originally very dense, so when imposed upon a normal-mixture prior, they all become really sparse:
| Factor | Factor_1 | Factor_2 | Factor_3 | Factor_4 | Factor_5 |
| Density | 0.0000 | 0.0004 | 0.0386 | 0.0837 | 0.0514 |
| Factor | Factor_6 | Factor_7 | Factor_8 | Factor_9 | Factor_10 |
| Density | 0.0087 | 0.1005 | 0.0378 | 0.0377 | 0.0480 |
| Factor | Factor_11 | Factor_12 | Factor_13 | Factor_14 | Factor_15 |
| Density | 0.0148 | 0.0012 | 0.0304 | 0.0418 | 0.0309 |
| Factor | Factor_16 | Factor_17 | Factor_18 | Factor_19 | Factor_20 |
| Density | 0.0656 | 0.0262 | 0.0185 | 0.0391 | 0.0422 |
We found that 5 somatic mutations, TP53_LOF, TP53_mis, CDH1_LOF, GATA3_LOF and PIK3CA_mis, has significant effects (PIP > 0.95) on at least 1 of the 20 inferred factors.
Effects of all targets on all factors: 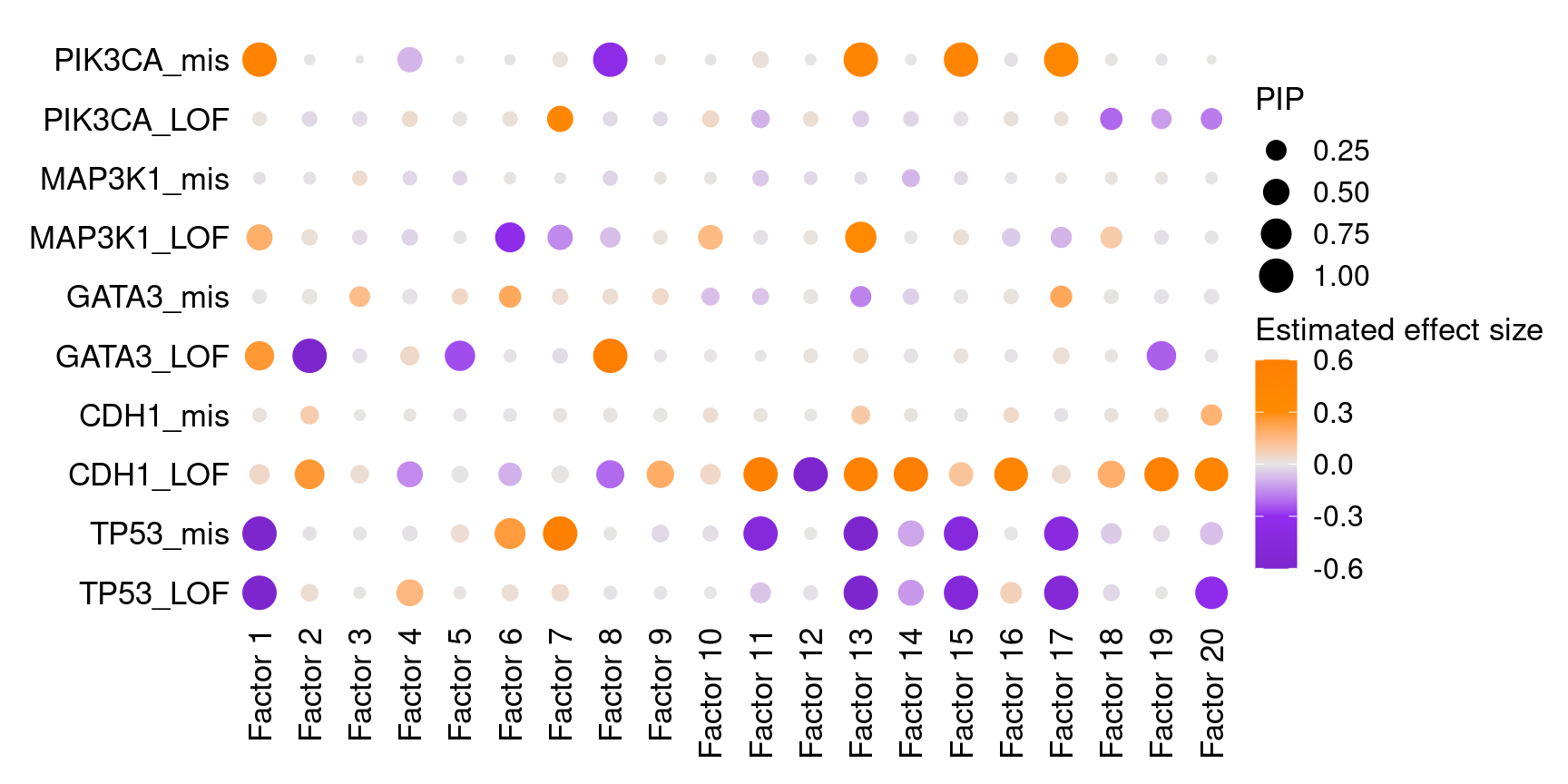
Here is a closer look at the estimated effects of selected mutations on selected factors:
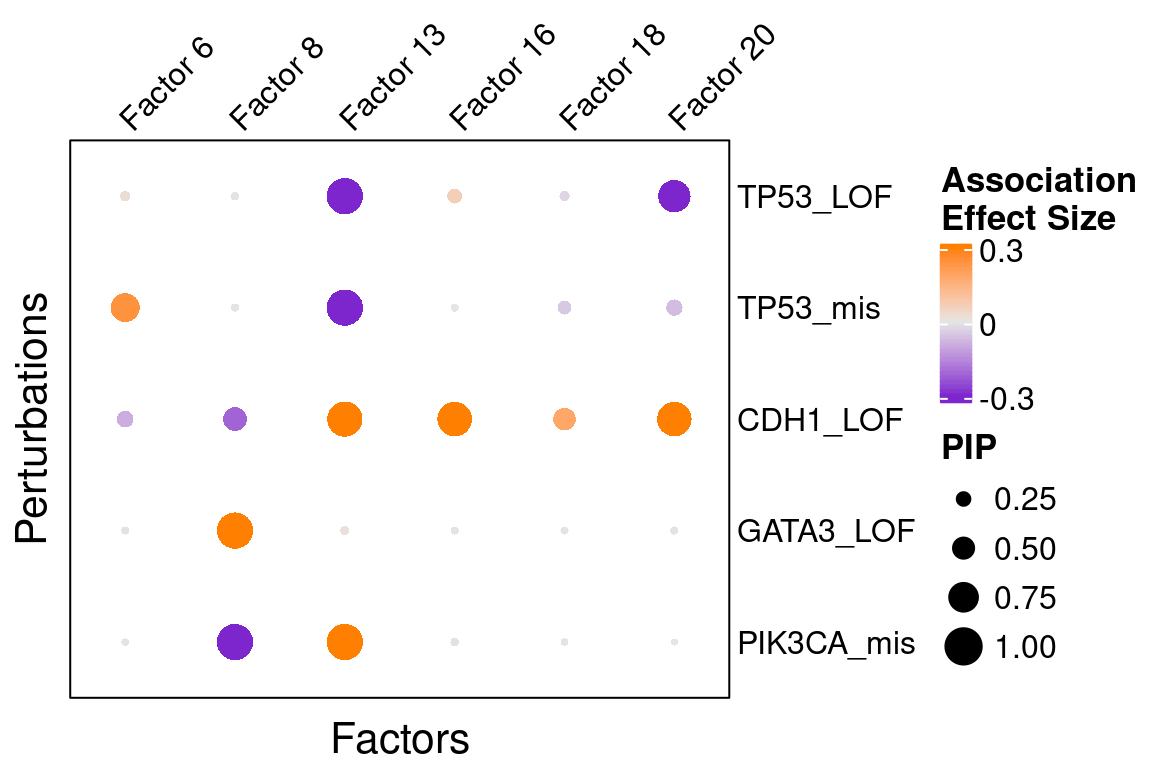
Enriched GO terms of selected factors:
4 (cell/tissue development), 5 (immune activation), 8 (neural development),
13 (cell cycle), 16 (immune response), 18 (immune response), 20 (vessel development).
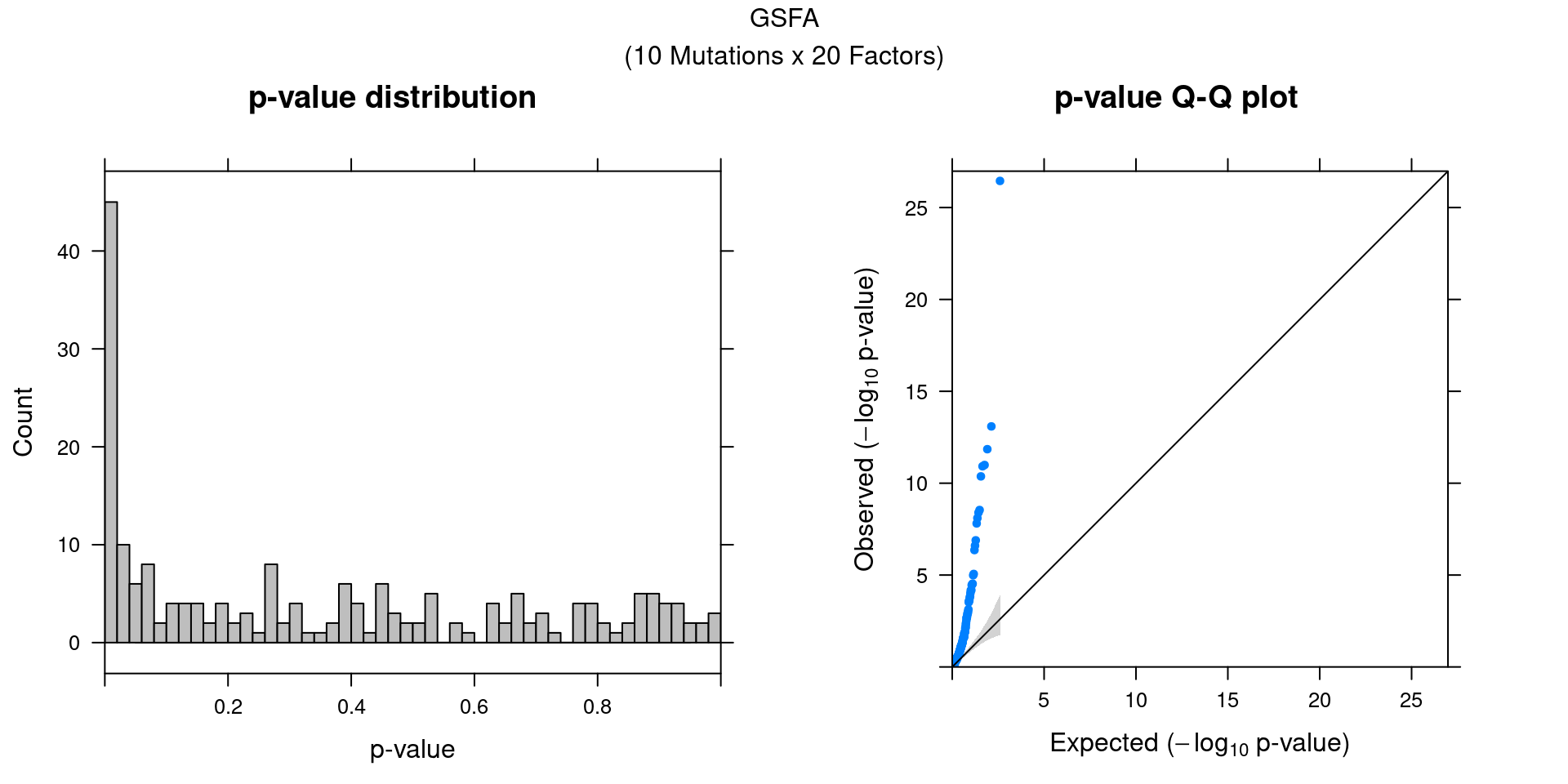
3.2 Factor-perturbation association p values
We can also assess the correlations between each pair of perturbation and inferred factor.
The distribution of correlation p values show significant signals.
4 Factor Interpretation
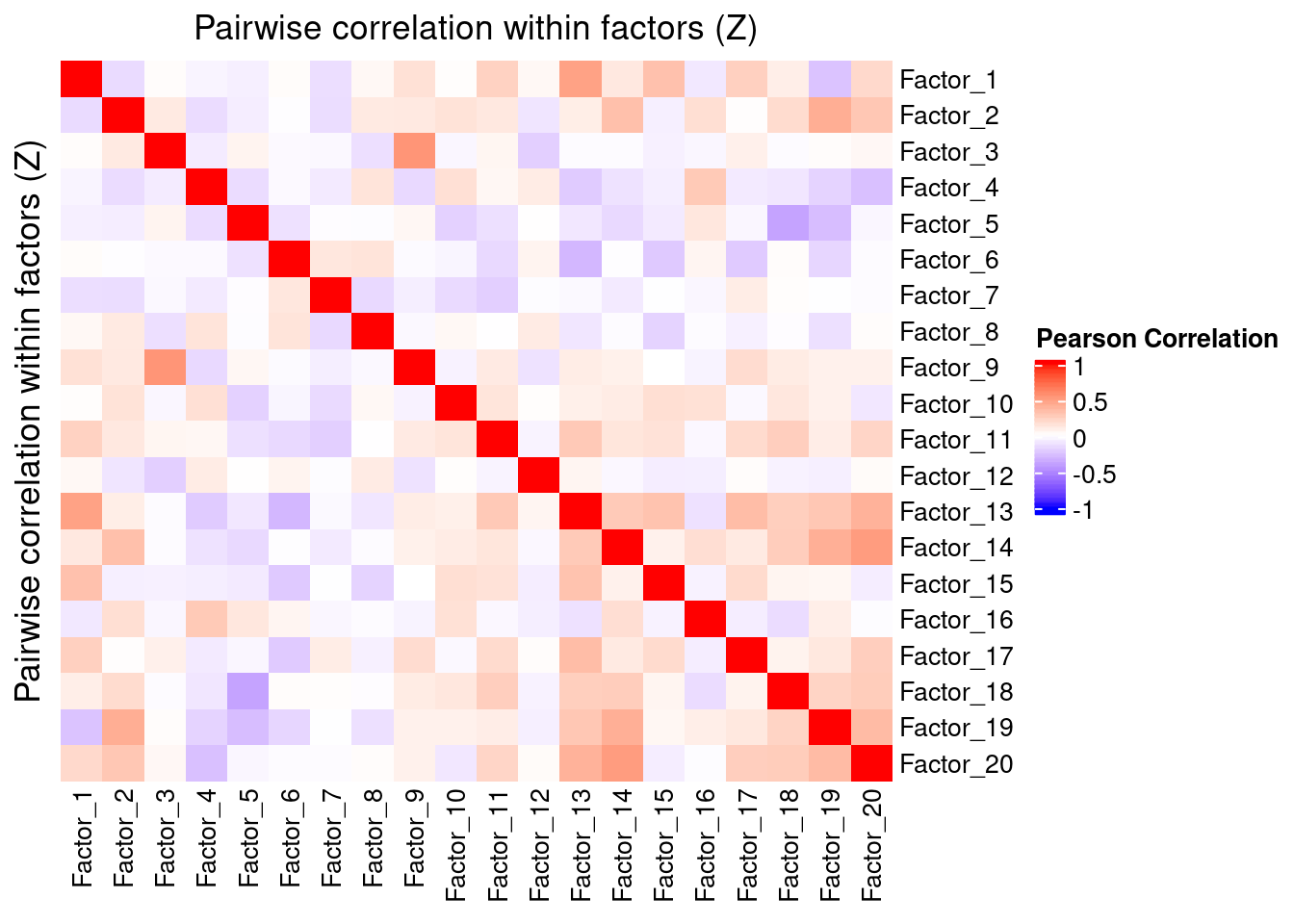
4.1 Correlation within factors
Since the GSFA model does not enforce orthogonality among factors, we also need to inspect the pairwise correlation within them to see if there is any redundancy.
As we can see below, the inferred factors are correlated with each other to some extent, especially among factors 13-20.
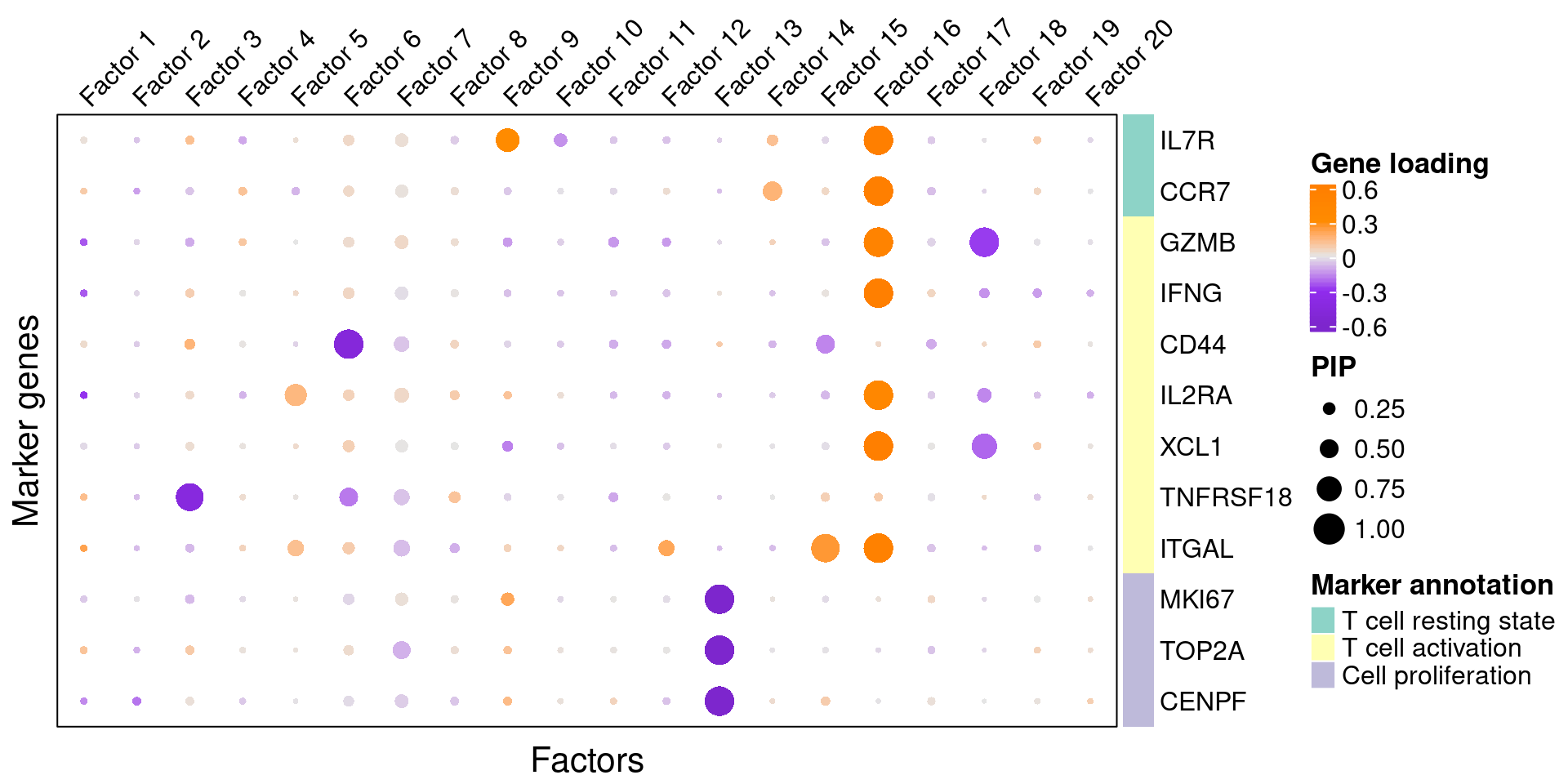
4.2 Gene loading in factors
To understand these latent factors, we inspect within them the loadings (weights) of several marker genes for cell proliferation and T cell activation/resting states.
| protein_name | gene_name | type |
|---|---|---|
| IL-7 receptor | IL7R | T cell resting state |
| C-C motif chemokine receptor 7 | CCR7 | T cell resting state |
| Granzyme B | GZMB | T cell activation |
| Interferon gamma | IFNG | T cell activation |
| CD44 | CD44 | T cell activation |
| IL-2 receptor | IL2RA | T cell activation |
| X-C motif chemokine ligand 1 | XCL1 | T cell activation |
| GITR | TNFRSF18 | T cell activation |
| LFA-1 | ITGAL | T cell activation |
| Marker of proliferation Ki-67 | MKI67 | Cell proliferation |
| DNA topoisomerase II alpha | TOP2A | Cell proliferation |
| Centromere protein F | CENPF | Cell proliferation |
We visualize both the gene PIPs (dot size) and gene weights (dot color) in all factors:
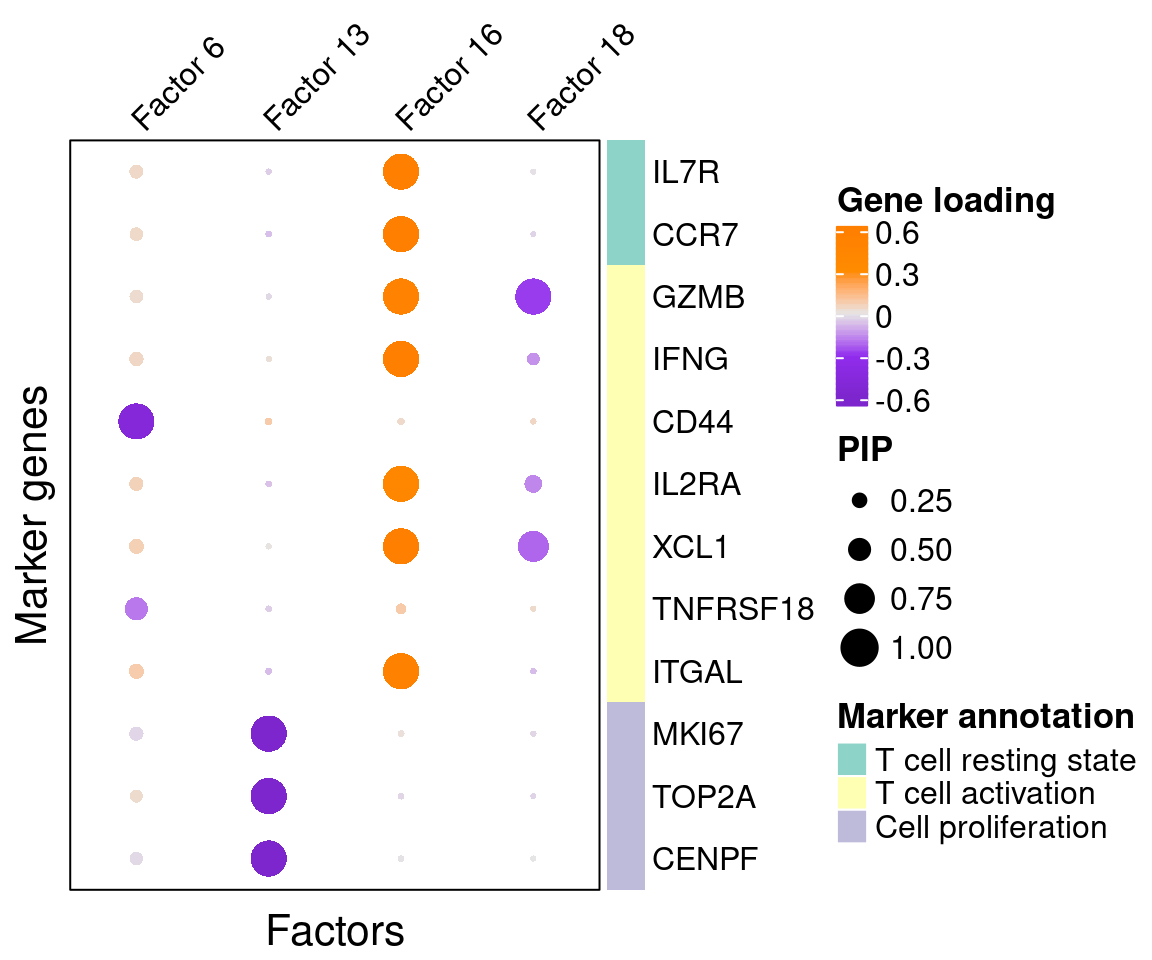
A closer look at some factors that are associated with mutations:
4.3 GO enrichment analysis in factors
To further characterize these latent factors, we perform GO (gene ontology) enrichment analysis of genes loaded on the factors using WebGestalt.
Foreground genes: genes w/ non-zero loadings in each factor (gene PIP > 0.95);
Background genes: all 10k genes used in GSFA;
Statistical test: hypergeometric test (over-representation test);
Gene sets: GO Slim "Biological Process" (non-redundant).
GO terms that passed over-representation test fold change \(\geq\) 2 and q value \(<\) 0.05:
Factor 3 : 12 significant GO terms| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0006399 | tRNA metabolic process | 10.20 | 2.19e-06 | 5.34e-04 | 7/17 | ADAT3; CTU1; CTU2; DUS1L; PUSL1; QTRT1; TRMT61A |
| GO:0006414 | translational elongation | 10.20 | 2.19e-06 | 5.34e-04 | 7/17 | AURKAIP1; EEF1D; GADD45GIP1; MRPL12; MRPL23; MRPL41; MRPS34 |
| GO:0033108 | mitochondrial respiratory chain complex assembly | 9.56 | 1.01e-04 | 9.10e-03 | 5/13 | UQCC3; NDUFB7; NDUFS6; NDUFS7; NDUFS8 |
| GO:0140053 | mitochondrial gene expression | 9.32 | 2.30e-05 | 3.12e-03 | 6/16 | AURKAIP1; GADD45GIP1; MRPL12; MRPL23; MRPL41; MRPS34 |
| GO:0010257 | NADH dehydrogenase complex assembly | 9.04 | 6.78e-04 | 4.60e-02 | 4/11 | NDUFB7; NDUFS6; NDUFS7; NDUFS8 |
| GO:0009451 | RNA modification | 8.28 | 2.62e-06 | 5.34e-04 | 8/24 | CTU1; CTU2; DUS1L; MRM1; PUSL1; QTRT1; RPUSD1; TRMT61A |
| GO:0034470 | ncRNA processing | 8.10 | 1.37e-10 | 1.12e-07 | 15/46 | ADAT3; BOP1; FAM207A; CTU1; CTU2; DUS1L; EXOSC4; NOP53; MRM1; PUSL1; QTRT1; RPS15; RRP7A; SART1; TRMT61A |
| GO:0016072 | rRNA metabolic process | 6.41 | 2.20e-05 | 3.12e-03 | 8/31 | BOP1; FAM207A; EXOSC4; NOP53; MRM1; RPS15; RRP7A; SART1 |
| GO:0007006 | mitochondrial membrane organization | 5.61 | 1.82e-04 | 1.48e-02 | 7/31 | ALKBH7; ATP5F1D; BBC3; UQCC3; HSPA1A; TIMM13; ZNF205 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 4.78 | 3.37e-05 | 3.92e-03 | 10/52 | BOP1; FAM207A; EXOSC4; NOP53; MRM1; PPAN; RPS15; RRP7A; SART1; ZNF593 |
| GO:0006839 | mitochondrial transport | 4.02 | 7.35e-05 | 7.48e-03 | 11/68 | ALKBH7; ATP5F1D; BBC3; DNLZ; HSPA1A; MICALL2; SLC25A10; SLC25A6; TIMM13; TSPO; ZNF205 |
| GO:0009141 | nucleoside triphosphate metabolic process | 2.91 | 4.83e-04 | 3.57e-02 | 13/111 | ABCD1; ATP5F1D; UQCC3; GALK1; GUK1; HSPA1A; NDUFB7; NDUFS6; NDUFS7; NDUFS8; NME3; NUDT1; TSPO |
Factor 4 : 49 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0060976 | coronary vasculature development | 4.74 | 7.08e-07 | 3.39e-05 | 13/29 | ADAMTS6; APLN; HAND2; LTBP1; NRP1; PCSK5; PDGFRB; PRDM1; PRICKLE1; PTK7; SGCD; TBX5; TGFBR1 |
| GO:0006929 | substrate-dependent cell migration | 4.23 | 2.77e-04 | 4.90e-03 | 8/20 | FN1; ITGA11; NRP1; NRP2; SEMA3A; SNAI2; TNFRSF12A; VEGFC |
| GO:1900115 | extracellular regulation of signal transduction | 4.23 | 1.67e-03 | 2.05e-02 | 6/15 | DKK3; FBN1; FBN2; GREM2; LTBP1; NBL1 |
| GO:0071526 | semaphorin-plexin signaling pathway | 4.15 | 2.40e-05 | 6.51e-04 | 11/28 | FLNA; GDNF; HAND2; NCAM1; NRP1; NRP2; PLXNC1; SEMA3A; SEMA3D; SEMA5A; SEMA7A |
| GO:0032963 | collagen metabolic process | 4.03 | 1.28e-11 | 1.30e-09 | 29/76 | ADAMTS14; ADAMTS2; COL15A1; COL1A1; COL5A1; CTGF; CTSK; EMILIN1; F2R; FAP; P3H3; MFAP4; MMP10; MMP11; MMP13; MMP14; MMP16; MMP1; MMP2; MMP3; MMP9; MRC2; PDGFRB; PLOD2; RCN3; SERPINB7; SERPINH1; TGFB3; VIM |
| GO:0043062 | extracellular structure organization | 3.82 | 0.00e+00 | 0.00e+00 | 119/329 | ADAM12; ADAM19; ADAMTS14; ADAMTS2; ADAMTS4; ADAMTS5; ADAMTSL2; AEBP1; ANTXR1; BGN; BMP1; CCDC80; COL10A1; COL11A1; COL12A1; COL15A1; COL16A1; COL18A1; COL1A1; COL1A2; COL22A1; COL24A1; COL3A1; COL4A1; COL4A2; COL5A1; COL5A2; COL5A3; COL6A1; COL6A2; COL6A3; COL7A1; COL8A1; COL8A2; COMP; CRISPLD2; CTGF; CTSK; CYR61; DCN; DDR2; DPP4; DPT; ECM2; EFEMP2; EGFL6; ELN; EMILIN1; FAP; FBLN1; FBLN2; FBN1; FBN2; FLRT2; FN1; FOXF2; FSCN1; GAS6; GREM1; HAPLN1; HAS2; HSPG2; HTRA1; ITGA11; ITGB5; JAM3; KLK2; KLK4; LAMA1; LAMA2; LAMA4; LAMB1; LAMC1; LCAT; LOXL2; LOX; LRP1; LUM; MATN3; MFAP2; MFAP4; MFAP5; MMP10; MMP11; MMP13; MMP14; MMP16; MMP1; MMP2; MMP3; MMP9; NID1; NID2; OLFML2A; PDGFRA; PDPN; PLA2G5; PLOD2; POSTN; PXDN; RECK; SDC1; SERPINE1; SERPINH1; SFRP2; SH3PXD2A; SH3PXD2B; SPARC; SULF1; SULF2; TGFB2; TGFBI; TGFBR1; THBS1; TIMP2; TLL1; TLL2; TNC; VCAN |
| GO:0060840 | artery development | 3.52 | 2.77e-07 | 1.50e-05 | 20/60 | ADAMTS6; AKT3; COL3A1; COMP; GJA5; HAND2; LOXL1; LOX; LRP1; LTBP1; MYLK; NOG; NRP1; PDGFRB; PKD2; PRDM1; PRICKLE1; PRRX1; TGFB2; TGFBR1 |
| GO:0007492 | endoderm development | 3.45 | 2.96e-06 | 9.64e-05 | 17/52 | COL11A1; COL12A1; COL4A2; COL5A1; COL5A2; COL6A1; COL7A1; COL8A1; FN1; INHBA; ITGB5; LAMB1; LAMC1; MMP14; MMP2; MMP9; NOG |
| GO:0060348 | bone development | 3.37 | 5.46e-14 | 8.34e-12 | 45/141 | COL12A1; COL1A1; COL6A1; COL6A2; COL6A3; COL7A1; COMP; CYP26B1; DCHS1; DLX5; RFLNA; RFLNB; FAT4; FBN1; FGF18; GPR68; GREM1; HAS2; HOXA11; LRRC17; MATN3; MMP13; MMP14; MMP16; MSX1; NPR2; PAX1; PDGFC; PITX2; PLS3; RAB23; RUNX2; SERPINH1; SFRP2; SFRP4; SH3PXD2B; SHOX2; SPARC; SULF1; SULF2; TGFB3; TMEM119; TNFSF11; TWIST1; XYLT1 |
| GO:0060324 | face development | 3.28 | 1.02e-03 | 1.36e-02 | 9/29 | COL1A1; CRISPLD2; DLX5; MMP2; MSX1; NOG; PDGFRA; TGFB3; WNT5A |
| GO:0090287 | regulation of cellular response to growth factor stimulus | 3.18 | 2.73e-14 | 5.56e-12 | 50/166 | ADAMTS12; ADAMTSL2; APLN; ASPN; LDLRAD4; NREP; CD109; CDKN2B; CHST11; CYR61; DAB2; DCN; DKK3; DOK5; EMILIN1; FBN1; FBN2; FGF16; FGF18; FGF1; FSTL3; FZD1; GPC1; ADGRA2; GREM1; GREM2; HTRA1; HTRA3; JCAD; LTBP1; MSX1; NBL1; NOG; PMEPA1; RASL11B; RUNX2; SFRP2; SFRP4; SHISA2; SULF1; SULF2; TGFB1I1; TGFB3; TGFBR1; THBS1; TMEM204; VEGFC; WNT5A; ZEB1; ZNF423 |
| GO:0010463 | mesenchymal cell proliferation | 3.14 | 4.32e-04 | 6.90e-03 | 11/37 | DCHS1; FAT4; FGF7; FOXP2; HAND2; MSX1; PRRX1; SHOX2; WNT2; WNT5A; ZEB1 |
| GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 3.14 | 4.32e-04 | 6.90e-03 | 11/37 | ADAMTS12; DCN; GAS1; ADGRA2; JCAD; NRP1; NRP2; PDGFRA; PDGFRB; PRKD1; VEGFC |
| GO:0003170 | heart valve development | 3.12 | 1.42e-04 | 2.95e-03 | 13/44 | CYR61; DCHS1; ELN; EMILIN1; GJA5; HEYL; PRDM1; SHOX2; SLIT3; SNAI2; TBX5; TGFB2; TWIST1 |
| GO:1904888 | cranial skeletal system development | 3.12 | 1.42e-04 | 2.95e-03 | 13/44 | CHST11; MMP14; MMP16; PDGFRA; PRRX1; RAB23; RUNX2; SH3PXD2B; TBX15; TGFB2; TGFB3; TGFBR1; TWIST1 |
| GO:0010171 | body morphogenesis | 3.07 | 1.72e-03 | 2.09e-02 | 9/31 | COL1A1; CRISPLD2; DLX5; GREM2; MMP2; MSX1; NOG; PDGFRA; TGFB3 |
| GO:0061448 | connective tissue development | 3.05 | 5.66e-15 | 1.54e-12 | 56/194 | ACTA2; ADAMTS12; ADAMTS7; BMP1; BMP8A; BMP8B; CHST11; COL11A1; COL12A1; COL1A1; COL5A1; COL6A1; COL6A2; COL6A3; COL7A1; COMP; CTGF; CTSK; CYR61; EVC; RFLNA; RFLNB; FGF18; GDF6; GLI2; GREM1; HAND2; HOXA11; HOXA3; KLF7; LOXL2; LUM; MAF; MATN3; MMP13; MSX1; NKX3-2; NOG; PDGFRB; PRRX1; PTH1R; ROR2; RUNX2; SERPINB7; SERPINH1; SFRP2; SH3PXD2B; SHOX2; SNAI2; SULF1; SULF2; TGFBI; TGFBR1; WNT5A; WNT5B; ZEB1 |
| GO:0071559 | response to transforming growth factor beta | 3.04 | 5.53e-11 | 5.00e-09 | 40/139 | ADAMTSL2; ANKRD1; ASPN; LDLRAD4; NREP; CD109; CDKN2B; CHST11; CILP; COL1A1; COL1A2; COL3A1; COL4A2; DAB2; DKK3; EMILIN1; FBN1; FBN2; FERMT2; FNDC4; FYN; HTRA1; HTRA3; ITGB5; LRRC32; LTBP1; LTBP2; MXRA5; NOX4; PMEPA1; POSTN; RASL11B; TGFB1I1; TGFB2; TGFB3; TGFBR1; THBS1; WNT2; WNT5A; ZEB1 |
| GO:0007178 | transmembrane receptor protein serine/threonine kinase signaling pathway | 2.90 | 6.15e-14 | 8.34e-12 | 56/204 | ADAMTSL2; ASPN; BMP8A; BMP8B; LDLRAD4; NREP; CD109; CDKN2B; CHST11; CILP; COL1A2; COL3A1; COMP; CYR61; DAB2; DACT1; DKK3; DLX5; DSG4; EMILIN1; FBN1; FBN2; FERMT2; FSTL1; FSTL3; FST; FZD1; GDF6; GREM1; GREM2; HTRA1; HTRA3; INHBA; ITGB5; LRRC32; LTBP1; LTBP2; MSX1; NBL1; NOG; PMEPA1; RASL11B; ROR2; RUNX2; SFRP2; SFRP4; SULF1; TGFB1I1; TGFB2; TGFB3; TGFBR1; THBS1; VIM; WNT5A; ZEB1; ZNF423 |
| GO:0048705 | skeletal system morphogenesis | 2.89 | 8.38e-12 | 9.75e-10 | 47/172 | BMP1; CHST11; COL11A1; COL12A1; COL1A1; COL6A1; COL6A2; COL6A3; COL7A1; COMP; CTGF; CYP26B1; DLX5; RFLNA; RFLNB; FBN2; FGF18; GREM1; HAS2; HOXA11; HOXA3; HOXA7; HOXC8; MATN3; MMP13; MMP14; MMP16; MMP2; MSX1; NKX3-2; NOG; PAX1; PDGFRA; PRRX1; RAB23; ROR2; RUNX2; SERPINH1; SFRP2; SFRP4; SH3PXD2B; SHOX2; TBX15; TGFB3; TGFBR1; TWIST1; ZEB1 |
| GO:0061383 | trabecula morphogenesis | 2.80 | 3.47e-03 | 3.76e-02 | 9/34 | COL1A1; FBN2; GREM1; HEG1; MMP2; NOG; NRG1; TGFB2; TGFBR1 |
| GO:0071772 | response to BMP | 2.79 | 3.26e-07 | 1.66e-05 | 28/106 | ADAMTS12; ADAMTS7; BMP8A; BMP8B; COMP; CYR61; DLX5; DSG4; FBN1; FSTL1; FSTL3; FZD1; GDF6; GREM1; GREM2; HEYL; HTRA1; HTRA3; MSX1; NBL1; NOG; ROR2; RUNX2; SFRP2; SFRP4; SULF1; WNT5A; ZNF423 |
| GO:0001503 | ossification | 2.79 | 1.55e-15 | 6.33e-13 | 67/254 | ANKH; ASPN; BMP1; BMP8A; BMP8B; CDH11; CLEC11A; COL11A1; COL1A1; COL5A2; COL6A1; COMP; CTGF; CTHRC1; CTSK; CYR61; DCHS1; DDR2; DLX5; RFLNA; RFLNB; FAT4; FBN2; FGF18; FSTL3; FZD1; GLI2; GPNMB; GREM1; HAND2; HOXA3; ID3; IGF2; ITGA11; LRRC17; MMP13; MMP14; MMP16; MMP2; MMP9; MN1; MRC2; NOG; NPR2; OMD; PDLIM7; PRKD1; PTH1R; ROR2; RUNX2; SEMA7A; SFRP2; SH3PXD2B; SHOX2; SNAI2; SPARC; TGFB2; TGFB3; TMEM119; TNC; TNFSF11; TNN; TWIST1; TWIST2; VCAN; WNT5A; XYLT1 |
| GO:0060021 | roof of mouth development | 2.76 | 7.77e-05 | 1.86e-03 | 17/65 | BNC2; DLX5; FOXF2; FZD1; HAND2; INHBA; LRRC32; MSC; MSX1; PDGFRA; PRRX1; SNAI2; TGFB2; TGFB3; TGFBR1; TWIST1; WNT5A |
| GO:0048706 | embryonic skeletal system development | 2.75 | 1.85e-06 | 7.94e-05 | 25/96 | CHST11; COL11A1; COL1A1; HOXA11; HOXA3; HOXA7; HOXC6; MMP14; MMP16; NKX3-2; NOG; PCSK5; PDGFRA; PRRX1; RUNX2; SHOX2; SULF1; SULF2; TBX15; TGFB3; TGFBR1; TWIST1; WNT5A; XYLT1; ZEB1 |
| GO:0042476 | odontogenesis | 2.72 | 2.28e-06 | 8.83e-05 | 25/97 | ADAMTS5; ASPN; COL1A1; COL1A2; FAM20A; FOXO1; GLI2; HAND2; HTRA1; ID3; INHBA; KLK4; LAMB1; LHX8; MSX1; PDGFRA; PITX2; RUNX2; SDC1; SERPINE1; TGFB2; TGFB3; TNC; TNFSF11; TWIST1 |
| GO:0050919 | negative chemotaxis | 2.72 | 4.28e-03 | 4.53e-02 | 9/35 | FLRT2; NRG1; NRP2; SEMA3A; SEMA3D; SEMA5A; SEMA7A; SLIT3; WNT5A |
| GO:0006022 | aminoglycan metabolic process | 2.62 | 2.08e-06 | 8.47e-05 | 27/109 | BGN; CHPF; CHST11; CHST3; CHST6; CHSY3; CSPG4; DCN; DSEL; DSE; GPC1; GPC6; HAS2; HS3ST3A1; HSPG2; KERA; CEMIP; LUM; OGN; OMD; PDGFRB; PRELP; SDC1; SDC2; CEMIP2; VCAN; XYLT1 |
| GO:0003205 | cardiac chamber development | 2.60 | 1.04e-06 | 4.71e-05 | 29/118 | ADAMTS6; COL11A1; CYR61; FZD1; GJA5; HAND2; HEG1; HEYL; LTBP1; MYL3; NOG; NRG1; NRP1; NRP2; PCSK5; PRDM1; PTK7; SALL1; SALL4; SFRP2; SHOX2; SLIT3; TBX5; TGFB2; TGFBR1; TPM1; WNT2; WNT5A; ZFPM2 |
| GO:0007164 | establishment of tissue polarity | 2.54 | 1.95e-03 | 2.30e-02 | 12/50 | CTHRC1; DAB2; DACT1; FOXF2; FZD1; GPC6; PRICKLE1; PRICKLE2; PTK7; ROR2; SFRP2; WNT5A |
| GO:0060485 | mesenchyme development | 2.53 | 2.64e-09 | 2.15e-07 | 45/188 | ACTA1; ACTA2; BNC2; LDLRAD4; COL1A1; DAB2; DCHS1; EDNRA; RFLNB; FN1; FOXF2; GDNF; GREM1; HAND2; HAS2; HEYL; LOXL2; MSX1; NOG; NRG1; NRP1; NRP2; PAX1; PDGFRB; PDPN; PITX2; PKD2; PTK7; S100A4; SEMA3A; SEMA3D; SEMA5A; SEMA7A; SFRP2; SNAI2; TBX5; TGFB1I1; TGFB2; TGFB3; TGFBR1; TWIST1; WNT2; WNT5A; ZEB2; ZFPM2 |
| GO:0071774 | response to fibroblast growth factor | 2.48 | 1.48e-04 | 3.01e-03 | 19/81 | APLN; COL1A1; CTGF; FAT4; FGF16; FGF18; FGF1; FGF7; FLRT2; GPC1; NOG; POSTN; RUNX2; SHISA2; SULF1; SULF2; THBS1; TNC; WNT5A |
| GO:0030323 | respiratory tube development | 2.45 | 5.67e-06 | 1.77e-04 | 28/121 | ADAMTS2; ADAMTSL2; CRISPLD2; CTGF; FGF18; FGF1; FGF7; FOXP2; FSTL3; GLI2; HEG1; LAMA1; LOX; MMP14; NOG; PCSK5; PDGFRA; PDGFRB; PDPN; PTK7; RCN3; SPARC; TBX5; TGFB3; TNC; WNT2; WNT5A; ZFPM2 |
| GO:0048736 | appendage development | 2.43 | 6.71e-06 | 2.02e-04 | 28/122 | CHST11; COMP; CYP26B1; DLX5; EN1; FBN2; GJA5; GLI2; GREM1; HAND2; HOXA10; HOXA11; MSX1; NOG; PCSK5; PITX2; PRRX1; RECK; ROR2; RUNX2; SALL1; SALL4; SFRP2; SHOX2; TBX5; TGFB2; TWIST1; WNT5A |
| GO:0007229 | integrin-mediated signaling pathway | 2.40 | 5.01e-04 | 7.84e-03 | 17/75 | BST1; CD177; COL16A1; COL3A1; CTGF; DAB2; FBLN1; FERMT2; FLNA; ITGA11; ITGB5; ITGBL1; LIMS2; NRP1; PRKD1; SEMA7A; THY1 |
| GO:0048645 | animal organ formation | 2.39 | 3.30e-03 | 3.63e-02 | 12/53 | FGF1; GDNF; GLI2; HAND2; HOXA11; HOXA3; NKX3-2; NOG; SULF1; TBX5; WNT2; WNT5A |
| GO:0031589 | cell-substrate adhesion | 2.33 | 3.42e-09 | 2.53e-07 | 52/236 | ADAMTS12; ANTXR1; AXL; BST1; CCDC80; COL16A1; COL1A1; COL3A1; COL5A3; COL8A1; CTGF; CYR61; DLC1; ECM2; EDIL3; EGFL6; EMILIN1; EPDR1; FBLN1; FBLN2; FERMT2; FLNA; FN1; GAS6; GREM1; HAS2; HOXA7; ITGA11; ITGB5; ITGBL1; JAM3; AJUBA; LAMB1; LAMC1; LGALS1; MMP14; NID1; NID2; NRP1; PDPN; PLAU; POSTN; PPFIA2; S100A10; SERPINE1; SGCE; SNED1; SPOCK1; THBS1; THY1; TNFRSF12A; TNN |
| GO:0060541 | respiratory system development | 2.27 | 1.80e-05 | 5.24e-04 | 29/135 | ADAMTS2; ADAMTSL2; CRISPLD2; CTGF; DLX5; FGF18; FGF1; FGF7; FOXP2; FSTL3; GLI2; HEG1; LAMA1; LOX; MMP14; MSC; NOG; PDGFRA; PDGFRB; PDPN; PTK7; RCN3; SPARC; TBX5; TGFB3; TNC; WNT2; WNT5A; ZFPM2 |
| GO:0031214 | biomineral tissue development | 2.23 | 1.74e-04 | 3.45e-03 | 23/109 | ANKH; ASPN; COL1A1; COMP; CYR61; DDR2; RFLNA; RFLNB; FAM20A; FBN2; FOXO1; GAS6; GPNMB; GREM1; HOXA3; KLK4; MMP13; OMD; PTH1R; ROR2; TGFB3; TMEM119; TWIST1 |
| GO:0021915 | neural tube development | 2.21 | 9.52e-04 | 1.29e-02 | 18/86 | CTHRC1; DACT1; DCHS1; DLC1; EN1; FZD1; GLI2; GPR161; NOG; PKD2; PRICKLE1; PTK7; SALL4; SFRP2; TGFB2; TWIST1; WNT5A; ZEB2 |
| GO:0003007 | heart morphogenesis | 2.20 | 2.94e-06 | 9.64e-05 | 37/178 | ANKRD1; COL11A1; COL5A1; CYR61; DCHS1; DLC1; ELN; FAT4; FLRT2; FZD1; GJA5; HAND2; HAS2; HEG1; HEYL; MICAL2; MICALCL; MSX1; MYL3; NOG; NRG1; NRP1; NRP2; PITX2; PKD2; SFRP2; SHOX2; SLIT3; SNAI2; TBX5; TGFB2; TGFB3; TGFBR1; TPM1; TWIST1; WNT2; WNT5A; ZFPM2 |
| GO:0198738 | cell-cell signaling by wnt | 2.13 | 6.09e-08 | 4.13e-06 | 53/263 | AMOTL2; APCDD1; AXL; BICC1; CDH2; COL1A1; CPZ; CTHRC1; DAB2; DACT1; DKK3; DLX5; FERMT2; FOXO1; FZD1; GNG2; GPC6; ADGRA2; GREM1; HECW1; HIC1; LGR5; LRP1; MITF; NKD2; NOG; NXN; PKD2; PRICKLE1; PRICKLE2; PTK7; RBMS3; RECK; ROR2; RSPO1; SALL1; SDC1; SEMA5A; SFRP2; SFRP4; SHISA2; SHISA6; SNAI2; SULF1; SULF2; TBX18; TGFB1I1; TNN; WISP1; WNT2; WNT5A; WNT5B; ZEB2 |
| GO:2000027 | regulation of animal organ morphogenesis | 2.11 | 3.12e-05 | 8.19e-04 | 32/160 | ASPN; CTHRC1; DAB2; DACT1; FGF1; FGF7; FOXP2; FZD1; GDNF; GPC6; GREM1; HAND2; HOXA11; LIMS2; MSX1; NOG; PRICKLE1; PRICKLE2; PTK7; ROR2; RUNX2; SFRP2; SNAI2; SULF1; SULF2; TBX5; TGFB2; TGFB3; TGFBR1; TWIST1; WNT2; WNT5A |
| GO:1901136 | carbohydrate derivative catabolic process | 2.11 | 5.28e-04 | 7.96e-03 | 22/110 | ADAMTS12; BGN; LINC00473; CSPG4; DCN; ENPP4; GPC1; GPC6; HSPG2; KERA; CEMIP; LUM; NT5E; NUDT10; NUDT11; OGN; OMD; PDE10A; PRELP; SDC1; SDC2; CEMIP2; VCAN |
| GO:0001763 | morphogenesis of a branching structure | 2.10 | 8.29e-05 | 1.93e-03 | 29/146 | ADAMTS16; CCL11; COL4A1; DCHS1; EDNRA; ERMN; FAT4; FGF1; FGF7; GDNF; GLI2; GREM1; HOXA11; HOXD11; LAMA1; MMP14; NOG; NRP1; PKD2; PRDM1; SALL1; SEMA3A; SFRP2; SHOX2; SNAI2; SULF1; TNC; WNT2; WNT5A |
| GO:0002576 | platelet degranulation | 2.09 | 2.50e-03 | 2.91e-02 | 17/86 | CD109; F13A1; FLNA; FN1; GAS6; IGF2; ISLR; PCDH7; RARRES2; SERPINE1; SERPING1; SPARC; TGFB2; TGFB3; THBS1; TIMP3; VEGFC |
| GO:0045995 | regulation of embryonic development | 2.06 | 3.80e-03 | 4.06e-02 | 16/82 | COL5A1; COL5A2; FZD1; GDNF; GPR161; LAMA1; LAMA2; LAMA4; NRK; TENM4; PTK7; SFRP2; SULF1; TBX18; WNT2; WNT5A |
| GO:0007369 | gastrulation | 2.04 | 1.16e-03 | 1.50e-02 | 21/109 | APLN; COL11A1; COL12A1; COL4A2; COL5A1; COL5A2; COL6A1; COL7A1; COL8A1; FN1; HOXA11; INHBA; ITGB5; LAMB1; MMP14; MMP2; MMP9; NOG; TENM4; SFRP2; WNT5A |
| GO:0055123 | digestive system development | 2.02 | 1.31e-03 | 1.67e-02 | 21/110 | CLMP; COL3A1; DACT1; DCHS1; FAT4; FOXF2; GLI2; IGF2; NKX3-2; PCSK5; PDGFC; PDGFRA; PRDM1; RARRES2; SALL1; SCT; SFRP2; SHOX2; TGFB2; TGFB3; WNT5A |
Factor 5 : 106 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0009595 | detection of biotic stimulus | 8.95 | 1.30e-07 | 2.20e-06 | 9/17 | CD1D; CRTAM; LY96; NLRP3; TLR1; TLR2; TLR4; TLR6; TREM2 |
| GO:0045730 | respiratory burst | 7.35 | 2.69e-07 | 4.30e-06 | 10/23 | CYBA; CYBB; HCK; NCF1; NCF2; NCF4; PIK3CD; PIK3CG; SLAMF8; SLC11A1 |
| GO:0097242 | amyloid-beta clearance | 7.25 | 8.26e-05 | 8.97e-04 | 6/14 | APOE; C3; ITGAM; ITGB2; MARCO; MSR1 |
| GO:0010324 | membrane invagination | 6.96 | 2.44e-09 | 5.08e-08 | 14/34 | ABCA1; AIF1; ARHGAP25; BIN2; C3; CD300A; FCER1G; FCGR1A; FCGR2B; ITGAM; ITGB2; MSR1; NCKAP1L; TREM2 |
| GO:0055094 | response to lipoprotein particle | 6.76 | 1.31e-04 | 1.38e-03 | 6/15 | ABCA1; APOE; FCER1G; ITGB2; TLR4; TLR6 |
| GO:0002269 | leukocyte activation involved in inflammatory response | 6.64 | 2.35e-07 | 3.82e-06 | 11/28 | AIF1; FPR2; ITGAM; ITGB2; MMP8; PTPRC; SNCA; TLR2; TLR6; TLR7; TLR8 |
| GO:0071402 | cellular response to lipoprotein particle stimulus | 6.34 | 1.99e-04 | 2.03e-03 | 6/16 | ABCA1; APOE; FCER1G; ITGB2; TLR4; TLR6 |
| GO:0032613 | interleukin-10 production | 6.11 | 5.99e-08 | 1.08e-06 | 13/36 | VSIR; CD28; CD83; FCER1G; FCGR2B; HGF; LGALS9; LILRB1; MMP8; PDCD1LG2; SASH3; TLR2; TLR4 |
| GO:0090382 | phagosome maturation | 6.04 | 9.06e-04 | 8.02e-03 | 5/14 | CORO1A; MYO7A; P2RX7; RAB39A; SLAMF8 |
| GO:0042116 | macrophage activation | 6.02 | 6.46e-12 | 1.81e-10 | 21/59 | AIF1; CD74; CEBPA; FCGR2B; FPR2; HAVCR2; ITGAM; ITGB2; MMP8; PTPRC; SLC11A1; SNCA; SUCNR1; TLR1; TLR2; TLR4; TLR6; TLR7; TLR8; TYROBP; VSIG4 |
| GO:0150076 | neuroinflammatory response | 5.90 | 9.77e-09 | 1.85e-07 | 15/43 | AIF1; CD200R1; FPR2; IL18; IL1B; ITGAM; ITGB2; MMP8; PTPRC; SNCA; TLR2; TLR6; TLR7; TLR8; TNFRSF1B |
| GO:0032623 | interleukin-2 production | 5.88 | 3.27e-09 | 6.48e-08 | 16/46 | CARD11; CARD9; CD28; CD4; CD80; CD83; CD86; FCER1G; HAVCR2; IL18; IL1A; IL1B; PTPRC; SASH3; SLC11A1; VSIG4 |
| GO:0045576 | mast cell activation | 5.88 | 3.27e-09 | 6.48e-08 | 16/46 | BTK; CD226; CD300A; CD300LF; CD84; ADGRE2; FCER1G; FCGR2B; FGR; HMOX1; LAT2; LCP2; LGALS9; LYN; PIK3CD; PIK3CG |
| GO:0006909 | phagocytosis | 5.87 | 0.00e+00 | 0.00e+00 | 50/144 | ABCA1; AIF1; ARHGAP25; AXL; BIN2; C2; C3; CCL2; CD14; CD300A; CD300LF; CEACAM4; CLEC7A; CORO1A; CYBA; DOCK2; FCER1G; FCGR1A; FCGR2A; FCGR2B; FCGR2C; FCGR3A; FGR; HCK; IL1B; IRF8; ITGAM; ITGB2; ITGB3; LYN; MARCO; MERTK; MSR1; MYO1G; MYO7A; NCF2; NCF4; NCKAP1L; P2RX7; P2RY6; PLD4; PTPRC; RAB39A; SIRPA; SIRPB1; SLC11A1; TLR2; TREM2; VAV1; WAS; WIPF1 |
| GO:0002448 | mast cell mediated immunity | 5.80 | 3.78e-07 | 5.80e-06 | 12/35 | BTK; CD300A; CD84; ADGRE2; FCER1G; FGR; HMOX1; LAT2; LGALS9; LYN; PIK3CD; PIK3CG |
| GO:0032418 | lysosome localization | 5.80 | 3.78e-07 | 5.80e-06 | 12/35 | BTK; CD300A; CD84; ADGRE2; FCER1G; FGR; HMOX1; LAT2; LGALS9; LYN; PIK3CD; PIK3CG |
| GO:0042107 | cytokine metabolic process | 5.33 | 1.16e-11 | 3.14e-10 | 23/73 | CARD11; CARD9; CD28; CD4; CD80; CD86; CX3CR1; CYBB; EBI3; HMOX1; IL18; IL1A; IL1B; INPP5D; LILRB1; PTAFR; PTPRC; SPN; TLR1; TLR4; TLR6; TLR7; TLR8 |
| GO:0050690 | regulation of defense response to virus by virus | 5.20 | 5.62e-03 | 3.75e-02 | 4/13 | CD28; CD4; DOCK2; HCK |
| GO:0071706 | tumor necrosis factor superfamily cytokine production | 5.18 | 1.84e-14 | 8.82e-13 | 30/98 | ACP5; ADAM8; AXL; VSIR; CARD9; CCL3; CD14; CD86; CLEC4A; CX3CR1; CYBA; CYBB; FCER1G; GPNMB; HAVCR2; LGALS9; LILRA2; LILRA4; LILRB1; LY96; MMP8; PTAFR; PTPN22; PTPRC; SASH3; SIRPA; SPN; TLR1; TLR2; TLR4 |
| GO:0036230 | granulocyte activation | 5.16 | 0.00e+00 | 0.00e+00 | 79/259 | ADAM8; ALDH3B1; ALOX5; ARHGAP9; ATP8B4; BIN2; MCEMP1; C3AR1; C3; C5AR1; CCL3; CD14; CD300A; CD33; CD53; CD68; ADGRE5; ADA2; CHIT1; CLEC5A; COTL1; CR1; CTSC; CTSD; CTSS; CTSZ; CYBA; CYBB; DOCK2; DOK3; ADGRE3; FCER1G; FCGR2A; FCGR2B; FCGR2C; FCGR3B; FGL2; FGR; FPR1; FPR2; FTL; GMFG; GPR84; HK3; HPSE; HVCN1; IL18; ITGAM; ITGAX; ITGB2; KCNAB2; LILRA2; LILRA3; LRMP; LYZ; MMP8; MNDA; NCKAP1L; NFAM1; OLR1; OSCAR; PLAUR; PLAU; PRAM1; PTAFR; PTPRC; RNASE2; SIGLEC14; SIGLEC5; SIGLEC9; SIRPA; SIRPB1; SLC11A1; SLC2A5; TCIRG1; TLR2; TNFRSF1B; TRPM2; TYROBP; VNN1 |
| GO:0006968 | cellular defense response | 5.11 | 6.48e-07 | 9.58e-06 | 13/43 | C5AR1; CCR5; CD300C; CLEC5A; CX3CR1; LSP1; LY96; MNDA; NCF1; NCF2; SPN; TCIRG1; TYROBP |
| GO:0002446 | neutrophil mediated immunity | 5.03 | 0.00e+00 | 0.00e+00 | 75/252 | ADAM8; ALDH3B1; ALOX5; ARHGAP9; ATP8B4; BIN2; MCEMP1; C3AR1; C3; C5AR1; CD14; CD300A; CD33; CD53; CD68; ADGRE5; ADA2; CHIT1; CLEC5A; COTL1; CR1; CTSC; CTSD; CTSS; CTSZ; CYBA; CYBB; DOCK2; DOK3; ADGRE3; FCER1G; FCGR2A; FCGR2C; FCGR3B; FGL2; FGR; FPR1; FPR2; FTL; GMFG; GPR84; HK3; HPSE; HVCN1; ITGAM; ITGAX; ITGB2; KCNAB2; LILRA3; LRMP; LYZ; MMP8; MNDA; NCKAP1L; NFAM1; OLR1; OSCAR; PLAUR; PLAU; PRAM1; PTAFR; PTPRC; RNASE2; SIGLEC14; SIGLEC5; SIGLEC9; SIRPA; SIRPB1; SLC11A1; SLC2A5; TCIRG1; TLR2; TNFRSF1B; TRPM2; TYROBP; VNN1 |
| GO:0032615 | interleukin-12 production | 4.97 | 1.71e-05 | 2.21e-04 | 10/34 | ACP5; CD40; CMKLR1; IRF5; IRF8; LGALS9; LILRB1; TLR2; TLR4; TLR8 |
| GO:0001562 | response to protozoan | 4.97 | 2.42e-03 | 1.88e-02 | 5/17 | CCDC88B; CD40; CLEC7A; IRF8; SLC11A1 |
| GO:0032633 | interleukin-4 production | 4.91 | 5.11e-05 | 5.85e-04 | 9/31 | CD28; CD83; CD86; CLECL1; FCER1G; HAVCR2; LGALS9; NLRP3; SASH3 |
| GO:0032637 | interleukin-8 production | 4.83 | 3.42e-06 | 4.81e-05 | 12/42 | CD14; IL1B; LGALS9; LILRA2; PTPN22; PTPRC; TLR1; TLR2; TLR4; TLR6; TLR7; TLR8 |
| GO:0019882 | antigen processing and presentation | 4.80 | 1.89e-11 | 4.80e-10 | 25/88 | CD1D; CD209; CD74; CLEC4A; CTSD; CTSL; CTSS; FCER1G; FCGR1A; FCGR1B; FCGR2B; HLA-DMB; HLA-DOA; HLA-DPA1; HLA-DPB1; HLA-DQA1; HLA-DQB1; HLA-DRA; HLA-DRB1; HLA-DRB5; IFI30; MARCH1; SLC11A1; TREM2; WAS |
| GO:0071216 | cellular response to biotic stimulus | 4.69 | 2.22e-16 | 1.29e-14 | 38/137 | ABCA1; AXL; BTK; CARD16; CASP1; CCL2; CCL3; CCR5; CD14; CD180; CD40; CD80; CD86; CX3CR1; FCGR2B; HAMP; HAVCR2; HCK; IL18; IL1B; IRF8; LILRA2; LILRB1; LY86; LY96; LYN; MRC1; NLRP3; PDCD1LG2; PTAFR; PTPN22; SIRPA; TLR1; TLR2; TLR4; TLR6; TNFRSF1B; TREM2 |
| GO:0070661 | leukocyte proliferation | 4.59 | 0.00e+00 | 0.00e+00 | 54/199 | AIF1; BTK; VSIR; CARD11; CCDC88B; CCL8; CD180; CD1D; CD209; CD28; CD300A; CD40; CD4; CD74; CD80; CD86; CLECL1; CORO1A; CRTAM; CSF1; DOCK2; DOCK8; EBI3; FCGR2B; GAPT; GPNMB; GPR183; HAVCR2; HLA-DMB; HLA-DPA1; HLA-DPB1; IL18; IL1B; INPP5D; LGALS9; LILRB1; LST1; LYN; MNDA; NCKAP1L; P2RX7; PDCD1LG2; PIK3CG; PTPN22; PTPRC; RASAL3; SASH3; SLC11A1; SPN; TGFBR2; TLR4; TNFRSF1B; TNFSF13B; VSIG4 |
| GO:0032635 | interleukin-6 production | 4.52 | 4.69e-10 | 1.03e-08 | 23/86 | CARD9; CD200R1; CYBA; FCER1G; HAVCR2; HGF; IL18; IL1B; INPP5D; LGALS9; LILRA2; MMP8; NCKAP1L; P2RX7; PTAFR; PTPN22; SIRPA; TLR1; TLR2; TLR4; TLR6; TLR7; TLR8 |
| GO:0002764 | immune response-regulating signaling pathway | 4.49 | 0.00e+00 | 0.00e+00 | 77/290 | BLK; BTK; THEMIS2; C3AR1; C5AR1; CARD11; CARD9; CD14; CD200R1; CD209; CD226; CD28; CD300A; CD300LF; CD33; CD40; CD4; CLEC4A; CLEC4E; CLEC7A; CMKLR1; CR1; CTSS; CYBA; FCER1G; FCGR1A; FCGR1B; FCGR2A; FCGR2B; FCGR2C; FCGR3A; FGR; FPR1; FPR2; FPR3; FYB1; HAVCR2; HCK; HLA-DPA1; HLA-DPB1; HLA-DQA1; HLA-DQB1; HLA-DRA; HLA-DRB1; HLA-DRB5; INPP5D; ITGAM; ITGB2; KLHL6; LAT2; LCP2; LGALS9; LILRA2; LILRA4; LILRB1; LILRB4; LPXN; LY96; LYN; MARCO; MNDA; MYO1G; NCKAP1L; NFAM1; PIK3AP1; PIK3CD; PRAM1; PTPN22; PTPRC; TLR1; TLR2; TLR4; TLR6; TLR7; TLR8; VAV1; WAS; WIPF1 |
| GO:0050867 | positive regulation of cell activation | 4.48 | 0.00e+00 | 0.00e+00 | 58/219 | ADAM8; AIF1; AXL; BTK; VSIR; CARD11; CCDC88B; CCL2; CD1D; CD209; CD226; CD28; CD40; CD4; CD74; CD80; CD83; CD86; CEBPA; CLECL1; CORO1A; DOCK8; EBI3; FCER1G; FGR; GPR183; HAVCR2; HLA-DMB; HLA-DPA1; HLA-DPB1; IL18; IL1B; INPP5D; ITGAM; ITGB2; LGALS9; LILRA2; LILRB1; LILRB4; LYN; MMP8; NCKAP1L; NLRP3; PDCD1LG2; PIK3R6; PLEK; PTAFR; PTPRC; RASAL3; SASH3; SIRPA; SIRPB1; TGFBR2; TLR4; TLR6; TNFSF13B; VAV1; VNN1 |
| GO:0071887 | leukocyte apoptotic process | 4.48 | 4.50e-08 | 8.32e-07 | 18/68 | ADAM8; AXL; BIRC7; BTK; CCR5; CD74; CTSL; DOCK8; FCER1G; GIMAP8; HCLS1; LGALS9; LILRB1; LYN; MERTK; P2RX7; PIK3CD; SLC46A2 |
| GO:0032612 | interleukin-1 production | 4.42 | 1.30e-07 | 2.20e-06 | 17/65 | ABCA1; ACP5; CARD16; CASP1; CCL3; CX3CR1; HAVCR2; IL1B; LGALS9; LILRA2; NLRP3; P2RX7; SIRPA; SUCNR1; TLR4; TLR6; TLR8 |
| GO:0035587 | purinergic receptor signaling pathway | 4.38 | 7.60e-04 | 6.80e-03 | 7/27 | ADORA3; GPR34; P2RX7; P2RY12; P2RY13; P2RY6; PTAFR |
| GO:0034341 | response to interferon-gamma | 4.33 | 2.71e-13 | 9.66e-12 | 33/129 | AIF1; CASP1; CCL2; CCL3L1; CCL3L3; CCL3; CCL4L1; CCL8; CD40; CIITA; CXCL16; DAPK1; FCGR1A; FCGR1B; HCK; HLA-DPA1; HLA-DPB1; HLA-DQA1; HLA-DQB1; HLA-DRA; HLA-DRB1; HLA-DRB5; IFI30; IRF5; IRF8; LGALS9; MRC1; PTAFR; SIRPA; SLC11A1; SNCA; TLR2; TLR4; WAS |
| GO:0002285 | lymphocyte activation involved in immune response | 4.31 | 2.45e-10 | 5.53e-09 | 25/98 | APBB1IP; CD180; CD28; CD40; CD80; CD86; CLEC4E; CORO1A; DOCK10; FCER1G; FCGR2B; GAPT; GPR183; HAVCR2; HLA-DMB; IL18; LGALS9; LILRB1; MFNG; NLRP3; PTGER4; PTPRC; SLC11A1; SPN; TLR4 |
| GO:0032609 | interferon-gamma production | 4.23 | 4.52e-09 | 8.76e-08 | 22/88 | AXL; VSIR; CD14; CD226; EBI3; HAVCR2; HLA-DPA1; HLA-DPB1; IL18; IL1B; IRF8; LGALS9; LILRB1; PDCD1LG2; PTPN22; SASH3; SIRPA; SLC11A1; SPN; TLR4; TLR7; TLR8 |
| GO:0032606 | type I interferon production | 4.23 | 3.53e-05 | 4.37e-04 | 11/44 | CD14; HAVCR2; IRF5; LILRA4; LILRB1; PTPN22; SIRPA; TLR2; TLR4; TLR7; TLR8 |
| GO:0001773 | myeloid dendritic cell activation | 4.23 | 5.23e-03 | 3.52e-02 | 5/20 | DOCK2; HAVCR2; SPI1; TGFBR2; DCSTAMP |
| GO:0033028 | myeloid cell apoptotic process | 4.23 | 5.23e-03 | 3.52e-02 | 5/20 | CCR5; CLEC5A; CTSL; FCER1G; PIK3CD |
| GO:0090077 | foam cell differentiation | 4.23 | 2.23e-03 | 1.78e-02 | 6/24 | ABCA1; CSF1; IL18; ITGB3; MSR1; SOAT1 |
| GO:0034113 | heterotypic cell-cell adhesion | 4.13 | 4.43e-05 | 5.23e-04 | 11/45 | CD1D; CD200R1; GLDN; IL1B; ITGA4; ITGAD; ITGAX; ITGB2; ITGB3; PTPRC; SIRPA |
| GO:0002532 | production of molecular mediator involved in inflammatory response | 4.12 | 1.01e-04 | 1.08e-03 | 10/41 | ADCY7; ALOX5; BTK; FCER1G; GPSM3; LYN; SIRPA; SLAMF8; TLR4; TLR6 |
| GO:0030865 | cortical cytoskeleton organization | 4.06 | 2.79e-03 | 2.09e-02 | 6/25 | FMNL1; FMNL2; NCKAP1L; PLEK; WAS; WIPF1 |
| GO:0050663 | cytokine secretion | 4.00 | 2.73e-13 | 9.66e-12 | 36/152 | ABCA1; CARD11; CARD16; CASP1; CCL3; CD14; CD200R1; CLEC4E; CLEC5A; CLECL1; CRTAM; CSF1R; FCGR2B; FGR; FFAR4; HAVCR2; IL1A; IL1B; LCP2; LGALS9; LILRA2; LILRB1; LYN; MMP8; NLRP3; OSM; P2RX7; PTGER4; PTPN22; SRGN; TLR1; TLR2; TLR4; TLR6; TLR8; TNFRSF1B |
| GO:0002694 | regulation of leukocyte activation | 3.98 | 0.00e+00 | 0.00e+00 | 79/336 | ADAM8; AIF1; AXL; BTK; VSIR; CARD11; CCDC88B; CCL2; CD1D; CD209; CD226; CD28; CD300A; CD300LF; CD40; CD4; CD74; CD80; CD83; CD84; CD86; CEBPA; CLECL1; CORO1A; CR1; DOCK8; EBI3; ADGRE2; FCER1G; FCGR2B; FGR; GPNMB; GPR183; HAVCR2; HLA-DMB; HLA-DOA; HLA-DPA1; HLA-DPB1; HMOX1; IL18; IL1B; INPP5D; ITGAM; ITGB2; LGALS9; LILRB1; LILRB4; LST1; LYN; MERTK; MMP8; MNDA; NCKAP1L; NFAM1; NLRP3; PDCD1LG2; PIK3R6; PRAM1; PTAFR; PTPN22; PTPRC; RASAL3; SAMSN1; SASH3; SIRPA; SIRPB1; SLAMF8; SLC46A2; SNCA; SPN; TGFBR2; TLR4; TLR6; TNFAIP8L2; TNFRSF1B; TNFSF13B; VAV1; VNN1; VSIG4 |
| GO:0002449 | lymphocyte mediated immunity | 3.96 | 1.83e-13 | 7.08e-12 | 37/158 | BTK; C1QA; C1QB; C1QC; C2; C3; CD1D; CD226; CD28; CD40; CD74; CORO1A; CR1; CRTAM; CTSC; FCER1G; FCGR2B; GAPT; HAVCR2; HLA-DQB1; IL18; IL1B; INPP5D; LGALS9; LILRB1; MYO1G; NLRP3; P2RX7; PIK3R6; POU2F2; PTPRC; SASH3; SLC11A1; TLR8; TNFRSF1B; VAV1; WAS |
| GO:0007159 | leukocyte cell-cell adhesion | 3.94 | 0.00e+00 | 0.00e+00 | 54/232 | ADAM8; AIF1; VSIR; CARD11; CCDC88B; CCL2; CD1D; CD209; CD28; CD300A; CD4; CD74; CD80; CD83; CD86; CLECL1; CORO1A; CX3CR1; DOCK8; EBI3; FCGR2B; FERMT3; GPNMB; HAVCR2; HLA-DMB; HLA-DPA1; HLA-DPB1; IL18; IL1B; ITGA4; ITGB2; LGALS9; LILRB1; LILRB4; LYN; NCKAP1L; NLRP3; PDCD1LG2; PIK3R6; PTAFR; PTPN22; PTPRC; RASAL3; SASH3; SELPLG; SIRPA; SIRPB1; SPN; TGFBR2; TNFAIP8L2; TNFSF13B; VAV1; VNN1; VSIG4 |
| GO:0002521 | leukocyte differentiation | 3.86 | 0.00e+00 | 0.00e+00 | 73/320 | ADAM8; AXL; BTK; VSIR; C1QC; CARD11; CCL3; CCR1; CD1D; CD28; CD4; CD74; CD80; CD83; CD86; CEBPA; CLEC4E; CR1; CSF1R; CSF1; DOCK10; DOCK2; EVI2B; FCER1G; FCGR2B; GAB3; GPR183; HCLS1; HLA-DOA; IKZF1; IL18; INPP5D; ITGA4; LGALS9; LILRB1; LILRB4; NRROS; LYL1; LYN; MAFB; MERTK; MFNG; MITF; NCKAP1L; NFAM1; NLRP3; OSCAR; PDE1B; PIK3CD; PIK3R6; POU2F2; PTGER4; PTPN22; PTPRC; RASSF2; SASH3; SLAMF8; SLC46A2; SPI1; SPN; TGFBR2; TLR2; TLR4; DCSTAMP; TMEM176A; TMEM176B; TNFSF8; TREM2; TRPM2; TYROBP; UBASH3B; VAV1; VNN1 |
| GO:0002250 | adaptive immune response | 3.81 | 0.00e+00 | 0.00e+00 | 60/266 | ADCY7; BTK; C1QA; C1QB; C1QC; C2; C3; CD1D; CD209; CD226; CD28; CD40; CD48; CD4; CD74; CD80; CD84; CD86; CLEC4A; CR1; CRTAM; CTSC; CTSL; CTSS; EBI3; ADGRE1; FCER1G; FCGR1B; FCGR2B; GAPT; GPR183; HAVCR2; HLA-DQB1; IL18; IL1B; INPP5D; KLHL6; LAT2; LILRA1; LILRA3; LILRB1; LILRB4; LILRB5; LYN; MYO1G; NLRP3; P2RX7; PIK3CD; PIK3CG; POU2F2; PTPRC; SAMSN1; SASH3; SLC11A1; SPN; TLR4; TLR8; TNFRSF1B; TNFSF13B; WAS |
| GO:0050866 | negative regulation of cell activation | 3.80 | 2.38e-10 | 5.53e-09 | 29/129 | APOE; AXL; BTK; VSIR; CD300A; CD300LF; CD74; CD80; CD84; CD86; FCGR2B; GPNMB; HAVCR2; HMOX1; INPP5D; LGALS9; LILRB1; LST1; LYN; MERTK; MNDA; PDCD1LG2; PTPN22; PTPRC; SAMSN1; SPN; TNFAIP8L2; UBASH3B; VSIG4 |
| GO:0042110 | T cell activation | 3.76 | 0.00e+00 | 0.00e+00 | 69/310 | ADAM8; AIF1; JAML; APBB1IP; VSIR; CARD11; CCDC88B; CCL2; CD1D; CD209; CD28; CD300A; CD4; CD74; CD80; CD83; CD86; CLEC4A; CLEC4E; CLEC7A; CLECL1; CORO1A; CR1; CRTAM; DOCK2; DOCK8; EBI3; FCER1G; FCGR2B; GPNMB; GPR183; HAVCR2; HLA-DMB; HLA-DOA; HLA-DPA1; HLA-DPB1; IL18; IL1B; LGALS9; LILRB1; LILRB4; LYN; MAFB; NCKAP1L; NLRP3; P2RX7; PDCD1LG2; PIK3CD; PIK3CG; PIK3R6; PTGER4; PTPN22; PTPRC; RASAL3; SASH3; SIRPA; SIRPB1; SLC11A1; SLC46A2; SPN; TGFBR2; TNFAIP8L2; TNFRSF1B; TNFSF13B; TNFSF8; VAV1; VNN1; VSIG4; WAS |
| GO:0001819 | positive regulation of cytokine production | 3.75 | 0.00e+00 | 0.00e+00 | 61/275 | ADAM8; C3AR1; C3; C5AR1; CARD11; CARD9; CASP1; CCDC88B; CCL3; CD14; CD226; CD28; CD40; CD4; CD74; CD80; CD83; CD86; CLEC4E; CLEC5A; CRTAM; CSF1R; CYBA; CYBB; EBI3; FCER1G; FGR; GPSM3; HAVCR2; HGF; HLA-DPA1; HLA-DPB1; HMOX1; HPSE; IL18; IL1A; IL1B; IRF5; IRF8; LGALS9; LILRA2; LILRB1; LY96; MMP8; NFAM1; NLRP3; OSM; P2RX7; PTAFR; PTGER4; PTPN22; PTPRC; SASH3; SLC11A1; SPN; TLR1; TLR2; TLR4; TLR6; TLR7; TLR8 |
| GO:0038061 | NIK/NF-kappaB signaling | 3.70 | 1.62e-05 | 2.13e-04 | 14/64 | ALK; CD14; HAVCR2; IL18; IL1B; LGALS9; MMP8; NLRP3; PTPN22; RASSF2; TLR2; TLR4; TLR6; TLR7 |
| GO:0042113 | B cell activation | 3.64 | 1.40e-09 | 2.99e-08 | 28/130 | BTK; CARD11; CD180; CD28; CD300A; CD40; CD74; DOCK10; FCGR2B; GAPT; GPR183; INPP5D; ITGA4; LAT2; LYL1; LYN; MFNG; MNDA; NCKAP1L; NFAM1; PIK3CD; POU2F2; PTPRC; SAMSN1; SASH3; SLAMF8; TLR4; TNFSF13B |
| GO:0002237 | response to molecule of bacterial origin | 3.64 | 1.33e-15 | 7.23e-14 | 48/223 | ABCA1; ACP5; AXL; C5AR1; CARD16; CARD9; CASP1; CCL2; CCL3; CCR5; CD14; CD180; CD40; CD80; CD86; CSF2RB; CX3CR1; FCGR2B; HAMP; HAVCR2; HCK; IL10RA; IL18; IL1B; IRF5; IRF8; LGALS9; LILRA2; LILRB1; LY86; LY96; LYN; MRC1; NLRP3; P2RX7; PDCD1LG2; PTAFR; PTGER4; PTPN22; SIRPA; SLC11A1; SNCA; TLR1; TLR2; TLR4; TLR6; TNFRSF1B; TREM2 |
| GO:0001818 | negative regulation of cytokine production | 3.64 | 2.93e-12 | 9.17e-11 | 37/172 | ACP5; ADCY7; AXL; BTK; VSIR; CARD16; CD200R1; CD83; CD84; CLEC4A; CMKLR1; CX3CR1; FCGR2B; GPNMB; FFAR4; HAVCR2; HGF; HMOX1; INPP5D; LGALS9; LILRA4; LILRB1; MERTK; MMP8; NCKAP1L; NLRP3; PDCD1LG2; PTGER4; PTPN22; PTPRC; SIRPA; SLC11A1; SRGN; TLR4; TLR6; TLR8; VSIG4 |
| GO:0010573 | vascular endothelial growth factor production | 3.62 | 5.11e-03 | 3.50e-02 | 6/28 | C3AR1; C3; C5AR1; HPSE; IL1A; IL1B |
| GO:0007229 | integrin-mediated signaling pathway | 3.61 | 5.72e-06 | 7.75e-05 | 16/75 | DAB2; FCER1G; FERMT3; FGR; FYB1; HCK; ITGA4; ITGAD; ITGAM; ITGAX; ITGB2; ITGB3; PLEK; PRAM1; TYROBP; VAV1 |
| GO:2001057 | reactive nitrogen species metabolic process | 3.58 | 1.81e-04 | 1.89e-03 | 11/52 | ACP5; AIF1; CX3CR1; GCHFR; IL1B; ITGB2; MMP8; SIRPA; TLR2; TLR4; TLR6 |
| GO:1903706 | regulation of hemopoiesis | 3.45 | 1.13e-14 | 5.76e-13 | 48/235 | ADAM8; AXL; BTK; VSIR; C1QC; CARD11; CCL3; CCR1; CD28; CD4; CD74; CD80; CD83; CD86; CR1; CSF1; CSF3R; EVI2B; FCGR2B; HCLS1; HLA-DOA; IL18; INPP5D; KAT2B; LGALS9; LILRB1; LILRB4; LYN; MAFB; MITF; NCKAP1L; NFAM1; NLRP3; PIK3R6; PTPRC; RASSF2; SASH3; SLAMF8; SLC46A2; SPI1; TGFBR2; TLR4; DCSTAMP; TMEM176A; TMEM176B; TYROBP; UBASH3B; VNN1 |
| GO:0045088 | regulation of innate immune response | 3.44 | 9.19e-14 | 3.74e-12 | 45/221 | ADAM8; APOE; BTK; CARD11; CARD9; CD14; CD1D; CD209; CD226; CD300A; CD300LF; CLEC4A; CLEC4E; CLEC7A; CR1; CRTAM; CTSS; CYBA; FCER1G; FCGR2B; FGR; HAVCR2; HCK; ITGAM; ITGB2; LGALS9; LILRA2; LILRA4; LILRB1; LY96; LYN; MARCO; MNDA; PIK3AP1; PIK3R6; PTPN22; SAMHD1; SLAMF8; TLR1; TLR2; TLR4; TLR6; TLR7; TLR8; VAV1 |
| GO:0002440 | production of molecular mediator of immune response | 3.41 | 8.23e-08 | 1.46e-06 | 24/119 | BTK; CARD11; CD226; CD28; CD40; CD74; FCER1G; FCGR2B; GAPT; HLA-DQB1; HMOX1; IL18; IL1B; LILRB1; NLRP3; POU2F2; PTPRC; SAMHD1; SASH3; SLC11A1; TLR2; TLR4; TNFRSF1B; TNFSF13B |
| GO:0071825 | protein-lipid complex subunit organization | 3.38 | 3.82e-03 | 2.70e-02 | 7/35 | ABCA1; APOC1; APOC2; APOE; PLA2G7; PLTP; SOAT1 |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 3.38 | 5.71e-04 | 5.28e-03 | 10/50 | AXL; CYBA; CYBB; IL1B; ITGB3; NCF1; NCF2; NCF4; NCKAP1L; VAV1 |
| GO:0050900 | leukocyte migration | 3.34 | 0.00e+00 | 0.00e+00 | 63/319 | ADAM8; AIF1; JAML; C3AR1; C5AR1; CCL2; CCL3L1; CCL3L3; CCL3; CCL4L1; CCL8; CCR1; CCR5; CD200R1; CD300A; CD48; CD74; CD84; CMKLR1; CORO1A; CSF1; CSF3R; CX3CR1; CXCL16; DOCK8; DOK2; ADGRE2; FCER1G; FOLR2; FPR2; GPR183; GPSM3; GYPC; HCK; HMOX1; IL1B; INPP5D; ITGA4; ITGAM; ITGAX; ITGB2; ITGB3; LGALS9; LYN; MERTK; MYO1G; NCKAP1L; OLR1; P2RY12; PIK3CD; PIK3CG; PLA2G7; PTAFR; PTGER4; PTPRO; SELPLG; SIRPA; SLAMF8; SLC16A3; SLC7A7; SPN; TREM1; TRPM2; VAV1 |
| GO:0030101 | natural killer cell activation | 3.31 | 1.26e-03 | 1.06e-02 | 9/46 | AXL; CORO1A; HAVCR2; IL18; ITGB2; MERTK; PIK3CD; PTPN22; PTPRC |
| GO:0032103 | positive regulation of response to external stimulus | 3.29 | 6.21e-12 | 1.81e-10 | 41/211 | ADAM8; AIF1; BTK; C3AR1; C3; C5AR1; CCL3L1; CCL3L3; CCL3; CCL4L1; CCR1; CD180; CD28; CD74; CEBPA; CMKLR1; CSF1; CYBA; FCER1G; FPR2; GPSM3; HAVCR2; IL18; IL1B; ITGAM; ITGB2; LGALS9; LY86; LY96; MMP8; NCKAP1L; OSM; P2RY12; PIK3CG; PLA2G7; PTGER4; SNCA; SUCNR1; TLR2; TLR4; TLR6; TLR7 |
| GO:0030099 | myeloid cell differentiation | 3.25 | 1.61e-11 | 4.22e-10 | 40/208 | C1QC; CCL3; CCR1; CD4; CD74; CEBPA; CLEC5A; CSF1R; CSF1; CSF3R; EVI2B; FCER1G; FLI1; GAB3; GPR183; HCLS1; IKZF1; INPP5D; IRF8; KAT2B; LGALS9; LILRB1; LILRB4; NRROS; LYN; MAFB; MITF; NCKAP1L; OSCAR; PDE1B; PIK3CD; RASSF2; SPI1; TGFBR2; TLR2; TLR4; DCSTAMP; TREM2; TYROBP; UBASH3B |
| GO:0022407 | regulation of cell-cell adhesion | 3.15 | 4.20e-14 | 1.90e-12 | 52/279 | ADAM8; AIF1; VSIR; CARD11; CCDC88B; CCL2; CD1D; CD209; CD28; CD300A; CD4; CD74; CD80; CD83; CD86; CLECL1; CORO1A; DOCK8; EBI3; FCGR2B; FERMT3; GPNMB; HAVCR2; HLA-DMB; HLA-DPA1; HLA-DPB1; IL18; IL1B; ITGA4; LGALS9; LILRB1; LILRB4; LYN; NCKAP1L; NLRP3; PDCD1LG2; PIK3R6; PTAFR; PTPN22; PTPRC; RASAL3; SASH3; SIRPA; SIRPB1; SPN; TGFBR2; TNFAIP8L2; TNFSF13B; UBASH3B; VAV1; VNN1; VSIG4 |
| GO:0097006 | regulation of plasma lipoprotein particle levels | 3.13 | 1.08e-03 | 9.22e-03 | 10/54 | ABCA1; APOBR; APOC1; APOC2; APOE; HMOX1; MSR1; PLA2G7; PLTP; SOAT1 |
| GO:0031349 | positive regulation of defense response | 3.13 | 5.67e-14 | 2.43e-12 | 52/281 | ADAM8; BTK; C3; CARD11; CARD9; CCL3L1; CCL3L3; CCL3; CD14; CD1D; CD209; CD226; CD28; CD300A; CD300LF; CEBPA; CLEC4A; CLEC4E; CLEC7A; CRTAM; CTSS; CYBA; FCER1G; GPSM3; HAVCR2; HCK; IL18; IL1B; ITGAM; ITGB2; LGALS9; LILRA2; LILRA4; LY96; LYN; MARCO; MMP8; MNDA; OSM; PIK3AP1; PIK3CG; PLA2G7; PTGER4; PTPN22; SNCA; SUCNR1; TLR1; TLR2; TLR4; TLR6; TLR7; TLR8; VAV1 |
| GO:0050727 | regulation of inflammatory response | 3.12 | 1.19e-12 | 4.05e-11 | 47/255 | ACP5; ADAM8; ADCY7; APOE; BTK; C1QA; C1QB; C1QC; C2; C3AR1; C3; C5AR1; CCL3L1; CCL3L3; CCL3; CD200R1; CD28; CEBPA; CR1; FCER1G; FCGR2B; FFAR4; GPSM3; HCK; HGF; IL18; IL1B; ITGAM; ITGB2; LYN; MMP8; NLRP3; OSM; PIK3AP1; PIK3CG; PLA2G7; PTGER4; PTPRC; SIRPA; SLAMF8; SNCA; SUCNR1; TLR2; TLR4; TLR6; TLR7; TNFAIP8L2; TNFRSF1B |
| GO:0032602 | chemokine production | 3.10 | 6.70e-04 | 6.12e-03 | 11/60 | CD74; CSF1R; HAVCR2; HMOX1; IL18; IL1B; LGALS9; SIRPA; TLR2; TLR4; TLR7 |
| GO:0060326 | cell chemotaxis | 3.09 | 4.57e-11 | 1.09e-09 | 41/224 | ADAM8; AIF1; JAML; BIN2; C3AR1; C5AR1; CCL2; CCL3L1; CCL3L3; CCL3; CCL4L1; CCL8; CCR1; CCR5; CCRL2; CD74; CMKLR1; CORO1A; CSF1; CSF3R; CX3CR1; CXCL16; DOCK4; ADGRE2; FCER1G; FOLR2; FPR2; GPR183; GPSM3; HGF; IL1B; ITGB2; LGALS9; LYN; NCKAP1L; PIK3CD; PIK3CG; PLA2G7; PTPRO; SLAMF8; TRPM2; VAV1 |
| GO:0002697 | regulation of immune effector process | 3.08 | 5.69e-12 | 1.72e-10 | 45/247 | BTK; C1QA; C1QB; C1QC; C2; C3AR1; C3; C5AR1; CD1D; CD226; CD28; CD300A; CD40; CD74; CD80; CD84; CD86; CR1; CRTAM; ADGRE2; FCER1G; FCGR2B; FGR; HAVCR2; HLA-DMB; HMOX1; IL18; IL1B; ITGAM; ITGB2; LGALS9; LILRB1; LYN; NLRP3; P2RX7; PIK3R6; PRAM1; PTAFR; PTPRC; SASH3; SLAMF8; TLR4; TNFRSF1B; VAV1; WAS |
| GO:0048872 | homeostasis of number of cells | 3.00 | 5.99e-07 | 9.02e-06 | 25/141 | AXL; CARD11; CD74; CORO1A; CSF1; DOCK10; FCER1G; GAPT; GPR183; HCLS1; HMOX1; IKZF1; INPP5D; LGALS9; LYN; MAFB; MERTK; NCKAP1L; P2RX7; PIK3CD; RASSF2; SASH3; SLC46A2; SPI1; TNFSF13B |
| GO:0002526 | acute inflammatory response | 2.98 | 5.21e-06 | 7.19e-05 | 21/119 | ADAM8; BTK; C1QA; C1QB; C1QC; C2; C3AR1; C3; C5AR1; CD163; CR1; FCER1G; FCGR2B; HAMP; IL1A; IL1B; NLRP3; OSM; PIK3CG; TREM1; VNN1 |
| GO:0002683 | negative regulation of immune system process | 2.96 | 2.87e-12 | 9.17e-11 | 49/280 | AXL; BTK; VSIR; C1QC; CCL2; CCL3; CD14; CD200R1; CD300A; CD300LF; CD74; CD80; CD84; CD86; CR1; FCER1G; FCGR2B; GPNMB; HAVCR2; HLA-DOA; HMOX1; INPP5D; LGALS9; LILRA2; LILRA4; LILRB1; LILRB4; LPXN; LST1; LY96; LYN; MAFB; MERTK; MNDA; PDCD1LG2; PTGER4; PTPN22; PTPRC; SAMHD1; SAMSN1; SLAMF8; SPN; TLR4; TLR6; TMEM176A; TMEM176B; TNFAIP8L2; UBASH3B; VSIG4 |
| GO:0033627 | cell adhesion mediated by integrin | 2.93 | 3.08e-03 | 2.26e-02 | 9/52 | FERMT3; ITGB2; ITGB3; LPXN; LYN; NCKAP1L; P2RY12; PIK3CG; PLAU |
| GO:0045785 | positive regulation of cell adhesion | 2.84 | 2.32e-11 | 5.72e-10 | 48/286 | ADAM8; AIF1; APBB1IP; VSIR; CARD11; CASS4; CCDC88B; CCL2; CD1D; CD209; CD28; CD4; CD74; CD80; CD83; CD86; CLECL1; CORO1A; CSF1; DOCK8; EBI3; EMILIN2; HAVCR2; HLA-DMB; HLA-DPA1; HLA-DPB1; IL18; IL1B; ITGA4; LGALS9; LILRB1; LILRB4; LYN; NCKAP1L; NLRP3; P2RY12; PDCD1LG2; PIK3R6; PTAFR; PTPRC; RASAL3; SASH3; SIRPA; SIRPB1; TGFBR2; TNFSF13B; VAV1; VNN1 |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 2.75 | 1.90e-05 | 2.42e-04 | 21/129 | BTK; CARD11; CARD16; CARD9; CASP1; CD14; CD40; CD4; CD74; CX3CR1; HMOX1; IL1B; LGALS9; LY96; S100A4; SIRPA; TLR2; TLR4; TLR6; TLR7; TLR8 |
| GO:0002831 | regulation of response to biotic stimulus | 2.74 | 1.96e-03 | 1.59e-02 | 11/68 | CARD16; CD180; CD226; CRTAM; CYBA; HAVCR2; IL1B; LILRA2; LILRB1; LY86; LY96 |
| GO:0031348 | negative regulation of defense response | 2.65 | 7.87e-05 | 8.66e-04 | 19/121 | ACP5; ADCY7; APOE; CD200R1; CR1; FCGR2B; FFAR4; HAVCR2; HGF; LGALS9; LILRB1; NLRP3; PTGER4; PTPRC; SAMHD1; SIRPA; SLAMF8; TNFAIP8L2; TNFRSF1B |
| GO:0043491 | protein kinase B signaling | 2.51 | 3.60e-05 | 4.37e-04 | 23/155 | ADAM8; AXL; CALCR; CCL2; CCL3; CD28; CD40; CD80; CD86; CX3CR1; HCLS1; HGF; HPSE; IL18; IL1B; MERTK; OSM; P2RY12; PIK3AP1; PIK3CD; PIK3CG; PIK3R5; VAV1 |
| GO:0001906 | cell killing | 2.50 | 5.60e-04 | 5.24e-03 | 16/108 | CD1D; CD226; CORO1A; CRTAM; CTSC; FCGR2B; HAMP; HAVCR2; IL18; LGALS9; LILRB1; LYZ; P2RX7; PIK3R6; PTPRC; VAV1 |
| GO:0048017 | inositol lipid-mediated signaling | 2.50 | 5.60e-04 | 5.24e-03 | 16/108 | CD28; CSF1R; FGR; HCLS1; HCST; HGF; IL18; LYN; NCF1; OSM; PIK3AP1; PIK3CD; PIK3CG; PIK3R5; PIK3R6; PTAFR |
| GO:0007200 | phospholipase C-activating G protein-coupled receptor signaling pathway | 2.50 | 2.68e-03 | 2.02e-02 | 12/81 | C3AR1; C5AR1; CMKLR1; CX3CR1; FPR1; FPR2; FPR3; GNA15; GPR65; GRM1; P2RY12; P2RY6 |
| GO:1990868 | response to chemokine | 2.50 | 2.68e-03 | 2.02e-02 | 12/81 | CCL2; CCL3L1; CCL3L3; CCL3; CCL4L1; CCL8; CCR1; CCR5; CCRL2; CMKLR1; CX3CR1; DOCK8; TREM2 |
| GO:0098542 | defense response to other organism | 2.47 | 1.51e-07 | 2.50e-06 | 38/260 | ACP5; APOBEC3C; C5AR1; CARD9; CCDC88B; CD40; CD4; CLEC4E; CLEC7A; COTL1; CYBA; FCER1G; FGR; HAMP; HAVCR2; IL1B; IRF5; IRF8; LILRB1; LYZ; MPEG1; NLRP3; P2RX7; PTPRC; RNASE1; RNASE2; RNASE6; SAMHD1; SLAMF8; SLC11A1; SPN; STAB1; TLR2; TLR4; TLR6; TLR7; TLR8; TNFSF8 |
| GO:0048771 | tissue remodeling | 2.46 | 6.88e-04 | 6.23e-03 | 16/110 | ACP5; ADAM8; AXL; CSF1R; GPNMB; IL18; IL1A; INPP5D; ITGB3; MERTK; MITF; P2RX7; RASSF2; SPP1; DCSTAMP; UBASH3B |
| GO:0072593 | reactive oxygen species metabolic process | 2.45 | 5.43e-05 | 6.13e-04 | 23/159 | ACP5; AIF1; CX3CR1; CYBA; CYBB; FPR2; GCHFR; IL1B; ITGAM; ITGB2; NRROS; MMP8; NCF1; NCF2; NCF4; P2RX7; SIRPA; SNCA; TGFBR2; TLR2; TLR4; TLR6; VAV1 |
| GO:0006959 | humoral immune response | 2.36 | 4.71e-05 | 5.48e-04 | 25/179 | C1QA; C1QB; C1QC; C2; C3AR1; C3; C5AR1; CCL2; CD28; CD83; CR1; EBI3; FCGR2B; GPR183; HLA-DQB1; LY86; LYZ; POU2F2; PTPRC; RNASE2; RNASE6; SLC11A1; TREM1; TREM2; VSIG4 |
| GO:0006898 | receptor-mediated endocytosis | 2.35 | 3.60e-05 | 4.37e-04 | 26/187 | APOBR; APOC1; APOC2; APOE; ASGR1; ASGR2; CD14; CD163; CXCL16; DAB2; FCER1G; FCGR1A; FCGR1B; FCGR2B; FOLR2; FPR2; ITGA4; ITGB2; ITGB3; LILRB1; MARCO; MRC1; MSR1; SNCA; STAB1; TGFBR2 |
| GO:0060627 | regulation of vesicle-mediated transport | 2.29 | 7.80e-07 | 1.11e-05 | 39/288 | APOC1; APOC2; APOE; AXL; C2; C3; CCL2; CD14; CD300A; CD300LF; CD84; CHRNA6; CORO1A; CYBA; DAB2; DOCK2; ADGRE2; FCER1G; FCGR2B; FGR; HCK; HMOX1; IL1B; ITGAM; ITGB2; ITGB3; LGALS9; LILRB1; LYN; MERTK; NCKAP1L; PRAM1; PTAFR; PTPRC; SIRPA; SIRPB1; SLC11A1; SNCA; TLR2 |
| GO:0070555 | response to interleukin-1 | 2.29 | 4.06e-03 | 2.85e-02 | 13/96 | CCL2; CCL3L1; CCL3L3; CCL3; CCL4L1; CCL8; CD40; CYBA; IL1A; IL1B; LGALS9; SIRPA; SNCA; ST18 |
| GO:0050920 | regulation of chemotaxis | 2.28 | 3.18e-04 | 3.19e-03 | 21/156 | AIF1; C3AR1; C5AR1; CCL2; CCL3; CCL4L1; CCR1; CD74; CMKLR1; CSF1; FPR2; GPR183; GPSM3; LGALS9; LYN; NCKAP1L; P2RY12; PLA2G7; PTPRO; SLAMF8; SUCNR1 |
| GO:0050817 | coagulation | 2.27 | 6.81e-05 | 7.59e-04 | 26/194 | APOE; AXL; CD40; DOCK8; FCER1G; FERMT3; GNA15; HPSE; ITGB3; LCP2; LYN; MERTK; P2RX7; P2RY12; PIK3CD; PIK3CG; PIK3R5; PIK3R6; PLAUR; PLAU; PLEK; TLR4; TREML1; UBASH3B; VAV1; WAS |
| GO:0070371 | ERK1 and ERK2 cascade | 2.25 | 1.20e-05 | 1.60e-04 | 32/241 | ALK; APOE; AXL; C5AR1; CALCR; CCL2; CCL3L1; CCL3L3; CCL3; CCL4L1; CCL8; CCR1; CD4; CD74; CSF1R; EPHB2; FPR2; GPNMB; FFAR4; GPR183; HAVCR2; IL1B; LGALS9; LYN; MARCO; MERTK; P2RY6; PTGER4; PTPN22; PTPRC; SIRPA; TLR4; TREM2 |
| GO:2000147 | positive regulation of cell motility | 2.18 | 7.78e-07 | 1.11e-05 | 43/333 | ADAM8; AIF1; VSIR; C3AR1; C5AR1; CASS4; CCL3; CCL4L1; CCL8; CCR1; CD40; CD74; CMKLR1; CORO1A; CSF1R; CSF1; CXCL16; DAB2; DOCK4; DOCK8; FERMT3; FGR; FPR2; GPNMB; GPSM3; HGF; HMOX1; IL1B; ITGA4; ITGB3; LGALS9; LYN; NCKAP1L; P2RY12; P2RY6; PIK3CD; PLA2G7; PLAU; PTAFR; PTPRC; SLC8A1; SPN; TGFBR2 |
| GO:0034612 | response to tumor necrosis factor | 2.14 | 9.73e-04 | 8.43e-03 | 20/158 | CARD16; CASP1; CCL2; CCL3L1; CCL3L3; CCL3; CCL4L1; CCL8; CD14; CD40; CEBPA; CRHBP; CXCL16; CYBA; GSDME; HAMP; ST18; DCSTAMP; TNFRSF1B; TNFSF13B; TNFSF8 |
| GO:0007162 | negative regulation of cell adhesion | 2.14 | 5.60e-04 | 5.24e-03 | 22/174 | ADAMDEC1; VSIR; CD300A; CD74; CD80; CD86; FCGR2B; GPNMB; HAVCR2; LGALS9; LILRB1; LPXN; PDCD1LG2; PLXNC1; PTPN22; PTPRC; PTPRO; SPN; TGFBI; TNFAIP8L2; UBASH3B; VSIG4 |
| GO:0051701 | interaction with host | 2.13 | 7.41e-03 | 4.86e-02 | 13/103 | AXL; CCR5; CD209; CD4; CD74; CD80; CD86; CLEC5A; CR1; ITGB3; LGALS9; MRC1; SELPLG |
| GO:0019058 | viral life cycle | 2.13 | 2.42e-03 | 1.88e-02 | 17/135 | APOBEC3C; APOE; AXL; CCL2; CCR5; CD209; CD28; CD4; CD74; CD80; CD86; CLEC5A; CR1; ITGB3; LGALS9; MRC1; SELPLG |
| GO:0051235 | maintenance of location | 2.11 | 3.77e-04 | 3.70e-03 | 24/192 | ABCA1; APOE; C3; CCL3; CCR5; CD4; CORO1A; CYBA; FTL; IL1B; ITGB3; NRROS; LYN; MSR1; P2RX7; P2RY6; PLIN2; PTPRC; SLC8A1; SNCA; SOAT1; SRGN; TRPM2; UBASH3B |
Factor 7 : 4 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0006575 | cellular modified amino acid metabolic process | 2.27 | 1.64e-05 | 0.0134 | 29/121 | ACADL; ALDH4A1; ASS1; CRAT; CRYM; EGLN3; ERO1A; G6PD; GGCT; GGT1; GGT6; GGTLC1; GGTLC2; GNMT; GSR; GSTZ1; IDH1; IYD; MBOAT1; MTHFD2; MTHFR; P4HA1; CPQ; PLA2G2A; PRODH; PTGES; SERINC2; VNN2; VNN3 |
| GO:0007586 | digestion | 2.17 | 2.35e-04 | 0.0382 | 23/100 | ADM2; AKR1D1; AQP5; INAVA; CLPSL2; CLPSL1; CCKBR; CHIA; CHRM1; LCT; MOGAT2; MUC6; OPRL1; PGC; PPARGC1A; PRSS1; PTGER3; SCARB1; SERPINA3; SLC26A6; SLC26A7; SLC5A1; SNX10 |
| GO:0042180 | cellular ketone metabolic process | 2.10 | 3.01e-04 | 0.0409 | 24/108 | ACADL; ADIPOR2; ADM; AKR1B10; C1QTNF2; CBR3; COMT; FABP5; FGFR4; GSTZ1; IRS1; KMO; KYNU; NQO1; OAZ3; PDK1; PDP1; PLIN5; PPARGC1A; SRD5A1; STARD3; TRIB3; UGT1A8; WNT4 |
| GO:0006520 | cellular amino acid metabolic process | 2.03 | 3.66e-05 | 0.0149 | 34/158 | ADSSL1; ALDH4A1; ASRGL1; ASS1; BCAT2; COMT; CRYM; DDC; EGLN3; ERO1A; FAH; GGT1; GLYATL1; GNMT; GSTZ1; HGD; HMGCLL1; HPD; IYD; KMO; KYNU; MTHFR; NAGS; NAT8L; NMNAT3; NQO1; OAT; OAZ3; P4HA1; PAH; PHGDH; PRODH; PSPH; TST |
Factor 8 : 27 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0140029 | exocytic process | 5.29 | 1.32e-05 | 9.14e-04 | 10/47 | CPLX2; KCNB1; NAPB; PPFIA3; RAB3A; RAB3C; SNAP25; STX1A; STXBP1; UNC13A |
| GO:0099504 | synaptic vesicle cycle | 4.85 | 7.42e-11 | 6.04e-08 | 24/123 | BRSK2; BSN; CACNB2; CDK5R1; CHRNB2; CPLX2; DDC; DNAJC6; DRD2; NAPB; NRXN1; PPFIA3; RAB3A; RIMS2; SNAP25; SNCB; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; UNC13A |
| GO:0017156 | calcium ion regulated exocytosis | 4.82 | 3.28e-09 | 1.01e-06 | 20/103 | CACNB2; CDK5R2; CHRNB2; CPLX2; DRD2; KCNB1; NAPB; PPFIA3; RAB3A; RIMS2; SCAMP5; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; UNC13A |
| GO:0048284 | organelle fusion | 4.52 | 3.13e-04 | 1.16e-02 | 8/44 | CPLX2; GDAP1; RAB39A; RAB3A; SNAP25; STX1A; STXBP1; SYT4 |
| GO:0099003 | vesicle-mediated transport in synapse | 4.46 | 5.85e-09 | 1.01e-06 | 21/117 | AP3B2; CACNB2; CHRNB2; CPLX2; DNAJC6; DRD2; KIF5A; NAPB; PPFIA3; RAB3A; RIMS2; SNAP25; SNCB; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; UNC13A |
| GO:0099565 | chemical synaptic transmission, postsynaptic | 4.40 | 2.52e-06 | 2.05e-04 | 14/79 | CELF4; CHRNA3; CHRNB2; DRD2; GRIK2; GRIN2C; IGSF9B; MAPK8IP2; MPP2; NRXN1; P2RX6; RIMS2; SEZ6; STX1A |
| GO:0035418 | protein localization to synapse | 4.35 | 9.46e-04 | 3.08e-02 | 7/40 | BSN; GRIN2C; IQSEC2; KIF5A; NRXN1; ARHGAP44; SNAP25 |
| GO:0022406 | membrane docking | 4.30 | 1.96e-04 | 8.87e-03 | 9/52 | CPLX2; KCNB1; PPFIA3; RAB3A; RAB3C; SNAP25; STX1A; STXBP1; UNC13A |
| GO:0015837 | amine transport | 4.09 | 1.35e-05 | 9.14e-04 | 13/79 | CARTPT; CHGA; CHRNA3; CHRNB2; DDC; DRD2; HRH3; KCNB1; STX1A; STXBP1; SYT13; SYT4; SYT5 |
| GO:0015844 | monoamine transport | 3.71 | 3.10e-04 | 1.16e-02 | 10/67 | CARTPT; CHGA; CHRNB2; DRD2; HRH3; KCNB1; STX1A; SYT13; SYT4; SYT5 |
| GO:0051648 | vesicle localization | 3.57 | 3.15e-07 | 3.21e-05 | 21/146 | AP3B2; BRSK2; CACNB2; CHRNB2; CPLX2; DRD2; GRIA1; KIF5A; NAPB; PPFIA3; RAB3A; RIMS2; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; UNC13A |
| GO:0006836 | neurotransmitter transport | 3.57 | 6.23e-09 | 1.01e-06 | 27/188 | ASIC1; CACNB2; CHRNA3; CHRNB2; CPLX2; DDC; DRD2; GRM4; HRH3; NAPB; NRXN1; PPFIA3; PTPRN2; RAB3A; RIMS2; SLC1A2; SLC6A12; SLC6A17; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; UNC13A |
| GO:0030534 | adult behavior | 3.47 | 7.98e-05 | 3.82e-03 | 13/93 | CARTPT; CHRNA3; CHRNB2; DRD2; FGF12; GABRG2; UNC79; NRXN1; SEZ6L; SEZ6; SLC1A2; SLITRK1; SPTBN4 |
| GO:0098739 | import across plasma membrane | 3.15 | 1.17e-03 | 3.68e-02 | 10/79 | ATP1A3; FAM155B; HCN2; HCN4; HFE; KCNH2; KCNJ6; SLC12A5; SLC1A2; SLC36A4 |
| GO:0099177 | regulation of trans-synaptic signaling | 3.00 | 3.75e-09 | 1.01e-06 | 35/290 | ASIC1; ACHE; CACNB2; CAMK2B; CELF4; CHRNA3; CHRNB2; CPLX2; DGKE; DRD2; GRIA1; GRIK2; GRIN2C; GRM4; IGSF9B; IQSEC2; JPH3; KCNB1; MAPK8IP2; MPP2; NAPB; NPTXR; NRXN1; PPFIA3; RAB3A; RIMS2; SCGN; SLC8A2; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT4; UNC13A |
| GO:0001764 | neuron migration | 2.95 | 6.95e-04 | 2.46e-02 | 12/101 | ASCL1; CAMK2B; CDK5R1; CDK5R2; CELSR3; DRD2; FGFR1; GPR173; KIAA0319; NAV1; NEUROD4; SCRT1 |
| GO:1990778 | protein localization to cell periphery | 2.91 | 2.46e-05 | 1.54e-03 | 19/162 | ANK1; ANK3; CACNB2; GRIN2C; IQSEC2; KCNB1; KCNIP3; LGALS3; NFASC; PIK3R1; RAB3A; RAB3C; ARHGAP44; SCN3B; SNAP25; SPTBN4; STXBP1; TUB; ZDHHC22 |
| GO:0042391 | regulation of membrane potential | 2.88 | 1.75e-08 | 2.38e-06 | 34/293 | ASIC1; ANK3; ATP1A3; CACNB2; CELF4; CHRNA3; CHRNB2; DRD2; FGF12; GABRG2; GRIA1; GRIK2; GRIN2C; HCN2; HCN3; HCN4; IGSF9B; KCNB1; KCNC1; KCNH2; KCNH3; KCNH4; KCNH6; KCNU1; MAPK8IP2; MPP2; NRXN1; NTSR2; P2RX6; RIMS2; SCN3B; SEZ6; SLC26A10; STX1A |
| GO:0001505 | regulation of neurotransmitter levels | 2.80 | 1.06e-06 | 9.58e-05 | 27/240 | ASIC1; ACHE; CACNB2; CHRNA3; CHRNB2; CPLX2; DDC; DRD2; GRM4; HRH3; NAPB; NRXN1; PAH; PPFIA3; PTPRN2; RAB3A; RIMS2; SLC1A2; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; UNC13A |
| GO:0006813 | potassium ion transport | 2.78 | 3.07e-05 | 1.78e-03 | 20/179 | ANK3; ATP1A3; DRD2; HCN2; HCN3; HCN4; KCNB1; KCNC1; KCNH2; KCNH3; KCNH4; KCNH6; KCNIP3; KCNJ6; KCNK3; KCNQ2; KCNU1; SLC12A5; SLC24A5; SNAP25 |
| GO:0023061 | signal release | 2.66 | 1.35e-07 | 1.57e-05 | 34/318 | ASIC1; BRSK2; CACNB2; CARTPT; CHGA; CHRNA3; CHRNB2; CPLX2; DRD2; FGFR1; GRM4; HFE; HRH3; IRS2; KCNB1; NAPB; NRXN1; PPFIA3; PTPRN2; RAB3A; RIMS2; SCG5; SMPD3; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; TCF7L2; UNC13A; VGF |
| GO:0032386 | regulation of intracellular transport | 2.64 | 4.26e-05 | 2.31e-03 | 21/198 | ABCA2; ANK3; BRSK2; CACNB2; CDK5R1; CHRNB2; CPLX2; DRD2; KCNB1; NAPB; PIK3R1; RAB3A; REEP2; ARHGAP44; RIMS2; STX1A; STXBP1; SYN1; SYP; SYT4; TCF7L2 |
| GO:0016358 | dendrite development | 2.61 | 1.38e-03 | 4.16e-02 | 13/124 | CAMK2B; CDK5R1; CHRNA3; CHRNB2; CTNNA2; EFNA1; KIAA0319; MAP6; MAPK8IP2; ARHGAP44; SARM1; SEZ6; SLC12A5 |
| GO:0051656 | establishment of organelle localization | 2.33 | 2.51e-04 | 1.02e-02 | 21/224 | AP3B2; CACNB2; CHGA; CHRNB2; CPLX2; DRD2; GRIA1; KIF5A; NAPB; PPFIA3; RAB3A; RIMS2; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; UNC13A |
| GO:0050808 | synapse organization | 2.28 | 6.47e-05 | 3.29e-03 | 26/283 | ACHE; ANK3; BSN; C1QL1; CACNB2; CAMK2B; CDK5R1; CHRNB2; CTNNA2; DLGAP3; DRD2; EFNA1; IGSF9B; L1CAM; LRTM2; NFASC; NRXN1; RAB39B; RAB3A; ARHGAP44; SEZ6L; SEZ6; SLITRK1; SNCB; SYN1; UNC13A |
| GO:0021700 | developmental maturation | 2.25 | 1.44e-03 | 4.20e-02 | 17/188 | ASCL1; C1QL1; CAMK2B; CDK5R1; CDK5R2; CDKN1A; FGFR1; NFASC; NRXN1; RAB3A; SEZ6L; SEZ6; SPTBN4; STXBP1; SYP; SYT4; UNC13A |
| GO:0060627 | regulation of vesicle-mediated transport | 2.16 | 2.21e-04 | 9.47e-03 | 25/288 | CACNB2; CDK5R2; CHRNB2; CPLX2; DNAJC6; DRD2; HFE; KCNB1; LGALS3; NAPB; PACSIN3; RAB3A; RAB3C; ARHGAP44; RIMS2; SCAMP5; SMPD3; STX1A; STXBP1; SYN1; SYP; SYT4; SYT5; TUB; VTN |
Factor 9 : 11 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0140053 | mitochondrial gene expression | 10.50 | 1.14e-05 | 1.86e-03 | 6/16 | CHCHD10; MRPL21; MRPL23; MRPL41; MRPS21; NDUFA7 |
| GO:0070972 | protein localization to endoplasmic reticulum | 10.30 | 2.85e-09 | 1.16e-06 | 11/30 | RPL13; RPL18; RPL28; RPL35; RPLP2; RPS16; RPS19; RPS21; RPS26; RPS28; SEC61G |
| GO:0010257 | NADH dehydrogenase complex assembly | 10.20 | 4.26e-04 | 3.15e-02 | 4/11 | NDUFAF8; NDUFA3; NDUFA7; NDUFS5 |
| GO:0006413 | translational initiation | 9.36 | 1.90e-09 | 1.16e-06 | 12/36 | AGO2; RPL13; RPL18; RPL28; RPL35; RPLP2; RPS16; RPS19; RPS21; RPS26; RPS28; UHMK1 |
| GO:0000726 | non-recombinational repair | 8.26 | 2.40e-04 | 2.17e-02 | 5/17 | BARD1; PRKDC; RIF1; RNF168; USP51 |
| GO:0006414 | translational elongation | 8.26 | 2.40e-04 | 2.17e-02 | 5/17 | MRPL21; MRPL23; MRPL41; MRPS21; RPLP2 |
| GO:0006401 | RNA catabolic process | 4.40 | 6.28e-06 | 1.28e-03 | 13/83 | AGO2; ERN1; HELZ2; RPL13; RPL18; RPL28; RPL35; RPLP2; RPS16; RPS19; RPS21; RPS26; RPS28 |
| GO:0070646 | protein modification by small protein removal | 4.14 | 2.75e-04 | 2.24e-02 | 9/61 | ASXL2; BARD1; CLSPN; MINDY2; MYSM1; PSMB3; PSME2; USP12; USP51 |
| GO:0006302 | double-strand break repair | 3.95 | 1.88e-04 | 2.17e-02 | 10/71 | BARD1; BRCA2; EME2; EYA3; HELB; PRKDC; RIF1; RNF168; SAMHD1; USP51 |
| GO:0090150 | establishment of protein localization to membrane | 3.94 | 5.04e-06 | 1.28e-03 | 15/107 | SYNE3; KIAA0754; KIF13A; MACF1; ROMO1; RPL13; RPL18; RPL28; RPL35; RPLP2; RPS16; RPS19; RPS21; RPS26; RPS28; ZDHHC20 |
| GO:0006605 | protein targeting | 3.40 | 3.09e-05 | 4.20e-03 | 15/124 | LEPROT; ROMO1; RPL13; RPL18; RPL28; RPL35; RPLP2; RPS16; RPS19; RPS21; RPS26; RPS28; SEC61G; VPS13C; ZDHHC20 |
Factor 11 : 13 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0018149 | peptide cross-linking | 17.70 | 1.14e-11 | 3.09e-09 | 11/38 | IVL; PI3; SPRR1A; SPRR1B; SPRR2A; SPRR2B; SPRR2D; SPRR2E; SPRR2F; SPRR3; SPRR4 |
| GO:0042440 | pigment metabolic process | 9.94 | 2.56e-05 | 2.60e-03 | 6/37 | UGT1A10; UGT1A1; UGT1A4; UGT1A7; UGT1A8; UGT1A9 |
| GO:0009620 | response to fungus | 9.43 | 7.63e-04 | 4.78e-02 | 4/26 | C10orf99; S100A12; S100A8; S100A9 |
| GO:0043588 | skin development | 7.88 | 0.00e+00 | 0.00e+00 | 31/241 | CASP14; EPHA2; GJB3; IVL; KRT13; KRT16; KRT3; KRT5; KRT6A; KRT6B; KRT6C; KRT78; CERS3; PI3; RPTN; S100A7; SCEL; SERPINB13; SNAI1; SOX21; SPRR1A; SPRR1B; SPRR2A; SPRR2B; SPRR2D; SPRR2E; SPRR2F; SPRR2G; SPRR3; SPRR4; TP63 |
| GO:0008544 | epidermis development | 7.18 | 0.00e+00 | 0.00e+00 | 32/273 | BNC1; CASP14; EPHA2; IVL; KRT13; KRT16; KRT3; KRT5; KRT6A; KRT6B; KRT6C; KRT78; KRTDAP; CERS3; PI3; RPTN; S100A7; SCEL; SERPINB13; SNAI1; SOX21; SPRR1A; SPRR1B; SPRR2A; SPRR2B; SPRR2D; SPRR2E; SPRR2F; SPRR2G; SPRR3; SPRR4; TP63 |
| GO:0032602 | chemokine production | 6.13 | 4.06e-04 | 2.75e-02 | 6/60 | CXCL6; EPHA2; IL4R; LBP; S100A8; S100A9 |
| GO:0006720 | isoprenoid metabolic process | 5.84 | 6.15e-05 | 5.57e-03 | 8/84 | ADH7; AKR1B10; CYP2C19; UGT1A1; UGT1A3; UGT1A7; UGT1A8; UGT1A9 |
| GO:0035821 | modification of morphology or physiology of other organism | 5.65 | 2.22e-04 | 1.65e-02 | 7/76 | CXCL6; DEFB4A; KRT6A; PGLYRP3; PGLYRP4; S100A12; S100A9 |
| GO:0006959 | humoral immune response | 4.45 | 6.14e-06 | 7.15e-04 | 13/179 | CXCL1; CXCL6; DEFB1; DEFB4A; CXCL8; KRT6A; PGLYRP3; PGLYRP4; PI3; S100A12; S100A7; S100A8; S100A9 |
| GO:0009410 | response to xenobiotic stimulus | 4.33 | 3.61e-06 | 4.90e-04 | 14/198 | ALDH3A1; CYP2C18; CYP2C19; GJB2; S100A12; UGT1A10; UGT1A1; UGT1A3; UGT1A4; UGT1A5; UGT1A6; UGT1A7; UGT1A8; UGT1A9 |
| GO:0060326 | cell chemotaxis | 4.10 | 3.12e-06 | 4.90e-04 | 15/224 | C10orf99; CCL20; CXCL1; CXCL6; DEFB4A; EPHA2; HBEGF; IL36G; IL1RN; CXCL8; LBP; S100A12; S100A7; S100A8; S100A9 |
| GO:0098542 | defense response to other organism | 4.01 | 9.01e-07 | 1.83e-04 | 17/260 | C10orf99; CCL20; CXCL6; DEFB1; DEFB4A; EPHA2; GBP6; IL4R; KRT6A; LBP; MMP12; PGLYRP3; PGLYRP4; S100A12; S100A7; S100A8; S100A9 |
| GO:0005996 | monosaccharide metabolic process | 4.01 | 1.85e-04 | 1.51e-02 | 10/153 | SLC2A1; UGT1A10; UGT1A1; UGT1A3; UGT1A4; UGT1A5; UGT1A6; UGT1A7; UGT1A8; UGT1A9 |
Factor 13 : 67 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0051383 | kinetochore organization | 26.70 | 1.84e-14 | 6.25e-13 | 10/11 | CDT1; CENPA; CENPE; CENPF; CENPK; CENPN; CENPW; NDC80; NUF2; SMC4 |
| GO:0061641 | CENP-A containing chromatin organization | 22.60 | 4.51e-13 | 1.31e-11 | 10/13 | KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; HJURP; CENPU; OIP5 |
| GO:0034502 | protein localization to chromosome | 20.10 | 0.00e+00 | 0.00e+00 | 15/22 | AURKB; BRCA2; BUB1B; KNL1; CDCA5; CDK1; CDT1; CENPA; ESCO2; EZH2; HASPIN; MTBP; NDC80; PLK1; TTK |
| GO:0036297 | interstrand cross-link repair | 19.60 | 7.02e-10 | 1.54e-08 | 8/12 | EME1; FANCA; FANCB; FANCE; FANCI; RAD51AP1; RAD51; UBE2T |
| GO:0050000 | chromosome localization | 18.50 | 0.00e+00 | 0.00e+00 | 17/27 | CCNB1; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; DLGAP5; FAM83D; KIF14; KIF18A; KIF2C; KIFC1; NDC80; NUF2; PSRC1; SPAG5 |
| GO:0006260 | DNA replication | 17.20 | 0.00e+00 | 0.00e+00 | 56/96 | ATAD5; BARD1; BLM; BRCA1; BRCA2; BRIP1; TICRR; RMI2; CCNA2; CCNE1; CCNE2; CDC45; CDC6; CDC7; CDK1; CDT1; CHAF1B; CHEK1; CHTF18; CLSPN; DBF4; DDX11; DNA2; DSCC1; DTL; E2F7; E2F8; EME1; ESCO2; EXO1; FBXO5; FEN1; GINS1; GINS2; GINS4; GMNN; PCLAF; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; TONSL; ORC1; PIF1; POLD1; POLE2; POLQ; RAD51; RECQL4; RFC4; RNASEH2A; RRM2; ALYREF; WDHD1 |
| GO:0007059 | chromosome segregation | 16.90 | 0.00e+00 | 0.00e+00 | 72/125 | AURKB; BIRC5; BLM; BRCA1; BRIP1; BUB1B; BUB1; KNSTRN; RMI2; KNL1; CCNB1; CCNE1; CCNE2; CDC20; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CENPN; CENPW; CEP55; DDX11; DLGAP5; DSCC1; ECT2; EME1; ESCO2; ESPL1; FAM83D; FBXO5; FEN1; HASPIN; HJURP; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; MAD2L1; MKI67; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NUF2; NUSAP1; OIP5; PLK1; PRC1; PSRC1; PTTG1; RACGAP1; RAD51C; SGO1; SGO2; SKA1; SKA2; SKA3; SMC4; SPAG5; SYCE2; TACC3; NDC1; TOP2A; TRIP13; TTK; ZWINT |
| GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 16.10 | 0.00e+00 | 0.00e+00 | 29/53 | AURKA; AURKB; SAPCD2; CCNB1; CDC20; CENPA; CENPE; ESPL1; GPSM2; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; NUSAP1; PLK1; POC1A; PRC1; PSRC1; RACGAP1; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0045023 | G0 to G1 transition | 15.80 | 6.95e-08 | 1.41e-06 | 7/13 | BRCA1; CDC7; CHEK1; E2F1; EZH2; RAD51; RRM2 |
| GO:0006301 | postreplication repair | 14.70 | 1.13e-06 | 2.05e-05 | 6/12 | BRCA1; DTL; PCLAF; POLD1; POLE2; RFC4 |
| GO:0000075 | cell cycle checkpoint | 14.50 | 0.00e+00 | 0.00e+00 | 36/73 | AURKA; AURKB; BLM; BRCA1; BRIP1; BUB1B; BUB1; TICRR; CCNB1; CDC20; CDC25C; CDC45; CDC6; CDK1; CDT1; CENPF; CHEK1; CLSPN; DNA2; DTL; E2F1; E2F7; E2F8; EME1; GTSE1; H2AFX; KNTC1; MAD2L1; NDC80; ORC1; PLK1; TOP2A; TRIP13; TTK; WDR76; ZWINT |
| GO:0007051 | spindle organization | 13.80 | 0.00e+00 | 0.00e+00 | 31/66 | ASPM; AURKA; AURKB; KNSTRN; AUNIP; CCNB1; CDC20; CENPE; ESPL1; FBXO5; GPSM2; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; PLK1; POC1A; PRC1; PSRC1; RACGAP1; SPAG5; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0032200 | telomere organization | 13.80 | 2.09e-14 | 6.80e-13 | 15/32 | AURKB; BLM; BRCA2; CCNE1; CCNE2; DNA2; EXO1; FEN1; NEK2; PIF1; POLD1; RAD51C; RAD51; RECQL4; RFC4 |
| GO:0006302 | double-strand break repair | 13.70 | 0.00e+00 | 0.00e+00 | 33/71 | BARD1; BLM; BRCA1; BRCA2; BRIP1; PARPBP; RMI2; AUNIP; CDC45; CDC7; CDCA5; CHEK1; DDX11; DEK; DNA2; EME1; ESCO2; EXO1; FANCB; FEN1; FOXM1; GINS2; GINS4; H2AFX; TONSL; POLQ; RAD51AP1; RAD51C; RAD51; RAD54B; RAD54L; RECQL4; TRIP13 |
| GO:0051653 | spindle localization | 12.90 | 4.24e-07 | 8.03e-06 | 7/16 | ASPM; SAPCD2; CENPA; ESPL1; GPSM2; NDC80; NUSAP1 |
| GO:0071103 | DNA conformation change | 12.60 | 0.00e+00 | 0.00e+00 | 42/98 | ASF1B; BLM; BRIP1; KNL1; CCNB1; CDC45; CDCA5; CDK1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; DDX11; DNA2; GINS1; GINS2; GINS4; H2AFX; HELLS; HJURP; HMGB2; MCM2; MCM4; MCM6; MCM7; CENPU; NCAPD2; NCAPG2; NCAPG; NCAPH; NUSAP1; OIP5; PIF1; RAD51; RAD54B; RAD54L; RECQL4; SMC4; TOP2A |
| GO:0006284 | base-excision repair | 12.60 | 3.47e-06 | 5.88e-05 | 6/14 | DNA2; FEN1; NEIL3; POLD1; POLQ; RECQL4 |
| GO:0000910 | cytokinesis | 12.50 | 0.00e+00 | 0.00e+00 | 25/59 | ANLN; AURKA; AURKB; BRCA2; CDC25B; CDC6; CDT1; CENPA; CEP55; CIT; CKAP2; E2F7; E2F8; ECT2; ESPL1; KIF14; KIF20A; KIF23; KIF4A; NUSAP1; PLK1; PRC1; RACGAP1; SEPT3; STMN1 |
| GO:0044839 | cell cycle G2/M phase transition | 12.40 | 0.00e+00 | 0.00e+00 | 35/83 | ATAD5; AURKA; AURKB; BLM; BRCA1; TICRR; CCNA2; CCNB1; CCNB2; CDC25A; CDC25B; CDC25C; CDC6; CDC7; CDK1; CENPF; CHEK1; CIT; CLSPN; DTL; FBXO5; FOXM1; GTSE1; HMMR; KIF14; MELK; NEK2; ORC1; PKMYT1; PLK1; PLK4; RAD51C; RECQL4; SKP2; TPX2 |
| GO:0000726 | non-recombinational repair | 12.10 | 7.01e-07 | 1.30e-05 | 7/17 | BARD1; BLM; BRCA1; AUNIP; DEK; H2AFX; POLQ |
| GO:0044772 | mitotic cell cycle phase transition | 11.80 | 0.00e+00 | 0.00e+00 | 77/192 | ANLN; ATAD5; AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; TICRR; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN2C; CDKN2D; CDKN3; CDT1; CENPE; CENPF; CIT; CKS1B; CKS2; CLSPN; DBF4; DLGAP5; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FBXO5; FOXM1; GMNN; GTSE1; HMMR; IQGAP3; KIF14; KNTC1; MAD2L1; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MELK; MTBP; NDC80; NEK2; ORC1; PKMYT1; PLK1; PLK4; POLE2; RAD51C; RBL1; RECQL4; RRM2; SKP2; TACC3; TPX2; TRIP13; TTK; TYMS; UBE2C; UBE2S |
| GO:0048285 | organelle fission | 11.00 | 0.00e+00 | 0.00e+00 | 81/216 | ANLN; ASPM; AURKA; AURKB; BLM; BRCA2; BRIP1; BUB1B; BUB1; KNSTRN; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CDC20; CDC25B; CDC25C; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; CHEK1; CKS2; DLGAP5; DSCC1; EME1; ESPL1; MTFR2; FANCA; FBXO5; HASPIN; KIF11; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; KNTC1; MAD2L1; MKI67; MND1; MTBP; MYBL1; MYBL2; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NUF2; NUSAP1; PKMYT1; PLK1; PRC1; PSRC1; PTTG1; RACGAP1; RAD51C; RAD51; RAD54B; SGO1; SGO2; SMC4; SPAG5; SYCE2; TACC3; NDC1; TOP2A; TPX2; TRIP13; TTK; UBE2C; UBE2S; ZWINT |
| GO:0031023 | microtubule organizing center organization | 10.90 | 2.30e-13 | 6.94e-12 | 16/43 | AURKA; BRCA1; BRCA2; CCNF; CDK1; CHEK1; CNTLN; PCLAF; KIF11; NEK2; PLK1; PLK4; POC1A; SGO1; STIL; WDR62 |
| GO:0044843 | cell cycle G1/S phase transition | 10.50 | 0.00e+00 | 0.00e+00 | 39/109 | AURKA; CCNA2; CCNB1; CCNE1; CCNE2; CDC25A; CDC25C; CDC45; CDC6; CDC7; CDK1; CDKN2C; CDKN2D; CDKN3; CDT1; DBF4; E2F1; E2F7; E2F8; EZH2; FAM83D; FBXO5; GMNN; GTSE1; IQGAP3; KIF14; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MTBP; ORC1; POLE2; RBL1; RRM2; SKP2; TYMS |
| GO:1901987 | regulation of cell cycle phase transition | 10.50 | 0.00e+00 | 0.00e+00 | 53/149 | ANLN; ATAD5; AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; TICRR; CCNB1; CCNE1; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN2D; CDT1; CENPE; CENPF; CHEK1; CLSPN; DLGAP5; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FAM83D; FBXO5; GTSE1; HMMR; KIF14; KNTC1; MAD2L1; MTBP; NDC80; NEK2; ORC1; PLK1; PLK4; RAD51C; RBL1; RECQL4; TPX2; TRIP13; TTK; UBE2C |
| GO:0010948 | negative regulation of cell cycle process | 10.30 | 0.00e+00 | 0.00e+00 | 39/111 | AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; TICRR; CCNB1; CCNF; CDC20; CDC25C; CDC6; CDC7; CDK1; CDKN2D; CDT1; CENPF; CHEK1; CLSPN; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FBXO5; GTSE1; MAD2L1; MTBP; NDC80; NEK2; ORC1; PLK1; PTTG1; RAD51; RBL1; RRM2; TRIP13; TTK |
| GO:0051321 | meiotic cell cycle | 9.73 | 0.00e+00 | 0.00e+00 | 43/130 | ASPM; AURKA; BLM; BRCA2; BRIP1; BUB1B; BUB1; CCNE1; CCNE2; CDC20; CDC25A; CDC25B; CDC25C; CKS2; EME1; ESPL1; EXO1; FANCA; FBXO5; H2AFX; KIF18A; MND1; MNS1; MYBL1; NCAPD2; NCAPH; NEK2; NUF2; PKMYT1; PLK1; PTTG1; RAD51C; RAD51; RAD54B; RAD54L; SGO1; SGO2; SMC4; SYCE2; NDC1; TOP2A; TRIP13; TTK |
| GO:0006310 | DNA recombination | 9.71 | 0.00e+00 | 0.00e+00 | 33/100 | ATAD5; BLM; BRCA1; BRCA2; BRIP1; PARPBP; RMI2; AUNIP; CDC45; CDC7; CHEK1; EME1; EXO1; FANCB; FEN1; GINS2; GINS4; H2AFX; HMGB2; KPNA2; MND1; TONSL; PIF1; POLQ; RAD51AP1; RAD51C; RAD51; RAD54B; RAD54L; RECQL4; ALYREF; TOP2A; TRIP13 |
| GO:1904029 | regulation of cyclin-dependent protein kinase activity | 9.63 | 9.35e-14 | 2.93e-12 | 18/55 | BLM; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CDC25A; CDC25C; CDC6; CDKN2C; CDKN2D; CDKN3; CKS1B; CKS2; PKMYT1; PLK1; PSRC1 |
| GO:0033044 | regulation of chromosome organization | 9.62 | 0.00e+00 | 0.00e+00 | 34/104 | ATAD2; AURKB; BRCA1; BUB1B; BUB1; RMI2; CCNB1; CDC20; CDC45; CDC6; CDCA5; CDT1; CENPE; CENPF; CHEK1; DDX11; DLGAP5; DNMT3B; ESPL1; FBXO5; FEN1; MAD2L1; MCM2; MKI67; NDC80; NEK2; PHF19; PIF1; PLK1; PTTG1; TACC3; TOP2A; TRIP13; TTK |
| GO:0045930 | negative regulation of mitotic cell cycle | 9.11 | 0.00e+00 | 0.00e+00 | 35/113 | AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; TICRR; CCNB1; CDC20; CDC25C; CDC6; CDK1; CDT1; CENPF; CHEK1; CLSPN; E2F1; E2F7; E2F8; EME1; EZH2; FBXO5; GTSE1; KNTC1; MAD2L1; MTBP; NDC80; ORC1; PLK1; PTTG1; RBL1; TOP2A; TRIP13; TTK; ZWINT |
| GO:0071166 | ribonucleoprotein complex localization | 9.05 | 7.31e-04 | 9.60e-03 | 4/13 | DDX39A; NUP210; ALYREF; NDC1 |
| GO:0006289 | nucleotide-excision repair | 8.65 | 1.93e-04 | 2.66e-03 | 5/17 | BRCA2; BRIP1; NEIL3; POLD1; RFC4 |
| GO:0042770 | signal transduction in response to DNA damage | 8.49 | 8.96e-11 | 2.21e-09 | 15/52 | ATAD5; AURKA; BRCA1; BRCA2; CCNB1; CDC25C; CDK1; CHEK1; DTL; E2F1; E2F7; E2F8; FOXM1; GTSE1; PLK1 |
| GO:0071897 | DNA biosynthetic process | 8.34 | 6.58e-12 | 1.73e-10 | 17/60 | AURKB; BLM; CDKN2D; CENPF; CHTF18; DSCC1; DTL; PCLAF; LIN9; NEK2; PIF1; POLD1; POLE2; POLQ; RFC4; TK1; TYMS |
| GO:0071824 | protein-DNA complex subunit organization | 8.25 | 1.33e-15 | 4.72e-14 | 23/82 | ANP32E; ASF1B; KNL1; CDC45; CDT1; CENPA; CENPE; CENPF; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; GMNN; H2AFX; HELLS; HJURP; HMGB2; MCM2; CENPU; OIP5; RAD51 |
| GO:0006338 | chromatin remodeling | 8.24 | 6.02e-10 | 1.36e-08 | 14/50 | ANP32E; KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHEK1; HELLS; HJURP; HMGB2; CENPU; OIP5 |
| GO:0045787 | positive regulation of cell cycle | 8.16 | 0.00e+00 | 0.00e+00 | 53/191 | ATAD5; AURKA; AURKB; BRCA1; BRCA2; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDT1; CHEK1; CIT; CKS1B; CKS2; DDX11; DLGAP5; DTL; E2F1; E2F7; E2F8; ECT2; ESPL1; EZH2; FAM83D; FBXO5; FEN1; GPSM2; GTSE1; KIF14; KIF23; MAD2L1; MTBP; NDC80; NUSAP1; PLK4; POC1A; PSRC1; RACGAP1; RAD51C; RECQL4; SPAG5; TP73; UBE2C |
| GO:0044380 | protein localization to cytoskeleton | 8.02 | 6.85e-05 | 9.96e-04 | 6/22 | ANLN; AURKA; FAM83D; SPAG5; STIL; TTK |
| GO:0051052 | regulation of DNA metabolic process | 7.61 | 0.00e+00 | 0.00e+00 | 38/147 | ATAD5; AURKB; BLM; BRCA1; BRCA2; PARPBP; TICRR; RMI2; AUNIP; CCNA2; CDC6; CDC7; CDK1; CDT1; CHEK1; CHTF18; DBF4; DDX11; DEK; DNA2; DSCC1; E2F7; E2F8; ESCO2; FANCB; FBXO5; FOXM1; GMNN; H2AFX; KPNA2; NEK2; PIF1; POLQ; RAD51AP1; RAD51; RECQL4; RFC4; ALYREF |
| GO:0006333 | chromatin assembly or disassembly | 7.47 | 1.73e-10 | 4.14e-09 | 16/63 | ASF1B; KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; H2AFX; HELLS; HJURP; HMGB2; MCM2; CENPU; OIP5 |
| GO:2001020 | regulation of response to DNA damage stimulus | 7.17 | 7.06e-12 | 1.80e-10 | 19/78 | ATAD5; BLM; BRCA1; DDIAS; PARPBP; RMI2; AUNIP; CDKN2D; CHEK1; DDX11; DEK; FANCB; FBXO5; FOXM1; H2AFX; POLQ; RAD51AP1; RAD51; WDR76 |
| GO:0031109 | microtubule polymerization or depolymerization | 6.30 | 2.70e-07 | 5.23e-06 | 12/56 | CKAP2; FBXO5; KIF14; KIF18A; KIF18B; KIF24; KIF2C; PSRC1; SKA1; SKA2; SKA3; STMN1 |
| GO:0006997 | nucleus organization | 6.24 | 9.65e-05 | 1.38e-03 | 7/33 | CCNB1; CCNB2; CDK1; CEP55; HMGB2; PLK1; NDC1 |
| GO:0032886 | regulation of microtubule-based process | 6.11 | 4.72e-12 | 1.28e-10 | 22/106 | AURKA; BRCA1; CCNF; CHEK1; CKAP2; CNIH2; GPSM2; KIF11; KIF18A; NEK2; PLK1; PLK4; POC1A; PSRC1; SKA1; SKA2; SKA3; SPAG5; STIL; STMN1; TACC3; TPX2 |
| GO:0072331 | signal transduction by p53 class mediator | 5.88 | 2.53e-09 | 5.43e-08 | 17/85 | ATAD5; AURKA; AURKB; BRCA1; BRCA2; CCNB1; CDC25C; CDK1; CHEK1; E2F1; E2F2; E2F7; E2F8; FOXM1; GTSE1; TP73; TPX2 |
| GO:0051302 | regulation of cell division | 5.82 | 3.41e-10 | 7.94e-09 | 19/96 | ASPM; AURKA; AURKB; BLM; BRCA2; CDC25B; CDC6; CIT; E2F7; E2F8; ECT2; KIF14; KIF18B; KIF20A; KIF23; PLK1; PRC1; RACGAP1; RBL1 |
| GO:0010639 | negative regulation of organelle organization | 5.05 | 7.15e-13 | 2.01e-11 | 28/163 | ATAD2; AURKB; BRCA1; BUB1B; BUB1; CCNB1; CCNF; CDC20; CDT1; CENPF; CHEK1; CKAP2; DNMT3B; ESPL1; FBXO5; MAD2L1; MCM2; MTBP; NDC80; NEK2; PIF1; PLK1; PTTG1; STMN1; TMSB15A; TMSB15B; TOP2A; TRIP13; TTK |
| GO:0007050 | cell cycle arrest | 4.86 | 2.09e-08 | 4.36e-07 | 18/109 | AURKA; BARD1; BRCA1; DDIAS; CCNB1; CDC25C; CDK1; CDKN2C; CDKN2D; CDKN3; E2F1; E2F7; E2F8; FOXM1; GTSE1; MTBP; PRR11; TP73 |
| GO:0007292 | female gamete generation | 4.84 | 2.11e-06 | 3.66e-05 | 13/79 | ASPM; AURKA; BRCA2; CCNB1; CDC25B; FBXO5; NCAPH; PLK1; RAD51C; TOP2A; TRIP13; TTK; YBX2 |
| GO:0040029 | regulation of gene expression, epigenetic | 4.75 | 3.89e-05 | 5.98e-04 | 10/62 | ATAD2; BRCA1; CDC45; CHEK1; DEK; DNMT3B; EZH2; HELLS; LHX2; PHF19 |
| GO:1903320 | regulation of protein modification by small protein conjugation or removal | 4.20 | 1.13e-04 | 1.59e-03 | 10/70 | BRCA1; CDC20; FANCI; FBXO5; MAD2L1; MTBP; PLK1; SKP2; UBE2C; UBE2S |
| GO:0010498 | proteasomal protein catabolic process | 4.02 | 1.83e-06 | 3.24e-05 | 16/117 | AURKA; AURKB; BUB1B; CCNB1; CCNF; CDC20; CDK1; FBXO5; KIF14; MAD2L1; PBK; PLK1; PTTG1; SKP2; UBE2C; UBE2S |
| GO:0070646 | protein modification by small protein removal | 3.86 | 9.86e-04 | 1.27e-02 | 8/61 | BARD1; BRCA1; CCNA2; CDC20; CDC25A; CDK1; CLSPN; SKP2 |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 3.82 | 2.54e-04 | 3.44e-03 | 10/77 | ATAD5; DNA2; EME1; EXO1; FEN1; HMGB2; REXO5; POLD1; RFC4; RNASEH2A |
| GO:0007018 | microtubule-based movement | 3.68 | 6.07e-06 | 1.01e-04 | 16/128 | CENPE; CNIH2; DLGAP5; KIF11; KIF14; KIF15; KIF18A; KIF18B; KIF20A; KIF23; KIF24; KIF2C; KIF4A; KIFC1; RACGAP1; WDR34 |
| GO:0051169 | nuclear transport | 3.41 | 3.50e-04 | 4.68e-03 | 11/95 | BARD1; CCHCR1; CDK1; DDX39A; ECT2; GTSE1; KPNA2; NUP210; PSIP1; ALYREF; NDC1 |
| GO:0016569 | covalent chromatin modification | 3.32 | 4.12e-05 | 6.22e-04 | 15/133 | AURKA; AURKB; BRCA1; BRCA2; CCNA2; CCNB1; CDK1; CHEK1; DNMT3B; EZH2; HASPIN; HELLS; PHF19; SAP30; UHRF1 |
| GO:0051656 | establishment of organelle localization | 3.28 | 1.30e-07 | 2.58e-06 | 25/224 | SAPCD2; CCNB1; CDCA5; CDCA8; CDT1; CENPA; CENPE; CENPF; CEP55; CNIH2; CSPG5; DLGAP5; ESPL1; FAM83D; GPSM2; KIF14; KIF18A; KIF23; KIF2C; KIFC1; NDC80; NUF2; NUSAP1; PSRC1; SPAG5 |
| GO:0031503 | protein-containing complex localization | 3.27 | 2.91e-03 | 3.59e-02 | 8/72 | BIRC5; CNIH2; DDX39A; HASPIN; NUP210; ALYREF; NDC1; WDR34 |
| GO:0009314 | response to radiation | 3.10 | 6.18e-06 | 1.01e-04 | 20/190 | AURKB; BLM; BRCA1; BRCA2; TICRR; CDC25A; CDKN2D; CHEK1; DNMT3B; DTL; ECT2; FEN1; H2AFX; PCLAF; NMU; PBK; POLD1; RAD51AP1; RAD51; RAD54L |
| GO:0019882 | antigen processing and presentation | 3.01 | 2.88e-03 | 3.59e-02 | 9/88 | CENPE; CTSV; KIF11; KIF15; KIF18A; KIF23; KIF2C; KIF4A; RACGAP1 |
| GO:0022412 | cellular process involved in reproduction in multicellular organism | 3.01 | 1.64e-05 | 2.61e-04 | 19/186 | ASPM; AURKA; BRCA2; BRIP1; KNL1; CCNB1; CDC25B; FANCA; FBXO5; HMGB2; KIF18A; MYBL1; NCAPH; PLK1; RAD51C; TOP2A; TRIP13; TTK; YBX2 |
| GO:0051493 | regulation of cytoskeleton organization | 2.57 | 1.87e-05 | 2.93e-04 | 24/275 | AURKA; BRCA1; CCNF; CHEK1; CIT; CKAP2; ECT2; GPSM2; KIF11; KIF18A; NEK2; PLK1; PLK4; POC1A; PSRC1; SKA1; SKA2; SKA3; SPAG5; STIL; STMN1; TACC3; TMSB15A; TMSB15B; TPX2 |
| GO:0071900 | regulation of protein serine/threonine kinase activity | 2.38 | 6.56e-05 | 9.71e-04 | 24/297 | BLM; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CDC25A; CDC25C; CDC6; CDK1; CDKN2C; CDKN2D; CDKN3; CKS1B; CKS2; DBF4; EZH2; IQGAP3; PKMYT1; PLK1; PSRC1; TIAM1; TP73 |
| GO:0060249 | anatomical structure homeostasis | 2.33 | 9.95e-04 | 1.27e-02 | 17/215 | AURKB; BARD1; BLM; BRCA2; CCNE1; CCNE2; DNA2; EXO1; FEN1; NEK2; PIF1; POLD1; RAD51C; RAD51; RECQL4; RFC4; TYRO3 |
| GO:0051098 | regulation of binding | 2.29 | 3.19e-03 | 3.88e-02 | 14/180 | AURKA; AURKB; CDCA5; CDT1; DDX11; E2F1; GMNN; HJURP; HMGB2; LHX2; NEK2; PLK1; STMN1; TIAM1 |
Factor 14 : 59 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0044321 | response to leptin | 6.39 | 2.67e-03 | 3.29e-02 | 4/13 | EDN1; LEPR; LEP; NR4A3 |
| GO:0090077 | foam cell differentiation | 5.19 | 7.66e-04 | 1.27e-02 | 6/24 | ADIPOQ; APOB; CD36; LPL; PLA2G2A; PPARG |
| GO:0010644 | cell communication by electrical coupling | 4.72 | 3.35e-03 | 3.97e-02 | 5/22 | ATP1A2; CASQ2; CAV1; KCNA1; TRDN |
| GO:0061614 | pri-miRNA transcription by RNA polymerase II | 4.45 | 1.82e-03 | 2.56e-02 | 6/28 | ATOH8; FOS; KLF4; NGFR; PPARG; PRL |
| GO:0042180 | cellular ketone metabolic process | 4.23 | 5.87e-09 | 1.49e-06 | 22/108 | ABCD2; ACACB; ACADL; ADH4; ADIPOQ; AKR1C1; AKR1C2; AKR1C3; BHMT; BMP6; CAV1; CNR1; CYP19A1; DGAT2; DKK3; EGR1; FABP5; GPD1; MLXIPL; NR4A3; PDK4; PPARG |
| GO:0050886 | endocrine process | 4.09 | 1.20e-05 | 5.42e-04 | 13/66 | AVPR2; BMP6; CMA1; CORIN; CRHBP; CTSG; EDN1; FZD4; HCAR2; LEP; MME; RETN; SUCNR1 |
| GO:2001057 | reactive nitrogen species metabolic process | 4.00 | 1.52e-04 | 3.64e-03 | 10/52 | CAV1; CD36; EDN1; HBB; KLF2; KLF4; MMP8; MARC1; PTGIS; PTX3 |
| GO:0006638 | neutral lipid metabolic process | 3.94 | 4.13e-06 | 1.98e-04 | 15/79 | ACSL1; GPAT3; APOB; CAV1; DGAT2; FABP12; FABP4; FABP5; GPAM; LIPE; LPL; PCK1; PNPLA2; THRSP; AGMO |
| GO:0016999 | antibiotic metabolic process | 3.78 | 1.46e-05 | 6.27e-04 | 14/77 | ADH1A; ADH1B; ADH1C; ADH4; AKR1C1; AKR1C2; AKR1C3; ALDH2; GPX3; HBA1; HBB; HP; PCK1; TPO |
| GO:0005976 | polysaccharide metabolic process | 3.53 | 8.46e-04 | 1.35e-02 | 9/53 | CHIT1; GYG2; GYS2; HAS1; IGF1; PHKG1; PPP1R1A; PYGM; SORBS1 |
| GO:0008643 | carbohydrate transport | 3.46 | 7.65e-05 | 2.01e-03 | 13/78 | ADIPOQ; AQP7; AQP9; EDN1; FABP5; GPC3; IGF1; KLF15; LEP; NR4A3; SLC2A4; SORBS1; TRARG1 |
| GO:0032602 | chemokine production | 3.46 | 5.14e-04 | 9.29e-03 | 10/60 | ADIPOQ; ACKR1; EGR1; ELANE; FFAR3; IL33; IL6; KLF4; LBP; LPL |
| GO:0019216 | regulation of lipid metabolic process | 3.45 | 2.01e-11 | 1.64e-08 | 37/223 | ABCD2; ACACB; ACADL; ACSL1; ADIPOQ; AKR1C3; ANGPTL4; APOB; BMP6; CAV1; CCL19; CD36; CIDEA; CNR1; DGAT2; DKK3; EGR1; FABP5; FGF2; FGR; G0S2; GPAM; HCAR2; KLF4; LEP; LGALS12; ANGPTL8; MLXIPL; NR4A3; PDE3B; PDK4; PLIN2; PNPLA2; PPARG; SOCS3; SORBS1; THRSP |
| GO:0055088 | lipid homeostasis | 3.42 | 4.66e-05 | 1.52e-03 | 14/85 | AKR1C1; ANGPTL4; APOB; CAV1; CEBPA; DGAT2; FABP4; GPAM; GPIHBP1; ANGPTL8; LPL; MLXIPL; PNPLA2; PPARG |
| GO:0002526 | acute inflammatory response | 3.32 | 3.37e-06 | 1.90e-04 | 19/119 | C2CD4B; C5AR1; C6; C7; CD163; CFP; CNR1; CR1; DNASE1L3; ELANE; FFAR3; HPR; HP; IL6; LBP; PPARG; SAA1; SAA2; SAA4 |
| GO:1990845 | adaptive thermogenesis | 3.21 | 5.37e-05 | 1.63e-03 | 15/97 | ACADL; ACSL1; ADIPOQ; ADRB1; ADRB3; CAV1; CD36; CIDEA; ELOVL3; FABP4; FABP5; G0S2; LEPR; LEP; VEGFA |
| GO:0097305 | response to alcohol | 3.21 | 5.76e-07 | 4.69e-05 | 23/149 | ADIPOQ; AKR1C2; AKR1C3; CA3; CCL19; CDO1; CNR1; CRHBP; CSF3; CSN1S1; FOSB; FOS; GSN; KLF2; KLF4; LEP; PENK; PPARG; PRL; PTGER4; PTGFR; RBP4; SLC2A4 |
| GO:0016051 | carbohydrate biosynthetic process | 3.13 | 2.48e-05 | 9.19e-04 | 17/113 | ADIPOQ; ATF3; DGAT2; GPD1; GYG2; GYS2; HAS1; IGF1; LEPR; LEP; LHCGR; PCK1; PFKFB1; PHKG1; PTH1R; RBP4; SORBS1 |
| GO:0006109 | regulation of carbohydrate metabolic process | 3.10 | 2.40e-04 | 5.37e-03 | 13/87 | ACACB; ADIPOQ; DGAT2; GPD1; IGF1; LEPR; LEP; LHCGR; MLXIPL; PDK4; PFKFB1; PTH1R; SORBS1 |
| GO:0050918 | positive chemotaxis | 3.08 | 3.97e-03 | 4.37e-02 | 8/54 | ANGPT2; FGF2; VEGFD; S1PR1; SAA1; SAA2; SAA4; VEGFA |
| GO:0097006 | regulation of plasma lipoprotein particle levels | 3.08 | 3.97e-03 | 4.37e-02 | 8/54 | ADIPOQ; APOB; CD36; DGAT2; GPIHBP1; LPL; PLA2G2A; PLA2G7 |
| GO:0046677 | response to antibiotic | 2.97 | 4.98e-07 | 4.69e-05 | 26/182 | ADIPOQ; ALPL; AOC3; CA3; CDO1; CNR1; CRHBP; CRYAB; CSF3; DUSP1; EDN1; EGR1; GSN; HBA1; HBB; HP; HYAL1; IL6; KLF2; KLF4; LEP; NR4A3; PENK; PRL; RBP4; SLC2A4 |
| GO:0050727 | regulation of inflammatory response | 2.93 | 3.90e-09 | 1.49e-06 | 36/255 | ABCD2; ADIPOQ; C2CD4B; C5AR1; C6; C7; CCL14; CCL18; CCL24; CEBPA; CFP; CMA1; CNR1; CR1; CYP19A1; DNASE1L3; ELANE; FABP4; FFAR3; IL33; IL6; KLF4; LBP; LEP; LPL; MMP8; PDE2A; PLA2G2A; PLA2G7; PPARG; PTGER4; PTGIS; SAA1; SOCS3; SUCNR1; TYRO3 |
| GO:0062012 | regulation of small molecule metabolic process | 2.92 | 4.12e-07 | 4.69e-05 | 27/192 | ABCD2; ACACB; ACADL; ADIPOQ; AKR1C3; APOB; BHMT; BMP6; CAV1; CNR1; DGAT2; DKK3; EGR1; FABP5; GPAM; GPD1; IGF1; LEPR; LEP; LHCGR; MLXIPL; NR4A3; PDK4; PFKFB1; PPARG; PTH1R; SORBS1 |
| GO:0010876 | lipid localization | 2.92 | 7.32e-09 | 1.49e-06 | 35/249 | ABCA10; ABCA6; ABCA8; ABCA9; ABCD2; ACACB; ACSL1; ACVR1C; ADIPOQ; AKR1C1; APOB; AQP9; BMP6; SPX; CAV1; CD36; CES1; CIDEA; CYP19A1; DGAT2; EDN1; FABP4; FZD4; IL6; LBP; LEP; LPL; NMB; PLA2G2A; PLIN2; PNPLA2; PPARG; RBP4; RETN; THRSP |
| GO:0046683 | response to organophosphorus | 2.90 | 4.69e-04 | 8.77e-03 | 13/93 | ADIPOQ; AKR1C1; AQP9; CDO1; CRHBP; DUSP1; FOSB; FOS; GPD1; PCK1; PDE2A; PENK; PFKFB1 |
| GO:0014074 | response to purine-containing compound | 2.88 | 3.12e-04 | 6.36e-03 | 14/101 | ADIPOQ; AQP9; CASQ2; CDO1; CRHBP; DUSP1; FOSB; FOS; GPD1; PCK1; PDE2A; PENK; PFKFB1; PPARG |
| GO:0070555 | response to interleukin-1 | 2.81 | 6.40e-04 | 1.11e-02 | 13/96 | CCL13; CCL14; CCL18; CCL19; CCL23; CCL24; EDN1; EGR1; HYAL1; IL6; KLF2; PCK1; PTGIS |
| GO:0016042 | lipid catabolic process | 2.78 | 1.74e-06 | 1.18e-04 | 26/194 | ABCD2; ACACB; ACADL; ADIPOQ; AKR1C3; APOB; CES1; CIDEA; CNR1; CYP19A1; ENPP2; FABP12; FABP4; FABP5; HCAR2; LEP; LGALS12; LIPE; LPL; PCK1; PDE3B; PLA2G2A; PLA2G7; PLCXD3; PLIN1; PNPLA2 |
| GO:0001101 | response to acid chemical | 2.78 | 2.73e-07 | 3.71e-05 | 30/224 | ACSL1; ADIPOQ; AKR1C1; AKR1C2; AKR1C3; ALDH1A2; APOB; BMP6; CCL19; CD36; CDO1; DGAT2; DUSP1; EDN1; EGR1; FFAR3; FOLR2; FZD4; GSN; KLF2; KLF4; LEP; OTC; PCK1; PDK4; PPARG; PTGER4; PTGFR; RBP4; VEGFA |
| GO:1901342 | regulation of vasculature development | 2.75 | 5.55e-07 | 4.69e-05 | 29/219 | ANGPT2; ANGPTL4; BMPER; C5AR1; C6; CCL24; CMA1; EGR1; ENPP2; FGF2; VEGFD; HSPB6; HYAL1; IL6; KLF2; KLF4; LEP; MEOX2; NGFR; NPR1; PDE3B; PPARG; PRL; PROK1; PTGIS; THBS4; TMEM100; TNMD; VEGFA |
| GO:0033500 | carbohydrate homeostasis | 2.74 | 5.50e-05 | 1.63e-03 | 19/144 | ADIPOQ; CEBPA; CMA1; CNR1; FABP5; GPR21; KCNB1; KLF15; LEPR; LEP; MLXIPL; NGFR; NMB; PCK1; PDK4; PPARG; RABGAP1; RBP4; SLC2A4; SUCNR1 |
| GO:0044262 | cellular carbohydrate metabolic process | 2.69 | 1.10e-04 | 2.72e-03 | 18/139 | ACACB; ADIPOQ; DGAT2; GYG2; GYS2; HAS1; IGF1; LEPR; LEP; LHCGR; PCK1; PDK4; PFKFB1; PHKG1; PPP1R1A; PTH1R; PYGM; SORBS1 |
| GO:0045444 | fat cell differentiation | 2.69 | 1.10e-04 | 2.72e-03 | 18/139 | ADIPOQ; ADRB1; MEDAG; CEBPA; EGR2; FABP4; GDF10; IL6; KLF4; LEP; LGALS12; ANGPTL8; NR4A1; NR4A3; PPARG; RETN; SLC2A4; ZFP36 |
| GO:0042445 | hormone metabolic process | 2.69 | 1.95e-05 | 7.56e-04 | 22/170 | ADH4; AKR1C1; AKR1C2; AKR1C3; ALDH1A2; BMP6; CMA1; CORIN; CRHBP; CTSG; CYP19A1; DGAT2; DKK3; EGR1; FFAR3; GHR; HSD11B1; LEP; MME; RBP4; RDH5; TPO |
| GO:0006091 | generation of precursor metabolites and energy | 2.67 | 3.74e-06 | 1.90e-04 | 26/202 | ADH1A; ADH1B; ADH1C; ADH4; ADIPOQ; ADRB3; ALDH2; AOC2; AOX1; AQP7; CEBPA; CYP19A1; DMGDH; GPD1; GYG2; GYS2; IGF1; LEPR; LEP; MLXIPL; NR4A3; PFKFB1; PHKG1; PPP1R1A; PYGM; SORBS1 |
| GO:1901652 | response to peptide | 2.67 | 7.29e-08 | 1.19e-05 | 35/272 | ACVR1C; ADIPOQ; CAV1; CD36; CDO1; CRHBP; EDN1; EGR1; EGR2; FFAR3; GHR; GLP2R; GPR21; IGF1; KLF15; KLF2; KLF4; LEP; LHCGR; NGFR; NR4A1; NR4A3; OTC; PCK1; PDE3B; PDK4; PFKFB1; PPARG; PRL; RABGAP1; RETN; SLC2A4; SOCS3; SORBS1; TIMP4; TRARG1 |
| GO:0032103 | positive regulation of response to external stimulus | 2.66 | 2.71e-06 | 1.70e-04 | 27/211 | BMP6; C2CD4B; C5AR1; CCL14; CCL18; CCL19; CCL24; CEBPA; CNR1; EDN1; FABP4; FFAR3; FGF2; VEGFD; IL33; IL6; LBP; LPL; MMP8; PDE2A; PLA2G2A; PLA2G7; PTGER4; S1PR1; SUCNR1; THBS4; VEGFA |
| GO:0002576 | platelet degranulation | 2.66 | 2.62e-03 | 3.29e-02 | 11/86 | CD36; CFD; CLEC3B; VEGFD; IGF1; LEFTY2; PECAM1; SELP; TF; VEGFA; VWF |
| GO:0034612 | response to tumor necrosis factor | 2.63 | 6.40e-05 | 1.80e-03 | 20/158 | ADIPOQ; APOB; CCL13; CCL14; CCL18; CCL19; CCL23; CCL24; CEBPA; CRHBP; EDN1; FABP4; GPD1; HYAL1; KLF2; PCK1; SLC2A4; DCSTAMP; TNFRSF8; ZFP36 |
| GO:0072593 | reactive oxygen species metabolic process | 2.48 | 2.11e-04 | 4.90e-03 | 19/159 | ABCD2; AKR1C3; CAV1; CD36; CRYAB; EDN1; GPX3; HBA1; HBB; HP; KLF2; KLF4; LEP; MMP8; PDK4; PTGIS; PTX3; SOD3; TPO |
| GO:1901654 | response to ketone | 2.47 | 1.00e-03 | 1.54e-02 | 15/126 | AKR1C2; AKR1C3; CAV1; CCL19; CSN1S1; DUSP1; EDN1; FOSB; FOS; KLF2; KLF4; PCK1; PPARG; PTGER4; PTGFR |
| GO:0050920 | regulation of chemotaxis | 2.40 | 4.74e-04 | 8.77e-03 | 18/156 | ANGPT2; C5AR1; CCL19; CYP19A1; DUSP1; EDN1; ELANE; FGF2; VEGFD; IL6; LBP; PLA2G7; PLXNA4; S1PR1; SEMA3G; SUCNR1; THBS4; VEGFA |
| GO:0006631 | fatty acid metabolic process | 2.38 | 3.11e-05 | 1.10e-03 | 26/227 | ABCD2; ACACB; ACADL; ACSL1; ACSM5; ADIPOQ; AKR1C2; AKR1C3; CAV1; CD36; CES1; CNR1; DGAT2; EDN1; ELOVL3; FABP5; GPAM; LEP; LIPE; LPL; MLXIPL; NR4A3; PCK1; PDK4; PPARG; PTGIS |
| GO:0009636 | response to toxic substance | 2.37 | 1.41e-06 | 1.04e-04 | 35/307 | ADIPOQ; ATP1A2; CA3; CD36; CDO1; CES1; CNR1; CRHBP; CRYAB; CSF3; DUSP1; EDN1; EGR1; FOS; GLYAT; GPX3; GSN; HBA1; HBB; HP; IL6; INMT; KLF2; KLF4; LEP; MARC1; MT1M; NR4A3; PDE1B; PENK; PRL; RBP4; SLC2A4; SOD3; TPO |
| GO:0060326 | cell chemotaxis | 2.32 | 6.81e-05 | 1.85e-03 | 25/224 | C5AR1; CCL13; CCL14; CCL18; CCL19; CCL23; CCL24; CH25H; CXCL2; CYP19A1; DUSP1; EDN1; FGF2; VEGFD; FOLR2; IL6; LBP; NR4A1; PLA2G7; S1PR1; SAA1; SAA2; SAA4; THBS4; VEGFA |
| GO:0048511 | rhythmic process | 2.31 | 7.47e-04 | 1.27e-02 | 18/162 | ADAMTS1; ADIPOQ; EGR1; EGR2; FZD4; HAS1; HS3ST2; LEP; LHCGR; NGFR; NLGN1; PPARG; PRL; PROK1; PTX3; RETN; TIMP4; TYRO3 |
| GO:0051235 | maintenance of location | 2.27 | 3.43e-04 | 6.81e-03 | 21/192 | ACACB; ACVR1C; ANK2; APOB; ATP1A2; SYNE3; CASQ2; CAV1; CCL19; CD36; CIDEA; DGAT2; FGF2; GSN; IL6; LEP; LPL; PLIN2; PNPLA2; PPARG; TRDN |
| GO:0016053 | organic acid biosynthetic process | 2.26 | 3.86e-05 | 1.31e-03 | 28/257 | ABCD2; ACACB; ACADL; ACSM5; ADIPOQ; AKR1C3; ALDH1A2; ASPA; BHMT2; BHMT; CDO1; CH25H; EDN1; ELOVL3; FABP5; FOLH1; GPD1; HAS1; HYAL1; IGF1; LPL; MLXIPL; NAALAD2; NAT8L; OTC; PDK4; PFKFB1; PTGIS |
| GO:1901615 | organic hydroxy compound metabolic process | 2.24 | 3.73e-06 | 1.90e-04 | 36/334 | ACACB; ACADL; ADH1A; ADH1B; ADH1C; ADH4; AKR1C1; AKR1C2; AKR1C3; ALDH1A2; ALDH2; AOC2; APOB; BMP6; CEBPA; CES1; CH25H; CYP19A1; DGAT2; DKK3; FGF2; GPAM; GPD1; IGF1; LEPR; LEP; LHCGR; LIPE; MAOA; NPR1; PCK1; PDE1B; PTH1R; RBP4; RDH5; TPO |
| GO:0006979 | response to oxidative stress | 2.18 | 2.52e-04 | 5.40e-03 | 24/229 | ADIPOQ; AKR1C3; ANGPTL7; CA3; CCL19; CD36; CRYAB; DUSP1; EDN1; FOS; GPX3; HBA1; HBB; HP; HYAL1; IL6; KLF2; KLF4; MCTP1; NR4A3; PENK; SGK2; SOD3; TPO |
| GO:0006909 | phagocytosis | 2.16 | 3.80e-03 | 4.29e-02 | 15/144 | ADIPOQ; CD36; ELANE; FCN1; FGR; GSN; LBP; LEPR; LEP; PEAR1; PECAM1; PPARG; PTX3; SRPX; TYRO3 |
| GO:0048545 | response to steroid hormone | 2.15 | 3.07e-04 | 6.36e-03 | 24/232 | ADIPOQ; AKR1C3; ALPL; ATP1A2; BMP6; CAV1; CDO1; CSN1S1; DUSP1; EDN1; FAM107A; FOSB; FOS; GPAM; IL6; LBH; NR4A1; NR4A3; PCK1; PFKFB1; PPARG; RXRG; S100B; ZFP36 |
| GO:0002237 | response to molecule of bacterial origin | 2.14 | 4.27e-04 | 8.27e-03 | 23/223 | ALPL; APOB; BMP6; C5AR1; CD36; CNR1; CSF3; CTSG; CXCL2; EDN1; ELANE; FOS; IL6; LBP; MRC1; PCK1; PENK; PRL; PTGER4; PTGFR; SELP; TIMP4; ZFP36 |
| GO:0001525 | angiogenesis | 2.11 | 1.95e-05 | 7.56e-04 | 35/345 | ANGPT2; ANGPTL4; BMPER; C5AR1; C6; CAV1; CCL24; CMA1; EDN1; ENPP2; ESM1; FGF2; VEGFD; HSPB6; HYAL1; IL6; KLF2; KLF4; LEPR; LEP; MEOX2; NGFR; NPR1; NR4A1; PDE3B; PECAM1; PPARG; PRL; PROK1; PTGIS; S1PR1; THBS4; TMEM100; TNMD; VEGFA |
| GO:0009410 | response to xenobiotic stimulus | 2.10 | 1.30e-03 | 1.96e-02 | 20/198 | ACSL1; AKR1C1; AOC2; AOC3; CASQ2; CDO1; CES1; CNR1; CRHBP; EDN1; EGR1; FMO2; FOSB; GHR; GLYAT; PCK1; PDE1B; PDE2A; PENK; PPARG |
| GO:0050900 | leukocyte migration | 2.08 | 5.61e-05 | 1.63e-03 | 32/319 | ANGPT2; APOB; C5AR1; CAV1; CCL13; CCL14; CCL18; CCL19; CCL23; CCL24; CH25H; CXCL2; CYP19A1; DUSP1; EDN1; ELANE; VEGFD; FOLR2; GYPC; IL33; IL6; LBP; LEP; PECAM1; PLA2G7; PTGER4; S1PR1; SAA1; SELP; SLC7A10; THBS4; VEGFA |
| GO:0006732 | coenzyme metabolic process | 2.08 | 1.91e-03 | 2.64e-02 | 19/190 | ACACB; ACSL1; ACSM5; ALDH1L1; BHMT2; BHMT; DGAT2; ELOVL3; FMO2; FOLH1; FOLR2; GLYAT; GPAM; GPD1; IGF1; MLXIPL; PDK4; PFKFB1; PTGIS |
| GO:0015850 | organic hydroxy compound transport | 2.08 | 2.50e-03 | 3.29e-02 | 18/180 | ADIPOQ; AKR1C1; APOB; AQP7; AQP9; BMP6; CAV1; CD36; CES1; CNR1; CYP19A1; FFAR3; KCNB1; LEP; PPARG; RBP4; SLC16A7; SLC22A3 |
Factor 15 : 6 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0001539 | cilium or flagellum-dependent cell motility | 18.90 | 1.88e-12 | 3.82e-10 | 11/22 | DRC1; CCDC65; DNAH2; DNAH3; DNAH6; DNAH7; SPEF1; TEKT1; TEKT2; TEKT3; CFAP57 |
| GO:0001578 | microtubule bundle formation | 15.60 | 0.00e+00 | 0.00e+00 | 24/58 | ARMC4; CFAP43; DRC1; CFAP206; CFAP157; CCDC103; CCDC114; CCDC65; DNAH7; DNAI1; DNAI2; DNAJB13; HYDIN; IQCG; LRGUK; LRRC49; DNAAF1; LRRC6; RSPH1; RSPH4A; TEKT2; TRIM46; TTLL6; ZMYND10 |
| GO:0021591 | ventricular system development | 12.60 | 2.01e-04 | 2.72e-02 | 4/12 | ARMC4; AK8; HYDIN; NME5 |
| GO:0007018 | microtubule-based movement | 10.60 | 0.00e+00 | 0.00e+00 | 36/128 | ARMC4; DRC1; MAATS1; CFAP206; CCDC103; CCDC114; CFAP53; CCDC65; DNAH12; DNAH2; DNAH3; DNAH6; DNAH7; DNAH9; DNAI1; DNAI2; HYDIN; KIF19; KIF24; KIF6; DNAAF1; LRRC6; MAK; NME5; CFAP221; ROPN1L; RSPH4A; SPA17; TEKT1; TEKT2; TEKT3; TRIM46; TTLL6; WDR63; WDR66; WDR78 |
| GO:0044782 | cilium organization | 9.85 | 0.00e+00 | 0.00e+00 | 41/157 | ARMC4; CFAP43; CFAP126; DRC1; CATIP; CFAP206; CFAP157; CCDC103; CCDC114; CFAP53; CCDC13; CCDC65; CCNO; DNAH7; DNAI1; DNAI2; DNAJB13; FAM92A; FOXJ1; HYDIN; IQCG; IQUB; KIF19; KIF24; LRGUK; LRRC49; DNAAF1; LRRC6; MAK; MAPK15; BBIP1; NME5; NPHP1; CFAP221; RSPH1; RSPH4A; TEKT1; TEKT2; TEKT3; WDR90; ZMYND10 |
| GO:0097722 | sperm motility | 7.68 | 1.21e-07 | 1.98e-05 | 11/54 | CFAP43; CFAP69; CFAP157; DNAI1; IQCG; LRRC6; ROPN1L; TCTE1; TEKT1; TEKT2; TEKT3 |
Factor 16 : 94 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0050690 | regulation of defense response to virus by virus | 8.44 | 7.01e-07 | 1.02e-05 | 8/13 | B2M; CD247; CD28; CD4; CD8B; DOCK2; FYN; LCK |
| GO:0006968 | cellular defense response | 7.66 | 0.00e+00 | 0.00e+00 | 24/43 | C5AR1; CCR2; CCR5; CCR6; CD19; CLEC5A; CXCL9; FCMR; GNLY; ITK; KIR2DL4; KIR3DL2; KLRC2; KLRC3; KLRG1; LSP1; NCF1; NCR1; PRF1; PTK2B; SH2D1A; SPN; TNFRSF4; TRAT1 |
| GO:0002507 | tolerance induction | 7.22 | 1.97e-07 | 3.21e-06 | 10/19 | CCR4; CD274; CD3E; FOXP3; HLA-E; ICOS; IDO1; IL2RA; PDCD1; TNFAIP3 |
| GO:0001773 | myeloid dendritic cell activation | 6.86 | 3.69e-07 | 5.66e-06 | 10/20 | BATF3; CAMK4; CD2; DOCK2; FLT3LG; IRF4; KLRK1; SLAMF1; TSPAN32; UBD |
| GO:0002250 | adaptive immune response | 6.60 | 0.00e+00 | 0.00e+00 | 128/266 | B2M; BTK; BTLA; BTN3A1; BTN3A2; BTN3A3; C1QA; C1S; C3; C7; CAMK4; CCL19; CCR2; CCR6; CD1A; CD1B; CD1C; CD1D; CD1E; CD226; CD244; CD247; CD274; CD27; CD28; CD3D; CD3E; CD3G; CD40LG; CD40; CD48; CD4; CD6; CD74; CD79A; CD79B; CD7; CD8A; CD8B; CLEC10A; CLEC4A; CLEC4C; CLEC6A; CR1; CR2; CRTAM; CTLA4; CTSS; CXCL13; EBI3; EOMES; FCAMR; FCER2; FCRL4; FOXP3; FYN; GAPT; GPR183; GZMM; HLA-DQB1; HLA-E; HLA-F; IFNG; JCHAIN; IGLL1; IL12B; IL18R1; IL18RAP; IL18; IL33; IL7R; INPP5D; IRF1; IRF4; ITK; JAK3; KLHL6; KLRK1; LAMP3; LAT; LAX1; LILRB1; LIME1; LTA; LY9; MCOLN2; MYO1G; PAG1; PIK3CD; PIK3CG; POU2F2; PRDM1; PRF1; PRKCB; PRKCQ; PTK2B; PTPRC; RFTN1; SAMSN1; SASH3; SH2D1A; SH2D1B; SIT1; SLA2; SLAMF1; SLAMF6; SLAMF7; SLC11A1; SPN; SYK; TAP1; TAP2; TBX21; THEMIS; TLR8; TNFAIP3; TNFRSF13B; TNFRSF13C; TNFRSF17; TNFRSF1B; TNFSF13B; TNFSF18; TRAT1; TXK; WAS; XCL1; ZAP70; ZNF683 |
| GO:0032620 | interleukin-17 production | 6.56 | 1.68e-07 | 2.79e-06 | 11/23 | VSIR; FOXP3; IFNG; IL12B; IL18; IL21; LY9; NCKAP1L; PRKCQ; RFTN1; SLAMF6 |
| GO:0098760 | response to interleukin-7 | 6.33 | 1.61e-04 | 1.79e-03 | 6/13 | BTK; IL2RG; IL7R; IL7; JAK3; SOCS1 |
| GO:0042113 | B cell activation | 6.23 | 0.00e+00 | 0.00e+00 | 59/130 | BANK1; BTK; CARD11; CCR6; CD22; CD27; CD28; CD38; CD40LG; CD40; CD74; CD79A; CD79B; CR2; CTLA4; CXCR5; DOCK11; FCRL1; FCRL3; FLT3; FOXP3; GAPT; GPR183; IGLL1; IKZF3; IL21; IL7R; IL7; INPP5D; ITGA4; ITM2A; JAK3; LAX1; LYL1; MFNG; MZB1; MS4A1; NCKAP1L; PIK3CD; PLCG2; PLCL2; POU2F2; PRKCB; PTK2B; PTPRC; SAMSN1; SASH3; SLA2; SLAMF8; SYK; TBC1D10C; TBX21; TNFAIP3; TNFRSF13B; TNFRSF13C; TNFRSF4; TNFSF13B; VCAM1; ZAP70 |
| GO:0002449 | lymphocyte mediated immunity | 6.08 | 0.00e+00 | 0.00e+00 | 70/158 | B2M; BTK; BTN3A2; BTN3A3; C1QA; C1S; C3; C7; CCR6; CD1A; CD1B; CD1C; CD1D; CD1E; CD226; CD27; CD28; CD40LG; CD40; CD74; CD8A; CD96; CORO1A; CR1; CR2; CRTAM; FCER2; FOXP3; GAPT; GZMB; GZMM; HLA-DQB1; HLA-E; HLA-F; IGLL1; IL12B; IL18R1; IL18RAP; IL18; IL21; IL7R; INPP5D; KIR2DL4; KIR3DL1; KLRC2; KLRK1; LAG3; LILRB1; LTA; MYO1G; NCR1; NCR3; POU2F2; PRF1; PTPRC; RFTN1; SASH3; SERPINB9; SH2D1A; SH2D1B; SLA2; SLAMF6; SLAMF7; SLC11A1; TBX21; TLR8; TNFRSF1B; VAV1; WAS; XCL1 |
| GO:0032615 | interleukin-12 production | 6.05 | 3.63e-09 | 7.21e-08 | 15/34 | CCL19; CCR7; CD40LG; CD40; IDO1; IFNG; IL12B; IRF1; IRF8; JAK3; LILRB1; LTB; SLAMF1; TIGIT; TLR8 |
| GO:0030101 | natural killer cell activation | 5.97 | 1.18e-11 | 2.83e-10 | 20/46 | CD244; CD2; CORO1A; FLT3LG; HLA-E; IL12B; IL18R1; IL18; IL21R; ITGB2; KLRK1; NCR1; NCR3; PIK3CD; PRDM1; PTPN22; PTPRC; SLAMF7; TOX; ZNF683 |
| GO:0045730 | respiratory burst | 5.97 | 1.87e-06 | 2.58e-05 | 10/23 | CD52; CYBB; JCHAIN; NCF1; NCF4; PIK3CD; PIK3CG; RAC2; SLAMF8; SLC11A1 |
| GO:0032609 | interferon-gamma production | 5.93 | 0.00e+00 | 0.00e+00 | 38/88 | BTN3A1; BTN3A2; VSIR; CCR2; CCR7; CD226; CD244; CD274; CD2; CD3E; CD96; EBI3; EOMES; FOXP3; HLA-DPA1; HLA-DPB1; IL12B; IL12RB2; IL18R1; IL18RAP; IL18; IL21; IL33; IRF8; ITK; KLRK1; LILRB1; LTA; PDCD1LG2; PTPN22; SASH3; SLAMF6; SLC11A1; SPN; TLR8; TNFRSF13C; TXK; XCL1 |
| GO:0031579 | membrane raft organization | 5.72 | 1.04e-03 | 1.01e-02 | 5/12 | CD2; DOCK2; MAL; PTPRC; RFTN1 |
| GO:0070661 | leukocyte proliferation | 5.65 | 0.00e+00 | 0.00e+00 | 82/199 | AIF1; BTK; BTN3A1; VSIR; CARD11; CCDC88B; CCL19; CCL5; CCR2; CD1D; CD22; CD274; CD28; CD38; CD3E; CD40LG; CD40; CD4; CD6; CD74; CD79A; CLECL1; CORO1A; CR2; CRTAM; CTLA4; DOCK2; DOCK8; EBI3; FCRL3; FLT3LG; FLT3; FOXP3; FYN; GAPT; GPR183; HLA-DMB; HLA-DPA1; HLA-DPB1; HLA-E; IDO1; IKZF3; IL12B; IL18; IL21; IL2RA; IL33; IL7R; IL7; INPP5D; IRF1; JAK3; LILRB1; LST1; MZB1; MS4A1; NCKAP1L; PDCD1LG2; PIK3CG; PLA2G2D; PLCL2; PRKCQ; PSMB10; PTPN22; PTPRC; RAC2; RASAL3; SASH3; SLC11A1; SPN; SYK; TNFAIP3; TNFRSF13B; TNFRSF13C; TNFRSF14; TNFRSF1B; TNFRSF4; TNFSF13B; TNFSF18; VCAM1; XCL1; ZAP70 |
| GO:0001562 | response to protozoan | 5.65 | 1.07e-04 | 1.24e-03 | 7/17 | CCDC88B; CD40; IL12B; IRF4; IRF8; SLC11A1; TSPAN32 |
| GO:0042110 | T cell activation | 5.58 | 0.00e+00 | 0.00e+00 | 126/310 | AIF1; JAML; APBB1IP; B2M; BCL11B; BTLA; BTN3A1; VSIR; CAMK4; CARD11; CCDC88B; CCL19; CCL21; CCL5; CCR2; CCR6; CCR7; CD1C; CD1D; CD274; CD27; CD28; CD2; CD3D; CD3E; CD3G; CD40LG; CD4; CD5; CD6; CD74; CD7; CD8A; CD8B; CLEC4A; CLECL1; CORO1A; CR1; CRTAM; CTLA4; DOCK2; DOCK8; EBI3; EOMES; RIPOR2; FOXP3; FUT7; FYN; GPR183; GPR18; GRAP2; HLA-DMB; HLA-DOA; HLA-DPA1; HLA-DPB1; HLA-E; ICOS; IDO1; IFNG; IL12B; IL18R1; IL18; IL21; IL2RA; IL7R; IL7; IRF1; IRF4; ITGAL; ITK; JAK3; TESPA1; KLRK1; LAG3; LAT; LAX1; LCK; LCP1; LILRB1; LY9; NCKAP1L; PAG1; PDCD1LG2; PDCD1; PIK3CD; PIK3CG; PLA2G2D; PRDM1; PRKCQ; PSMB10; PTGER4; PTPN22; PTPRC; RAC2; RASAL3; RHOH; RUNX3; SASH3; SIRPG; SIT1; SLA2; SLAMF6; SLC11A1; SOCS1; SPN; SYK; TBX21; TCF7; THEMIS; TIGIT; TNFAIP8L2; TNFRSF13C; TNFRSF14; TNFRSF1B; TNFRSF4; TNFSF13B; TNFSF18; TNFSF8; TREML2; VAV1; VCAM1; WAS; WNT1; XCL1; ZAP70; ZNF683 |
| GO:0032623 | interleukin-2 production | 5.37 | 1.05e-09 | 2.13e-08 | 18/46 | CARD11; CCR2; CD247; CD28; CD3E; CD4; FOXP3; GBP1; IL18; IRF4; LAG3; PRKCQ; PTPRC; SASH3; SLC11A1; TBX21; TNFAIP3; XCL1 |
| GO:0032613 | interleukin-10 production | 5.34 | 8.55e-08 | 1.48e-06 | 14/36 | VSIR; CD274; CD28; CD40LG; FOXP3; IDO1; IL12B; IRF4; JAK3; LILRB1; PDCD1LG2; SASH3; TIGIT; XCL1 |
| GO:0002285 | lymphocyte activation involved in immune response | 5.32 | 0.00e+00 | 0.00e+00 | 38/98 | APBB1IP; CCL19; CCR6; CD1C; CD244; CD28; CD40LG; CD40; CORO1A; DOCK11; EOMES; FOXP3; GAPT; GPR183; HLA-DMB; IFNG; IL12B; IL18R1; IL18; IRF4; ITGAL; ITM2A; JAK3; LCP1; LILRB1; LY9; MFNG; PLCG2; PLCL2; PTGER4; PTK2B; PTPRC; SLAMF6; SLC11A1; SPN; TBX21; TNFSF18; ZNF683 |
| GO:0032633 | interleukin-4 production | 5.31 | 7.76e-07 | 1.11e-05 | 12/31 | CD28; CD3E; CD40LG; CLECL1; FOXP3; HLA-E; IL33; IRF4; ITK; PRKCQ; SASH3; SYK |
| GO:0019882 | antigen processing and presentation | 5.30 | 0.00e+00 | 0.00e+00 | 34/88 | B2M; CCL19; CCL21; CCR7; CD1A; CD1B; CD1C; CD1D; CD1E; CD74; CD8A; CLEC4A; CTSS; HLA-DMB; HLA-DOA; HLA-DOB; HLA-DPA1; HLA-DPB1; HLA-DQA1; HLA-DQA1; HLA-DQA2; HLA-DQB1; HLA-DQB2; HLA-DRA; HLA-DRB1; HLA-DRB5; HLA-E; HLA-F; LAG3; RAB33A; RFTN1; SLC11A1; TAP1; TAP2; WAS |
| GO:0001906 | cell killing | 5.21 | 0.00e+00 | 0.00e+00 | 41/108 | APOL1; B2M; CCL13; CD1A; CD1B; CD1C; CD1D; CD1E; CD226; CORO1A; CRTAM; DNASE1L3; FCER2; GNLY; GZMB; GZMM; HLA-E; HLA-F; IFNG; IL12B; IL18RAP; IL18; IL21; IL7R; KIR2DL4; KIR3DL1; KLRK1; LAG3; LILRB1; LYZ; NCR1; NCR3; PRF1; PTPRC; SERPINB9; SH2D1A; SLAMF6; SLAMF7; STAP1; VAV1; XCL1 |
| GO:0007159 | leukocyte cell-cell adhesion | 5.20 | 0.00e+00 | 0.00e+00 | 88/232 | AIF1; BTLA; VSIR; CARD11; CCDC88B; CCL19; CCL21; CCL25; CCL5; CCR2; CCR7; CD1D; CD274; CD27; CD28; CD3E; CD40LG; CD4; CD5; CD6; CD74; CLECL1; CORO1A; CTLA4; DOCK8; EBI3; ETS1; RIPOR2; FERMT3; FOXP3; FYN; GRAP2; HLA-DMB; HLA-DPA1; HLA-DPB1; HLA-E; ICOS; IDO1; IFNG; IL12B; IL18; IL21; IL2RA; IL7R; IL7; IRF1; ITGA4; ITGAL; ITGB2; ITGB7; JAK3; TESPA1; KLRK1; LAG3; LAX1; LCK; LILRB1; NCKAP1L; PAG1; PDCD1LG2; PDCD1; PLA2G2D; PRKCQ; PTAFR; PTPN22; PTPRC; RAC2; RASAL3; RUNX3; SASH3; SELE; SELL; SELPLG; SELP; SIRPG; SOCS1; SPN; SYK; TBX21; TIGIT; TNFAIP8L2; TNFRSF13C; TNFRSF14; TNFSF13B; VAV1; VCAM1; XCL1; ZAP70 |
| GO:0002347 | response to tumor cell | 5.15 | 6.20e-04 | 6.31e-03 | 6/16 | CD226; CD274; CRTAM; IL12B; KLRK1; PRF1 |
| GO:0050867 | positive regulation of cell activation | 5.14 | 0.00e+00 | 0.00e+00 | 82/219 | AIF1; BTK; BTLA; VSIR; CARD11; CCDC88B; CCL19; CCL21; CCL5; CCR2; CCR7; CD1D; CD226; CD274; CD27; CD28; CD2; CD38; CD3E; CD40LG; CD40; CD4; CD5; CD6; CD74; CLECL1; CORO1A; CTLA4; DOCK8; EBI3; FCRL3; FLT3LG; FOXP3; FYN; GPR183; GRAP2; HLA-DMB; HLA-DPA1; HLA-DPB1; HLA-E; ICOS; IFNG; IGLL1; IL12B; IL18; IL21; IL2RA; IL33; IL7R; IL7; INPP5D; ITGB2; JAK3; TESPA1; KLRK1; LCK; LILRB1; NCKAP1L; PDCD1LG2; PDCD1; PLEK; PRKCQ; PTAFR; PTPRC; RASAL3; RUNX3; SASH3; SELP; SIRPG; SOCS1; STAP1; SYK; TBX21; TNFRSF13C; TNFRSF14; TNFRSF4; TNFSF13B; TOX; VAV1; VCAM1; XCL1; ZAP70 |
| GO:0042107 | cytokine metabolic process | 4.89 | 2.48e-12 | 6.51e-11 | 26/73 | CARD11; CCR2; CD28; CD3E; CD4; CYBB; EBI3; FOXP3; GHRL; IL12B; IL18; IL21; INPP5D; IRF1; IRF4; LAG3; LILRB1; LTB; PRKCQ; PTAFR; PTPRC; SPN; SYK; TLR8; TNFRSF13C; TNFRSF8 |
| GO:0033622 | integrin activation | 4.84 | 8.99e-04 | 8.93e-03 | 6/17 | CXCL13; FERMT3; MZB1; PLEK; PTGER4; SELP |
| GO:0002694 | regulation of leukocyte activation | 4.74 | 0.00e+00 | 0.00e+00 | 116/336 | AIF1; BANK1; BTK; BTLA; VSIR; CAMK4; CARD11; CCDC88B; CCL19; CCL21; CCL5; CCR2; CCR7; CD1D; CD226; CD22; CD274; CD27; CD28; CD2; CD300LF; CD38; CD3E; CD40LG; CD40; CD4; CD5; CD6; CD74; CLECL1; CNR2; CORO1A; CR1; CTLA4; DOCK8; EBI3; RIPOR2; FCRL3; FLT3LG; FOXP3; FYN; GPR183; GRAP2; HLA-DMB; HLA-DOA; HLA-DPA1; HLA-DPB1; HLA-E; ICOS; IDO1; IFNG; IGLL1; IKZF3; IL12B; IL18; IL21; IL2RA; IL33; IL7R; IL7; INPP5D; IRF1; IRF4; ITGB2; JAK3; TESPA1; KLRK1; LAG3; LAT; LAX1; LCK; LILRB1; LST1; MZB1; NCKAP1L; PAG1; PDCD1LG2; PDCD1; PLA2G2D; PRDM1; PRKCQ; PTAFR; PTPN22; PTPRC; RAC2; RASAL3; RUNX3; SAMSN1; SASH3; SIRPG; SIT1; SLA2; SLAMF8; SOCS1; SPN; STAP1; SYK; TBC1D10C; TBX21; TIGIT; TNFAIP3; TNFAIP8L2; TNFRSF13B; TNFRSF13C; TNFRSF14; TNFRSF1B; TNFRSF4; TNFSF13B; TNFSF18; TOX; TSPAN32; VAV1; VCAM1; XCL1; ZAP70; ZNF683 |
| GO:0002437 | inflammatory response to antigenic stimulus | 4.73 | 2.19e-05 | 2.83e-04 | 10/29 | BTK; C3; CCR7; CD28; CD6; HLA-E; IL12B; IL2RA; IL5RA; LTA |
| GO:0019835 | cytolysis | 4.73 | 2.19e-05 | 2.83e-04 | 10/29 | APOL1; C7; CFHR1; GZMA; GZMB; GZMH; GZMM; LILRB1; LYZ; PRF1 |
| GO:0034341 | response to interferon-gamma | 4.68 | 0.00e+00 | 0.00e+00 | 44/129 | AIF1; B2M; CASP1; CCL13; CCL17; CCL19; CCL21; CCL22; CCL25; CCL5; CD40; CIITA; GBP1; GBP2; GBP4; GBP5; HLA-DPA1; HLA-DPB1; HLA-DQA1; HLA-DQA1; HLA-DQA2; HLA-DQB1; HLA-DQB2; HLA-DRA; HLA-DRB1; HLA-DRB5; HLA-E; HLA-F; IFNG; IL12B; IRF1; IRF4; IRF8; NLRC5; PTAFR; SLC11A1; SOCS1; STAT1; TRIM22; TXK; UBD; VCAM1; WAS; XCL1; XCL2 |
| GO:0071887 | leukocyte apoptotic process | 4.64 | 1.56e-10 | 3.44e-09 | 23/68 | BTK; CCL19; CCL21; CCL5; CCR5; CCR7; CD274; CD27; CD3G; CD74; DOCK8; FASLG; GIMAP8; HCLS1; IDO1; IL2RA; IL7R; JAK3; LILRB1; PDCD1; PIK3CD; PRKCQ; TNFRSF4 |
| GO:0002764 | immune response-regulating signaling pathway | 4.64 | 0.00e+00 | 0.00e+00 | 98/290 | BIRC3; BLK; BTK; BTLA; BTN1A1; BTN3A1; BTN3A2; BTN3A3; C5AR1; CARD11; CCR7; CD19; CD200R1; CD226; CD22; CD247; CD28; CD300LF; CD38; CD3D; CD3E; CD3G; CD40; CD4; CD79A; CD79B; CLEC10A; CLEC4A; CLEC4C; CLEC6A; CR1; CR2; CTLA4; CTSS; FCN1; FCRL3; FOXP3; FYB1; FYN; GBP1; GCSAM; GFI1; GRAP2; HLA-DPA1; HLA-DPB1; HLA-DQA1; HLA-DQA1; HLA-DQA2; HLA-DQB1; HLA-DQB2; HLA-DRA; HLA-DRB1; HLA-DRB5; ICAM3; IGLL1; INPP5D; IRF1; IRF4; ITGB2; ITK; TESPA1; KIR2DL1; KLHL6; KLRK1; LAT; LCK; LCP2; LILRA4; LILRB1; LIME1; LPXN; MYO1G; NCKAP1L; NCR3; PAG1; PAX5; PIK3CD; PLCG2; PLCL2; PRKCB; PRKCQ; PTPN22; PTPRC; PVRIG; RFTN1; SLA2; STAP1; SYK; THEMIS; TLR8; TNFAIP3; TNIP3; TRAT1; TXK; UBASH3A; VAV1; WAS; WIPF1; ZAP70 |
| GO:0002440 | production of molecular mediator of immune response | 4.61 | 0.00e+00 | 0.00e+00 | 40/119 | B2M; BTK; CAMK4; CARD11; CCR6; CD226; CD22; CD244; CD28; CD40LG; CD40; CD74; CD96; FCRL3; FOXP3; GAPT; HLA-DQB1; HLA-E; IL12B; IL18R1; IL18RAP; IL18; IL33; IL7R; ITM2A; JAK3; KIR2DL4; LILRB1; MZB1; POU2F2; PTPRC; SASH3; SLC11A1; TBX21; TNFRSF14; TNFRSF1B; TNFRSF4; TNFSF13B; VPREB3; XCL1 |
| GO:1990868 | response to chemokine | 4.57 | 5.71e-12 | 1.41e-10 | 27/81 | CCL13; CCL17; CCL19; CCL21; CCL22; CCL25; CCL5; CCR2; CCR4; CCR5; CCR6; CCR7; CCR8; CXCL10; CXCL11; CXCL13; CXCL9; CXCR3; CXCR5; CXCR6; ACKR1; DOCK8; RIPOR2; PTK2B; XCL1; XCL2; XCR1 |
| GO:0002697 | regulation of immune effector process | 4.50 | 0.00e+00 | 0.00e+00 | 81/247 | AIM2; APOBEC3F; APOBEC3G; B2M; BIRC3; BTK; C1QA; C1S; C3; C5AR1; C7; CCL19; CCR2; CD19; CD1A; CD1B; CD1C; CD1D; CD1E; CD226; CD22; CD244; CD28; CD40LG; CD40; CD74; CD96; CFHR1; CFH; CFP; CR1; CR2; CRTAM; DNASE1L3; FCER2; FCRL3; FOXP3; HLA-DMB; HLA-E; HLA-F; IFNG; IL12B; IL18R1; IL18RAP; IL18; IL21; IL2RA; IL33; IL7R; IRF4; ITGB2; JAK3; KIR2DL4; KLRK1; LAG3; LILRB1; LTA; MZB1; NCR1; NCR3; PTAFR; PTPRC; RAC2; SASH3; SERPINB9; SH2D1A; SH2D1B; SLAMF6; SLAMF8; STAT1; SYK; TBX21; TNFAIP3; TNFRSF14; TNFRSF1B; TNFRSF4; TSPAN32; VAV1; WAS; XCL1; ZNF683 |
| GO:0002200 | somatic diversification of immune receptors | 4.41 | 1.07e-04 | 1.24e-03 | 9/28 | BCL11B; CCR6; CD28; CD40LG; CD40; FOXP3; PTPRC; TBX21; TCF7 |
| GO:0050866 | negative regulation of cell activation | 4.15 | 4.00e-15 | 1.25e-13 | 39/129 | BANK1; BTK; VSIR; CCR2; CD274; CD300LF; CD74; CNR2; CTLA4; RIPOR2; FOXP3; IDO1; IL2RA; INPP5D; IRF1; JAK3; LAG3; LAX1; LILRB1; LST1; PAG1; PDCD1LG2; PLA2G2D; PTPN22; PTPRC; RUNX3; SAMSN1; SLA2; SOCS1; SPN; TBC1D10C; TBX21; TIGIT; TNFAIP3; TNFAIP8L2; TNFRSF13B; TNFRSF14; TSPAN32; XCL1 |
| GO:0022407 | regulation of cell-cell adhesion | 4.03 | 0.00e+00 | 0.00e+00 | 82/279 | AIF1; BTLA; VSIR; CARD11; CCDC88B; CCL19; CCL21; CCL25; CCL5; CCR2; CCR7; CD1D; CD274; CD27; CD28; CD3E; CD40LG; CD4; CD5; CD6; CD74; CLECL1; CORO1A; CTLA4; CXCL13; DOCK8; EBI3; ETS1; RIPOR2; FERMT3; FOXP3; FYN; GRAP2; HLA-DMB; HLA-DPA1; HLA-DPB1; HLA-E; ICOS; IDO1; IFNG; IL12B; IL18; IL21; IL2RA; IL7R; IL7; IRF1; ITGA4; JAK3; TESPA1; KLRK1; LAG3; LAX1; LCK; LILRB1; NCKAP1L; PAG1; PDCD1LG2; PDCD1; PLA2G2D; PRKCQ; PTAFR; PTPN22; PTPRC; RASAL3; RUNX3; SASH3; SIRPG; SOCS1; SPN; SYK; TBX21; TIGIT; TNFAIP8L2; TNFRSF13C; TNFRSF14; TNFSF13B; VAV1; VCAM1; WNT1; XCL1; ZAP70 |
| GO:0002521 | leukocyte differentiation | 4.03 | 0.00e+00 | 0.00e+00 | 94/320 | B2M; BATF3; BCL11B; BTK; VSIR; CAMK4; CARD11; CCL19; CCR6; CCR7; CD1D; CD27; CD28; CD2; CD3D; CD3E; CD3G; CD40LG; CD4; CD74; CD79A; CD79B; CD8A; CR1; CR2; CTLA4; DOCK11; DOCK2; EOMES; EVI2B; FCRL3; FLT3LG; FLT3; FOXP3; FUT7; GAB3; GPR183; GPR18; GPR55; HCLS1; HLA-DOA; IFNG; IKZF1; IKZF3; IL12B; IL18R1; IL18; IL2RA; IL7R; IL7; INPP5D; IRF1; IRF4; ITGA4; ITK; ITM2A; JAK3; TESPA1; LCK; LILRB1; LY9; LYL1; MFNG; NCKAP1L; PIK3CD; PLA2G2D; PLCG2; PLCL2; POU2F2; PRDM1; PTGER4; PTK2B; PTPN22; PTPRC; RASSF2; RHOH; RUNX3; SASH3; SLAMF6; SLAMF8; SOCS1; SPN; SYK; TBX21; TCF7; THEMIS; TNFSF8; TOX; UBD; VAV1; VCAM1; WNT1; ZAP70; ZNF683 |
| GO:0001819 | positive regulation of cytokine production | 3.74 | 0.00e+00 | 0.00e+00 | 75/275 | AIM2; B2M; BIRC3; C3; C5AR1; CARD11; CASP1; CCDC88B; CCL19; CCR2; CCR7; CD226; CD244; CD274; CD28; CD2; CD3E; CD40LG; CD40; CD4; CD6; CD74; CLEC5A; CLEC6A; CLEC9A; CRTAM; CYBB; EBI3; FCN1; FOXP3; GBP5; GPSM3; HLA-DPA1; HLA-DPB1; HLA-E; IDO1; IFNG; IL12B; IL12RB2; IL18R1; IL18; IL21; IL33; IL7; IRF1; IRF4; IRF8; KIR2DL4; KLRK1; LILRB1; LOC728392; LTA; LTB; LY9; MCOLN2; NLRP1; PLCG2; PRKCQ; PTAFR; PTGER4; PTPN22; PTPRC; SASH3; SLAMF6; SLC11A1; SPN; STAT1; SYK; TIGIT; TLR8; TNFRSF13C; TNFRSF14; TNFRSF8; TXK; XCL1; ZBP1 |
| GO:0070670 | response to interleukin-4 | 3.74 | 3.96e-03 | 3.29e-02 | 6/22 | CD300LF; CORO1A; IL2RG; JAK3; TCF7; XCL1 |
| GO:0042092 | type 2 immune response | 3.69 | 2.03e-03 | 1.86e-02 | 7/26 | CCR2; CD74; IDO1; IL18; IL33; TBX21; XCL1 |
| GO:0002683 | negative regulation of immune system process | 3.58 | 0.00e+00 | 0.00e+00 | 73/280 | BANK1; BTK; VSIR; CCL21; CCL25; CCR2; CD200R1; CD22; CD274; CD300LF; CD74; CD96; CNR2; CR1; CTLA4; RIPOR2; FCRL3; FOXP3; GBP1; GCSAM; GPR171; GPR18; GPR55; HLA-DOA; HLA-DOB; HLA-E; IDO1; IL12B; IL2RA; IL33; IL7R; INPP5D; IRF1; IRF4; JAK3; KIR2DL4; KLRK1; LAG3; LAX1; LILRA4; LILRB1; LPXN; LST1; NLRC5; PAG1; PDCD1LG2; PDCD1; PLA2G2D; PLCL2; PTGER4; PTK2B; PTPN22; PTPRC; PVRIG; RUNX3; SAMSN1; SERPINB9; SLA2; SLAMF8; SOCS1; SPN; STAP1; TBC1D10C; TBX21; TIGIT; TNFAIP3; TNFAIP8L2; TNFRSF13B; TNFRSF14; TNFRSF4; TSPAN32; UBASH3A; XCL1 |
| GO:0034612 | response to tumor necrosis factor | 3.56 | 2.43e-13 | 7.02e-12 | 41/158 | AIM2; BIRC3; CARD16; CASP1; CCL13; CCL17; CCL19; CCL21; CCL22; CCL25; CCL5; CD27; CD40LG; CD40; CD70; GBP1; GBP2; LTA; LTB; PTK2B; SELE; STAT1; SYK; TCL1A; TNFAIP3; TNFRSF13B; TNFRSF13C; TNFRSF14; TNFRSF17; TNFRSF1B; TNFRSF4; TNFRSF8; TNFRSF9; TNFSF13B; TNFSF18; TNFSF8; TRAF1; UBD; VCAM1; XCL1; XCL2 |
| GO:0035587 | purinergic receptor signaling pathway | 3.56 | 2.58e-03 | 2.31e-02 | 7/27 | GPR171; P2RX1; P2RY10; P2RY13; P2RY14; P2RY8; PTAFR |
| GO:0045785 | positive regulation of cell adhesion | 3.55 | 0.00e+00 | 0.00e+00 | 74/286 | ABI3BP; AIF1; APBB1IP; BTLA; VSIR; CARD11; CASS4; CCDC88B; CCL19; CCL21; CCL25; CCL5; CCR2; CCR7; CD1D; CD274; CD27; CD28; CD3E; CD40LG; CD4; CD5; CD6; CD74; CLECL1; CORO1A; CTLA4; CXCL13; DOCK8; EBI3; ETS1; FOXP3; FYN; GRAP2; HLA-DMB; HLA-DPA1; HLA-DPB1; HLA-E; ICOS; IFNG; IL12B; IL18; IL21; IL2RA; IL7R; IL7; ITGA4; JAK3; TESPA1; KLRK1; LCK; LILRB1; NCKAP1L; PDCD1LG2; PDCD1; PRKCQ; PTAFR; PTK2B; PTPRC; RASAL3; RUNX3; SASH3; SIRPG; SOCS1; SPOCK2; SYK; TNFRSF13C; TNFRSF14; TNFSF13B; TNFSF18; VAV1; VCAM1; XCL1; ZAP70 |
| GO:0050663 | cytokine secretion | 3.52 | 1.41e-12 | 3.83e-11 | 39/152 | AIM2; BANK1; BTN3A1; BTN3A2; CARD11; CARD16; CARD17; CASP1; CCL19; CCR7; CD200R1; CD244; CD274; CD2; CLEC5A; CLEC6A; CLEC9A; CLECL1; CRTAM; FCN1; FOXP3; GBP1; GBP5; GHRL; IL33; LCP2; LILRB1; LOC728392; MCOLN2; NLRP1; PTGER4; PTPN22; SOCS1; SRGN; SYK; TLR8; TNFAIP3; TNFRSF14; TNFRSF1B; TNFRSF4 |
| GO:0045088 | regulation of innate immune response | 3.48 | 0.00e+00 | 0.00e+00 | 56/221 | AIM2; BIRC3; BTK; CARD11; CCL5; CD1D; CD226; CD300LF; CD96; CLEC10A; CLEC4A; CLEC4C; CLEC6A; CR1; CRTAM; CTSS; FCN1; FCRL3; FYN; GBP5; GFI1; HLA-E; ICAM3; IFNG; IL12B; IL18RAP; IL21; IRF1; IRF4; ITGB2; KIR2DL4; KLRK1; LAG3; LILRA4; LILRB1; NCR1; NCR3; NLRC5; PLCG2; PTPN22; PYHIN1; RFTN1; SERPINB9; SH2D1A; SH2D1B; SLAMF6; SLAMF8; SOCS1; STAT1; SYK; TLR8; TNFAIP3; TNIP3; TXK; VAV1; ZBP1 |
| GO:0032606 | type I interferon production | 3.43 | 2.33e-04 | 2.56e-03 | 11/44 | IRF1; LILRA4; LILRB1; NLRC5; PLCG2; PTPN22; STAT1; SYK; TLR8; TNFAIP3; ZBP1 |
| GO:0048872 | homeostasis of number of cells | 3.41 | 5.70e-11 | 1.29e-09 | 35/141 | B2M; CARD11; CCR2; CCR4; CCR7; CD74; CORO1A; DOCK11; ETS1; FLT3LG; FLT3; FOXP3; GAPT; GPR174; GPR183; HCLS1; IKZF1; IL2RA; IL7R; IL7; INPP5D; JAK3; LAT; NCKAP1L; PIK3CD; RASSF2; SASH3; SIT1; STAT1; TNFAIP3; TNFRSF13B; TNFRSF13C; TNFRSF17; TNFRSF4; TNFSF13B |
| GO:0050900 | leukocyte migration | 3.40 | 0.00e+00 | 0.00e+00 | 79/319 | AIF1; JAML; ANGPT2; C5AR1; CCL13; CCL17; CCL19; CCL21; CCL22; CCL25; CCL5; CCR2; CCR5; CCR6; CCR7; CD200R1; CD244; CD2; CD48; CD74; CNR2; CORO1A; CXCL10; CXCL11; CXCL13; CXCL9; CXCR3; CXCR5; DBH; DOCK8; DOK2; RIPOR2; FCAMR; FUT7; FYN; GCSAM; GPR15; GPR183; GPR18; GPSM3; JCHAIN; IGLL1; IL16; IL33; INPP5D; ITGA4; ITGAL; ITGB2; ITGB7; KLRK1; LCK; MCOLN2; MYO1G; NCKAP1L; PIK3CD; PIK3CG; PTAFR; PTGER4; PTK2B; RAC2; SELE; SELL; SELPLG; SELP; SIRPG; SLAMF8; SPN; STAP1; SYK; TBX21; TNFRSF14; TNFSF18; TREM1; VAV1; VCAM1; VPREB3; XCL1; XCL2; ZAP70 |
| GO:0006959 | humoral immune response | 3.37 | 2.50e-13 | 7.02e-12 | 44/179 | C1QA; C1S; C3; C5AR1; C7; CCL13; CCR2; CCR6; CCR7; CD19; CD28; CFHR1; CFH; CFP; CR1; CR2; CXCL10; CXCL11; CXCL13; CXCL9; EBI3; FCER2; FCN1; GNLY; GPR183; HLA-DQB1; HLA-E; IFNG; JCHAIN; IGLL1; IL7; LTA; LYZ; MS4A1; PAX5; PDCD1; POU2AF1; POU2F2; PSMB10; PTPRC; RNASE6; SH2D1A; SLC11A1; TREM1 |
| GO:0002532 | production of molecular mediator involved in inflammatory response | 3.35 | 5.53e-04 | 5.69e-03 | 10/41 | ABCD2; BTK; CD6; CD96; GBP5; GPSM3; IDO1; P2RX1; SLAMF8; SYK |
| GO:0045576 | mast cell activation | 3.28 | 3.54e-04 | 3.75e-03 | 11/46 | BTK; CD226; CD300LF; CNR2; LAT; LCP2; PIK3CD; PIK3CG; RAC2; RHOH; SYK |
| GO:0010324 | membrane invagination | 3.23 | 2.52e-03 | 2.28e-02 | 8/34 | AIF1; ARHGAP25; BIN2; C3; IGLL1; ITGB2; NCKAP1L; STAP1 |
| GO:0071706 | tumor necrosis factor superfamily cytokine production | 3.22 | 3.58e-07 | 5.61e-06 | 23/98 | VSIR; CCL19; CCR2; CD274; CD2; CLEC4A; CYBB; FOXP3; GHRL; GPR18; HLA-E; IFNG; IL12B; LILRA4; LILRB1; PTAFR; PTPN22; PTPRC; SASH3; SLAMF1; SPN; TNFAIP3; TNFRSF8 |
| GO:1903706 | regulation of hemopoiesis | 3.09 | 3.62e-14 | 1.09e-12 | 53/235 | B2M; BTK; VSIR; CAMK4; CARD11; CCL19; CD27; CD28; CD2; CD4; CD74; CR1; CTLA4; ETS1; EVI2B; FCRL3; FLT3LG; FOXP3; GP1BA; GPR171; GPR55; HCLS1; HLA-DOA; IFNG; IKZF3; IL12B; IL18; IL2RA; IL7R; IL7; INPP5D; IRF1; IRF4; JAK3; TESPA1; LILRB1; NCKAP1L; PRDM1; PRKCB; PRKCQ; PTK2B; PTPRC; RASSF2; RUNX3; SASH3; SLAMF8; SOCS1; STAT1; SYK; TBX21; TOX; ZAP70; ZNF683 |
| GO:0001818 | negative regulation of cytokine production | 3.03 | 3.41e-10 | 7.31e-09 | 38/172 | ABCD2; BANK1; BTK; VSIR; CARD16; CARD17; CD200R1; CD274; CD96; CLEC4A; FOXP3; GBP1; GHRL; GPR18; IDO1; IFNG; IL12B; IL33; INPP5D; JAK3; LAG3; LILRA4; LILRB1; NCKAP1L; NLRC5; PDCD1LG2; PTGER4; PTPN22; PTPRC; SLAMF1; SLC11A1; SRGN; TBX21; TIGIT; TLR8; TNFAIP3; TNFRSF4; XCL1 |
| GO:0031349 | positive regulation of defense response | 3.03 | 6.66e-16 | 2.17e-14 | 62/281 | AIM2; BIRC3; BTK; C3; CARD11; CCL5; CCR2; CCR7; CD1D; CD226; CD28; CD300LF; CD6; CLEC10A; CLEC4A; CLEC4C; CLEC6A; CREB3L3; CRTAM; CTSS; ETS1; FCN1; FCRL3; FYN; GBP5; GFI1; GPSM3; HLA-E; ICAM3; IDO1; IL12B; IL18RAP; IL18; IL21; IL33; IRF1; IRF4; ITGB2; KIR2DL4; KLRK1; LAG3; LILRA4; LTA; NCR3; NLRC5; PIK3CG; PLCG2; PTGER4; PTPN22; PYHIN1; RFTN1; SH2D1A; SH2D1B; SLAMF6; STAP1; SYK; TLR8; TNFAIP3; TNIP3; TXK; VAV1; ZBP1 |
| GO:0002831 | regulation of response to biotic stimulus | 3.03 | 8.50e-05 | 1.02e-03 | 15/68 | AIM2; APOBEC3F; APOBEC3G; BIRC3; CARD16; CD226; CD274; CRTAM; IL12B; IL2RA; KLRK1; LILRB1; STAT1; TNFAIP3; TSPAN32 |
| GO:0060326 | cell chemotaxis | 2.94 | 4.65e-12 | 1.18e-10 | 48/224 | AIF1; JAML; BIN2; C5AR1; CCL13; CCL17; CCL19; CCL21; CCL22; CCL25; CCL5; CCR2; CCR4; CCR5; CCR6; CCR7; CCR8; CD74; CNR2; CORO1A; CXCL10; CXCL11; CXCL13; CXCL9; CXCR3; CXCR5; CXCR6; RIPOR2; GPR183; GPR18; GPSM3; IL16; ITGB2; KLRK1; NCKAP1L; PIK3CD; PIK3CG; PRKCQ; PTK2B; RAC2; SLAMF8; STAP1; SYK; VAV1; VCAM1; XCL1; XCL2; XCR1 |
| GO:0033627 | cell adhesion mediated by integrin | 2.90 | 1.08e-03 | 1.02e-02 | 11/52 | CCL21; CCL5; CD3E; CXCL13; FERMT3; ITGB2; ITGB7; LPXN; NCKAP1L; PIK3CG; SYK |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 2.87 | 4.20e-07 | 6.34e-06 | 27/129 | APOL3; BIRC3; BTK; CARD11; CARD16; CASP10; CASP1; CCL19; CCL21; CCR7; CD40; CD4; CD74; CLEC6A; FASLG; FYN; LIME1; PIM2; PRKCB; RHOH; STAT1; TIFAB; TLR8; TNFAIP3; TNIP3; TRIM22; UBD |
| GO:0150076 | neuroinflammatory response | 2.87 | 3.26e-03 | 2.85e-02 | 9/43 | AIF1; CD200R1; IL18; IL33; ITGB2; PTPRC; STAP1; TLR8; TNFRSF1B |
| GO:0006909 | phagocytosis | 2.86 | 1.06e-07 | 1.80e-06 | 30/144 | AIF1; ARHGAP25; BIN2; C3; CD247; CD300LF; CD3G; CORO1A; DOCK2; FCN1; FYN; ICAM3; IGLL1; IRF8; ITGAL; ITGB2; MYO1G; NCF4; NCKAP1L; PLCG2; PLD4; PTPRC; SIRPG; SLAMF1; SLC11A1; STAP1; SYK; VAV1; WAS; WIPF1 |
| GO:0060759 | regulation of response to cytokine stimulus | 2.83 | 2.68e-05 | 3.36e-04 | 19/92 | BIRC3; CARD16; CASP1; CCL5; CD300LF; CD74; GFI1; IFNG; IL7; NLRC5; PTPRC; SOCS1; STAP1; STAT1; SYK; TNFAIP3; TRAF1; TXK; ZBP1 |
| GO:0009615 | response to virus | 2.80 | 4.30e-08 | 7.78e-07 | 33/162 | AIM2; APOBEC3C; APOBEC3F; APOBEC3G; APOBEC3H; BATF3; BIRC3; CCL19; CCL22; CCL5; CD40; CXCL10; CXCL9; FOXP3; GBP1; IFNG; IL12B; IL2RA; IL33; IRF1; LILRB1; NLRC5; PIM2; PRF1; PTPRC; RNASE6; STAT1; TBX21; TLR8; TNFAIP3; TRIM22; TSPAN32; XCL1 |
| GO:0072376 | protein activation cascade | 2.74 | 4.31e-04 | 4.50e-03 | 14/70 | C1QA; C1S; C3; C5AR1; C7; CD19; CFHR1; CFH; CFP; CR1; CR2; FCN1; GP1BA; IGLL1 |
| GO:0097696 | STAT cascade | 2.74 | 1.08e-04 | 1.24e-03 | 17/85 | CCL5; CCR2; CD40; FLT3; FYN; HCLS1; IFNG; IL12B; IL18; IL21; IL7R; JAK3; PTPRC; SOCS1; STAT1; STAT4; TNFSF18 |
| GO:0034113 | heterotypic cell-cell adhesion | 2.74 | 4.49e-03 | 3.70e-02 | 9/45 | CD1D; CD200R1; CD2; ITGA4; ITGAD; ITGB2; ITGB7; LCK; PTPRC |
| GO:0071216 | cellular response to biotic stimulus | 2.70 | 1.47e-06 | 2.07e-05 | 27/137 | BTK; CARD16; CARD17; CASP1; CCL5; CCR5; CD274; CD40; CD6; CXCL10; CXCL11; CXCL13; CXCL9; GFI1; IL12B; IL18; IRF8; KLRK1; LILRB1; PDCD1LG2; PTAFR; PTPN22; STAP1; SYK; TNFAIP3; TNFRSF1B; TNIP3 |
| GO:0098542 | defense response to other organism | 2.69 | 3.23e-11 | 7.52e-10 | 51/260 | AIM2; APOBEC3C; APOBEC3F; APOBEC3G; APOBEC3H; BIRC3; C5AR1; CCDC88B; CD40; CD4; CFP; CLEC6A; COTL1; CXCL10; CXCL13; CXCL9; GBP1; GNLY; HLA-E; IFNG; JCHAIN; IGLL1; IL12B; IL2RA; IL33; IRF1; IRF4; IRF8; KLRK1; LILRB1; LOC728392; LTA; LYZ; MPEG1; NLRC5; NLRP1; PLAC8; PRF1; PTPRC; RNASE6; SELP; SLAMF8; SLC11A1; SPN; STAT1; SYK; TLR8; TNFAIP3; TNFRSF14; TNFSF8; TRIM22; TSPAN32 |
| GO:0007162 | negative regulation of cell adhesion | 2.68 | 7.87e-08 | 1.39e-06 | 34/174 | ADAMDEC1; ANGPT2; VSIR; CCL21; CCL25; CD274; CD74; CTLA4; RIPOR2; FOXP3; GBP1; IDO1; IL2RA; IRF1; JAK3; LAG3; LAX1; LILRB1; LPXN; PAG1; PDCD1LG2; PLA2G2D; PTPN22; PTPRC; RUNX3; SOCS1; SPN; TBX21; TGFBI; TIGIT; TNFAIP8L2; TNFRSF14; WNT1; XCL1 |
| GO:0002237 | response to molecule of bacterial origin | 2.58 | 6.98e-09 | 1.35e-07 | 42/223 | B2M; C5AR1; CARD16; CARD17; CASP1; CCL5; CCR5; CCR7; CD274; CD40; CD6; CD96; CNR2; CSF2RB; CXCL10; CXCL11; CXCL13; CXCL9; FASLG; GFI1; IDO1; IL10RA; IL12B; IL12RB2; IL18; IRF8; KLRK1; LILRB1; LTA; PDCD1LG2; PLCG2; PTAFR; PTGER4; PTPN22; SELE; SELP; SLC11A1; STAP1; TNFAIP3; TNFRSF1B; TNIP3; VCAM1 |
| GO:0050727 | regulation of inflammatory response | 2.58 | 5.61e-10 | 1.17e-08 | 48/255 | ABCD2; BIRC3; BTK; C1QA; C1S; C3; C5AR1; C7; CCL5; CCR2; CCR7; CD19; CD200R1; CD28; CD6; CFHR1; CFH; CFP; CNR2; CR1; CR2; CREB3L3; DNASE1L3; ETS1; FOXP3; GBP5; GHRL; GPSM3; HLA-E; IDO1; IL12B; IL18; IL21; IL2RA; IL33; ITGB2; LOC728392; LTA; NLRP1; PIK3CG; PTGER4; PTPRC; SELE; SLAMF8; STAP1; TNFAIP3; TNFAIP8L2; TNFRSF1B; XCL1 |
| GO:0032103 | positive regulation of response to external stimulus | 2.54 | 4.22e-08 | 7.78e-07 | 39/211 | AIF1; BTK; C3; C5AR1; CCL19; CCL21; CCL5; CCR2; CCR4; CCR6; CCR7; CD28; CD6; CD74; CREB3L3; CXCL10; CXCL13; ETS1; RIPOR2; GBP5; GHRL; GPSM3; HLA-E; IDO1; IL12B; IL16; IL18; IL21; IL33; ITGB2; LTA; NCKAP1L; PIK3CG; PTGER4; PTK2B; RAC2; STAP1; XCL1; XCL2 |
| GO:0036230 | granulocyte activation | 2.44 | 9.08e-09 | 1.72e-07 | 46/259 | ARHGAP9; ATP8A1; B2M; BIN2; C3; C5AR1; CCL5; CCR2; CD53; ADGRE5; ADA2; CFP; CLEC12A; CLEC4C; CLEC5A; COTL1; CR1; CTSS; CYBB; DNASE1L3; DOCK2; FCN1; FGL2; GMFG; ARHGAP45; HVCN1; IL18RAP; IL18; ITGAL; ITGB2; KCNAB2; LRMP; LYZ; NCKAP1L; P2RX1; PGM1; PLAC8; PTAFR; PTPRC; RAB37; RHOF; SELL; SLC11A1; SYK; TBC1D10C; TNFRSF1B |
| GO:0031348 | negative regulation of defense response | 2.38 | 1.51e-04 | 1.71e-03 | 21/121 | ABCD2; CD200R1; CD96; CNR2; CR1; ETS1; FOXP3; GHRL; HLA-E; IL12B; IL2RA; KIR2DL4; LILRB1; NLRC5; PTGER4; PTPRC; SERPINB9; SLAMF8; TNFAIP3; TNFAIP8L2; TNFRSF1B |
| GO:0007229 | integrin-mediated signaling pathway | 2.38 | 2.71e-03 | 2.39e-02 | 13/75 | FERMT3; FYB1; ITGA4; ITGAD; ITGAL; ITGB2; ITGB7; LAT; PLEK; PTK2B; SYK; TSPAN32; VAV1 |
| GO:0002526 | acute inflammatory response | 2.31 | 3.37e-04 | 3.60e-03 | 20/119 | BTK; C1QA; C1S; C3; C5AR1; C7; CCR7; CD19; CD6; CFHR1; CFH; CFP; CR1; CR2; CREB3L3; DNASE1L3; HLA-E; PIK3CG; TREM1; VCAM1 |
| GO:0002446 | neutrophil mediated immunity | 2.29 | 2.63e-07 | 4.19e-06 | 42/252 | ARHGAP9; ATP8A1; B2M; BIN2; C3; C5AR1; CD53; ADGRE5; ADA2; CFP; CLEC12A; CLEC4C; CLEC5A; COTL1; CR1; CTSS; CYBB; DNASE1L3; DOCK2; FCN1; FGL2; GMFG; ARHGAP45; HVCN1; ITGAL; ITGB2; KCNAB2; LRMP; LYZ; NCKAP1L; P2RX1; PGM1; PLAC8; PTAFR; PTPRC; RAB37; RHOF; SELL; SLC11A1; SYK; TBC1D10C; TNFRSF1B |
| GO:0050920 | regulation of chemotaxis | 2.29 | 5.23e-05 | 6.36e-04 | 26/156 | AIF1; ANGPT2; C5AR1; CCL19; CCL21; CCL5; CCR2; CCR4; CCR6; CCR7; CD74; CXCL10; CXCL13; RIPOR2; GPR183; GPR18; GPSM3; IL16; KLRK1; NCKAP1L; PTK2B; RAC2; SLAMF8; STAP1; XCL1; XCL2 |
| GO:1904950 | negative regulation of establishment of protein localization | 2.26 | 1.14e-03 | 1.06e-02 | 17/103 | BANK1; CARD16; CARD17; CD200R1; CD22; DERL3; FOXP3; GBP1; GHRL; IL33; LILRB1; PTGER4; PTPN22; SRGN; TLR8; TNFAIP3; TNFRSF4 |
| GO:0032635 | interleukin-6 production | 2.23 | 3.45e-03 | 2.99e-02 | 14/86 | BANK1; CD200R1; FOXP3; GHRL; IL18RAP; IL18; IL33; INPP5D; NCKAP1L; PTAFR; PTPN22; SLAMF1; TLR8; TNFAIP3 |
| GO:0030099 | myeloid cell differentiation | 2.18 | 1.53e-05 | 2.04e-04 | 33/208 | B2M; BATF3; CAMK4; CCL19; CCR7; CD4; CD74; CLEC5A; ETS1; EVI2B; FLI1; GAB3; GP1BA; GPR171; GPR183; GPR55; HCLS1; IFNG; IKZF1; IL12B; INPP5D; IRF4; IRF8; JAK3; LILRB1; NCKAP1L; PIK3CD; PRKCB; PRKCQ; PTK2B; RASSF2; STAT1; UBD |
| GO:0048017 | inositol lipid-mediated signaling | 2.16 | 1.94e-03 | 1.80e-02 | 17/108 | CCL5; CD28; FLT3; FYN; HCLS1; HCST; IL18; LIME1; NCF1; PIK3CD; PIK3CG; PIK3R5; PPP1R16B; PRR5L; PTAFR; SELP; WNT16 |
| GO:0070555 | response to interleukin-1 | 2.14 | 3.77e-03 | 3.16e-02 | 15/96 | CCL13; CCL17; CCL19; CCL21; CCL22; CCL25; CCL5; CD38; CD40; ETS1; GBP1; GBP2; SELE; XCL1; XCL2 |
| GO:0019058 | viral life cycle | 2.13 | 7.05e-04 | 7.08e-03 | 21/135 | APOBEC3C; APOBEC3F; APOBEC3G; APOBEC3H; CCL5; CCR5; CD28; CD4; CD74; CLEC5A; CR1; CR2; CXCR6; FCN1; GPR15; ITGB7; LAMP3; SELPLG; SLAMF1; TNFRSF14; TNFRSF4 |
| GO:0018212 | peptidyl-tyrosine modification | 2.12 | 4.74e-06 | 6.43e-05 | 39/252 | ABI3; BANK1; BLK; BTK; CASS4; CCL5; CD3E; CD40; CD4; CD74; CSF2RB; FLT3; FYN; GHRL; HCLS1; IFNG; IL12B; IL12RB2; IL18; IL21; IL3RA; IL5RA; IL7; ITGB2; ITK; JAK3; LCK; NCF1; PTK2B; PTPRC; SAMSN1; SLA; SOCS1; STAP1; SYK; TNFRSF14; TNFSF18; TXK; ZAP70 |
| GO:0007163 | establishment or maintenance of cell polarity | 2.06 | 5.57e-03 | 4.45e-02 | 15/100 | CCL19; CCL21; CCR7; CD3G; DOCK2; DOCK8; RIPOR2; NCKAP1L; PARVG; PTK2B; RHOF; RHOH; CARMIL2; SPN; WNT7A |
| GO:0043087 | regulation of GTPase activity | 2.05 | 2.46e-05 | 3.13e-04 | 36/241 | ACAP1; AGAP2; ALDH1A1; ARHGAP15; ARHGAP25; ARHGAP30; ARHGAP4; ARHGAP9; CCL13; CCL17; CCL19; CCL21; CCL22; CCL25; CCL5; CCR7; CD40; CXCL13; DOCK11; DOCK2; DOCK8; FGD2; GPR65; ARHGAP45; KLRK1; NCKAP1L; PTK2B; RASAL3; RASGRP2; RGS18; RHOH; TAGAP; TBC1D10C; VAV1; XCL1; XCL2 |
| GO:0072507 | divalent inorganic cation homeostasis | 2.03 | 5.20e-07 | 7.70e-06 | 52/352 | C5AR1; CCL13; CCL19; CCL21; CCL5; CCR2; CCR4; CCR5; CCR6; CCR7; CCR8; CD19; CD38; CD40; CD4; CD52; CLIC2; CORO1A; CXCL10; CXCL11; CXCL13; CXCL9; CXCR3; CXCR5; CXCR6; FASLG; FYN; GHRL; GPR174; GPR18; GPR55; GPR65; JSRP1; LCK; LIME1; MCOLN2; P2RX1; P2RY10; P2RY8; PIK3CG; PLCG2; PRKCB; PTGDR; PTGER4; PTK2B; PTPRC; S1PR4; SLC11A1; SLC24A4; TMC8; XCL1; XCR1 |
Factor 17 : 2 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0022613 | ribonucleoprotein complex biogenesis | 6.12 | 4.02e-05 | 1.64e-02 | 8/52 | BOP1; DCAF13; AGO2; EIF3H; EXOSC4; MTERF3; PRKAA2; RRS1 |
| GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules | 3.96 | 1.31e-08 | 1.06e-05 | 23/231 | AMIGO2; CDH7; CNTN2; PCDHB14; PCDHGA10; PCDHGA11; PCDHGA1; PCDHGA2; PCDHGA3; PCDHGA4; PCDHGA5; PCDHGA6; PCDHGA7; PCDHGA8; PCDHGA9; PCDHGB1; PCDHGB2; PCDHGB3; PCDHGB4; PCDHGB5; PCDHGB6; PCDHGC4; PCDHGC5 |
Factor 18 : 63 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0035456 | response to interferon-beta | 33.80 | 0.00e+00 | 0.00e+00 | 13/19 | AIM2; BST2; IFI16; IFIT1; IFIT3; IFITM1; IFITM3; IFNB1; IRF1; PLSCR1; STAT1; TLR3; XAF1 |
| GO:0034340 | response to type I interferon | 32.90 | 0.00e+00 | 0.00e+00 | 32/48 | BST2; HLA-E; HLA-F; HLA-G; IFI27; IFI35; IFI6; IFIT1; IFIT2; IFIT3; IFITM1; IFITM3; IFNB1; IRF1; IRF7; IRF9; ISG15; ISG20; MX1; MX2; NLRC5; OAS1; OAS2; OAS3; OASL; PSMB8; RSAD2; SAMHD1; STAT1; USP18; XAF1; ZBP1 |
| GO:0035455 | response to interferon-alpha | 30.40 | 2.78e-11 | 1.62e-09 | 8/13 | BST2; EIF2AK2; IFIT2; IFIT3; IFITM1; IFITM3; LAMP3; MX2 |
| GO:0032069 | regulation of nuclease activity | 19.70 | 3.09e-05 | 6.62e-04 | 4/10 | OAS1; OAS2; OAS3; OASL |
| GO:0009615 | response to virus | 17.10 | 0.00e+00 | 0.00e+00 | 56/162 | AIM2; APOBEC3A; APOBEC3F; APOBEC3G; APOBEC3H; BST2; CCL5; CCL8; CXCL10; DDX58; DDX60; DHX58; EIF2AK2; GBP1; HERC5; IFI16; IFI27; IFI44L; IFI44; IFI6; IFIH1; IFIT1; IFIT2; IFIT3; IFIT5; IFITM1; IFITM3; IFNB1; IL15; IL27; IFNL2; IFNL3; IFNL1; IRF1; IRF7; IRF9; ISG15; ISG20; LGALS9; MX1; MX2; NLRC5; OAS1; OAS2; OAS3; OASL; PARP9; PLSCR1; PRF1; RSAD2; RTP4; SAMHD1; STAT1; TLR3; TRIM22; TRIM34 |
| GO:0032606 | type I interferon production | 16.80 | 2.66e-15 | 2.17e-13 | 15/44 | DDX58; DHX58; HERC5; IFI16; IFIH1; IRF1; IRF7; ISG15; NLRC5; NMI; STAT1; TLR3; UBA7; UBE2L6; ZBP1 |
| GO:0002507 | tolerance induction | 15.60 | 1.36e-06 | 4.44e-05 | 6/19 | CD274; HLA-E; HLA-G; ICOS; IDO1; LGALS9 |
| GO:0006471 | protein ADP-ribosylation | 14.10 | 1.38e-04 | 2.56e-03 | 4/14 | PARP10; PARP14; PARP9; WARS |
| GO:0043331 | response to dsRNA | 13.70 | 2.21e-05 | 4.86e-04 | 5/18 | DDX58; IFIH1; IFIT1; IFNB1; TLR3 |
| GO:0034341 | response to interferon-gamma | 11.90 | 0.00e+00 | 0.00e+00 | 31/129 | B2M; BST2; CASP1; CCL5; CCL7; CCL8; GBP1; GBP4; GBP5; HLA-E; HLA-F; HLA-G; IFITM1; IFITM3; IRF1; IRF7; IRF9; LGALS9; NLRC5; NMI; OAS1; OAS2; OAS3; OASL; PARP14; PARP9; SOCS1; STAT1; TLR3; TRIM22; TRIM34 |
| GO:0002831 | regulation of response to biotic stimulus | 10.90 | 3.45e-12 | 2.55e-10 | 15/68 | AIM2; APOBEC3F; APOBEC3G; CARD16; CD274; DDX58; DDX60; DHX58; HERC5; IFIT1; IL15; IL27; OPTN; PARP9; STAT1 |
| GO:0098542 | defense response to other organism | 10.80 | 0.00e+00 | 0.00e+00 | 57/260 | AIM2; APOBEC3A; APOBEC3F; APOBEC3G; APOBEC3H; BATF2; BST2; CXCL10; DDX58; DDX60; DHX58; EIF2AK2; GBP1; GNLY; HERC5; HLA-E; IFI16; IFI27; IFI44L; IFI6; IFIH1; IFIT1; IFIT2; IFIT3; IFIT5; IFITM1; IFITM3; IFNB1; IL15; IL22RA1; IL27; IFNL2; IFNL3; IFNL1; IRF1; IRF7; IRF9; ISG15; ISG20; MX1; MX2; NLRC5; OAS1; OAS2; OAS3; OASL; OPTN; PARP9; PLSCR1; PRF1; RSAD2; RTP4; SAMHD1; STAT1; TLR3; TRIM22; TRIM34 |
| GO:0044764 | multi-organism cellular process | 10.70 | 8.00e-05 | 1.59e-03 | 5/23 | DDX58; DDX60; DHX58; IFIH1; IRF7 |
| GO:0032196 | transposition | 10.40 | 4.95e-04 | 7.74e-03 | 4/19 | APOBEC3A; APOBEC3F; APOBEC3G; APOBEC3H |
| GO:0019058 | viral life cycle | 9.87 | 0.00e+00 | 0.00e+00 | 27/135 | APOBEC3A; APOBEC3F; APOBEC3G; APOBEC3H; BST2; CCL5; CD80; EIF2AK2; IFI16; IFI27; IFIT1; IFIT5; IFITM1; IFITM3; IFNB1; IFNL3; ISG15; ISG20; LAMP3; LGALS9; MX1; OAS1; OAS3; OASL; PARP10; PLSCR1; RSAD2 |
| GO:0043900 | regulation of multi-organism process | 9.67 | 0.00e+00 | 0.00e+00 | 38/194 | AIM2; APOBEC3A; APOBEC3F; APOBEC3G; APOBEC3H; BST2; CARD16; CCL5; DDX58; DDX60; DHX58; EIF2AK2; HERC5; IFI16; IFI27; IFIT1; IFIT5; IFITM1; IFITM3; IFNB1; IL15; IL27; IFNL3; ISG15; ISG20; LAMP3; LGALS9; MX1; OAS1; OAS3; OASL; OPTN; PARP10; PARP9; PLSCR1; PRF1; RSAD2; STAT1 |
| GO:0070988 | demethylation | 8.58 | 1.06e-03 | 1.57e-02 | 4/23 | APOBEC3A; APOBEC3F; APOBEC3G; APOBEC3H |
| GO:0009451 | RNA modification | 8.23 | 1.25e-03 | 1.82e-02 | 4/24 | APOBEC3A; APOBEC3F; APOBEC3G; APOBEC3H |
| GO:0060759 | regulation of response to cytokine stimulus | 8.05 | 3.36e-10 | 1.61e-08 | 15/92 | CARD16; CASP1; CCL5; DDX58; IFIH1; IRF7; NLRC5; PARP14; PARP9; SAMHD1; SOCS1; STAP1; STAT1; USP18; ZBP1 |
| GO:0042092 | type 2 immune response | 7.59 | 1.71e-03 | 2.40e-02 | 4/26 | IDO1; IFNB1; IFNL1; RSAD2 |
| GO:0032615 | interleukin-12 production | 7.26 | 5.53e-04 | 8.49e-03 | 5/34 | HLA-G; IDO1; IRF1; LGALS9; TLR3 |
| GO:0045088 | regulation of innate immune response | 6.70 | 0.00e+00 | 0.00e+00 | 30/221 | AIM2; CCL5; DDX58; DDX60; DHX58; GBP5; HLA-E; HLA-G; IFI16; IFIH1; IFNB1; IL18RAP; IRF1; IRF7; KIR2DL4; LAG3; LGALS9; NLRC5; NMI; PARP14; PARP9; PLSCR1; RSAD2; SAMHD1; SERPING1; SOCS1; STAT1; TLR3; USP18; ZBP1 |
| GO:0072527 | pyrimidine-containing compound metabolic process | 6.44 | 3.00e-04 | 5.09e-03 | 6/46 | APOBEC3A; APOBEC3F; APOBEC3G; APOBEC3H; CMPK2; TYMP |
| GO:0097696 | STAT cascade | 6.39 | 9.60e-07 | 3.26e-05 | 11/85 | CCL5; IFNB1; IL15; IFNL2; IFNL3; IFNL1; NMI; PARP14; PARP9; SOCS1; STAT1 |
| GO:0070206 | protein trimerization | 6.17 | 3.74e-03 | 4.83e-02 | 4/32 | HLA-G; MLKL; TRIM22; TRIM34 |
| GO:0002697 | regulation of immune effector process | 6.00 | 8.88e-16 | 8.03e-14 | 30/247 | AIM2; APOBEC3F; APOBEC3G; B2M; BST2; C2; CD80; CFB; DDX58; DDX60; DHX58; HERC5; HLA-E; HLA-F; HLA-G; IFIT1; IFNB1; IL15; IL18RAP; IL27; IFNL1; KIR2DL4; LAG3; LGALS9; PARP9; RSAD2; SERPING1; STAT1; TLR3; ZNF683 |
| GO:0001906 | cell killing | 5.94 | 2.26e-07 | 8.35e-06 | 13/108 | APOL1; B2M; GNLY; GZMB; HLA-E; HLA-F; HLA-G; IL18RAP; KIR2DL4; LAG3; LGALS9; PRF1; STAP1 |
| GO:0001818 | negative regulation of cytokine production | 5.74 | 1.85e-10 | 9.40e-09 | 20/172 | BST2; CARD16; CARD17; CD274; DDX58; DHX58; GBP1; HERC5; IDO1; IFIH1; IFNB1; IFNL1; ISG15; LAG3; LGALS9; NLRC5; NMI; PDCD1LG2; UBA7; UBE2L6 |
| GO:0006968 | cellular defense response | 5.74 | 1.65e-03 | 2.36e-02 | 5/43 | GNLY; HLA-G; KIR2DL4; LGALS3BP; PRF1 |
| GO:0070646 | protein modification by small protein removal | 5.66 | 2.12e-04 | 3.76e-03 | 7/61 | DDX58; IFIH1; PSMB10; PSMB8; PSMB9; PSME2; USP18 |
| GO:0032609 | interferon-gamma production | 5.61 | 1.00e-05 | 2.58e-04 | 10/88 | BTN3A1; BTN3A2; CD274; IL18RAP; IL27; IFNL1; ISG15; LGALS9; PDCD1LG2; TLR3 |
| GO:0061025 | membrane fusion | 5.57 | 2.35e-04 | 4.08e-03 | 7/62 | MX1; MX2; OTOF; SAMD9L; SAMD9; TAP1; TAP2 |
| GO:0032612 | interleukin-1 production | 5.32 | 3.17e-04 | 5.26e-03 | 7/65 | AIM2; CARD16; CARD17; CASP1; GBP5; IFI16; LGALS9 |
| GO:0002449 | lymphocyte mediated immunity | 5.31 | 1.56e-08 | 6.34e-07 | 17/158 | B2M; BTN3A2; BTN3A3; C2; GZMB; HLA-E; HLA-F; HLA-G; IFNB1; IL18RAP; IRF7; KIR2DL4; LAG3; LGALS9; PRF1; RSAD2; SERPING1 |
| GO:0002440 | production of molecular mediator of immune response | 4.98 | 4.52e-06 | 1.31e-04 | 12/119 | B2M; BST2; DDX58; HLA-E; HLA-G; IFNB1; IL18RAP; KIR2DL4; RSAD2; SAMHD1; TLR3; TNFSF13B |
| GO:0031348 | negative regulation of defense response | 4.90 | 5.38e-06 | 1.51e-04 | 12/121 | DHX58; HLA-E; HLA-G; IFI16; IFIT1; KIR2DL4; LGALS9; NLRC5; NMI; PARP14; SAMHD1; SERPING1 |
| GO:0001819 | positive regulation of cytokine production | 4.85 | 4.95e-12 | 3.35e-10 | 27/275 | AIM2; B2M; CASP1; CD274; CD80; DDX58; DDX60; DHX58; EIF2AK2; GBP5; HLA-E; HLA-G; HPSE; IDO1; IFI16; IFIH1; IL15; IL27; IFNL1; IRF1; IRF7; KIR2DL4; LGALS9; RSAD2; STAT1; TLR3; ZBP1 |
| GO:0031349 | positive regulation of defense response | 4.74 | 8.25e-12 | 5.17e-10 | 27/281 | AIM2; CCL5; DDX58; DDX60; DHX58; GBP5; HLA-E; HLA-G; IDO1; IFI16; IFIH1; IFNB1; IL15; IL18RAP; IRF1; IRF7; KIR2DL4; LAG3; LGALS9; NLRC5; OPTN; PARP9; PLSCR1; RSAD2; STAP1; TLR3; ZBP1 |
| GO:0042107 | cytokine metabolic process | 4.73 | 6.48e-04 | 9.76e-03 | 7/73 | CD80; IL27; IRF1; IRF7; LAG3; NMI; TLR3 |
| GO:0007249 | I-kappaB kinase/NF-kappaB signaling | 4.59 | 1.05e-05 | 2.58e-04 | 12/129 | APOL3; BST2; CARD16; CASP1; IFIT5; LGALS9; OPTN; SECTM1; STAT1; TLR3; TNFSF10; TRIM22 |
| GO:0050866 | negative regulation of cell activation | 4.59 | 1.05e-05 | 2.58e-04 | 12/129 | CD274; CD80; CTLA4; HLA-G; IDO1; IFNB1; IFNL1; IRF1; LAG3; LGALS9; PDCD1LG2; SOCS1 |
| GO:0002683 | negative regulation of immune system process | 4.58 | 4.60e-11 | 2.50e-09 | 26/280 | BST2; CD274; CD80; CTLA4; DHX58; GBP1; HLA-E; HLA-G; IDO1; IFI16; IFIT1; IFNB1; IFNL1; IRF1; KIR2DL4; LAG3; LGALS9; NLRC5; NMI; PARP14; PDCD1LG2; SAMHD1; SERPING1; SOCS1; STAP1; TLR3 |
| GO:0019882 | antigen processing and presentation | 4.49 | 3.84e-04 | 6.25e-03 | 8/88 | B2M; HLA-E; HLA-F; HLA-G; LAG3; PSMB8; TAP1; TAP2 |
| GO:0002250 | adaptive immune response | 4.45 | 5.11e-10 | 2.31e-08 | 24/266 | B2M; BTN3A1; BTN3A2; BTN3A3; C2; CD274; CD80; CTLA4; HLA-E; HLA-F; HLA-G; IFNB1; IL18RAP; IL27; IRF1; IRF7; LAMP3; PRF1; RSAD2; SERPING1; TAP1; TAP2; TNFSF13B; ZNF683 |
| GO:0071887 | leukocyte apoptotic process | 4.36 | 2.43e-03 | 3.36e-02 | 6/68 | CCL5; CD274; HSH2D; IDO1; IRF7; LGALS9 |
| GO:0050663 | cytokine secretion | 4.22 | 1.12e-05 | 2.67e-04 | 13/152 | AIM2; BTN3A1; BTN3A2; CARD16; CARD17; CASP1; CD274; DDX58; GBP1; GBP5; IFIH1; LGALS9; SOCS1 |
| GO:0070661 | leukocyte proliferation | 4.22 | 4.67e-07 | 1.65e-05 | 17/199 | BTN3A1; CCL5; CCL8; CD274; CD80; CTLA4; HLA-E; HLA-G; IDO1; IFNB1; IL15; IL27; IRF1; LGALS9; PDCD1LG2; PSMB10; TNFSF13B |
| GO:1903706 | regulation of hemopoiesis | 3.99 | 2.23e-07 | 8.35e-06 | 19/235 | B2M; CCR1; CD80; CTLA4; EIF2AK2; HLA-G; IFNB1; IL15; IL27; IFNL1; IRF1; IRF7; ISG15; LGALS9; RASSF2; SOCS1; STAT1; TLR3; ZNF683 |
| GO:0071216 | cellular response to biotic stimulus | 3.96 | 9.67e-05 | 1.83e-03 | 11/137 | CARD16; CARD17; CASP1; CCL5; CD274; CD80; CMPK2; CXCL10; CXCL11; PDCD1LG2; STAP1 |
| GO:0042110 | T cell activation | 3.82 | 1.12e-08 | 4.78e-07 | 24/310 | B2M; BTN3A1; CCL5; CD274; CD80; CTLA4; HLA-E; HLA-G; HSH2D; ICOS; IDO1; IFNB1; IL15; IL27; IFNL1; IRF1; LAG3; LGALS9; PDCD1LG2; PSMB10; RSAD2; SOCS1; TNFSF13B; ZNF683 |
| GO:0007162 | negative regulation of cell adhesion | 3.69 | 4.72e-05 | 9.61e-04 | 13/174 | CD274; CD80; CTLA4; GBP1; HLA-G; IDO1; IFNB1; IFNL1; IRF1; LAG3; LGALS9; PDCD1LG2; SOCS1 |
| GO:0007159 | leukocyte cell-cell adhesion | 3.62 | 3.96e-06 | 1.20e-04 | 17/232 | CCL5; CD274; CD80; CTLA4; HLA-E; HLA-G; ICOS; IDO1; IFNB1; IL15; IFNL1; IRF1; LAG3; LGALS9; PDCD1LG2; SOCS1; TNFSF13B |
| GO:0032103 | positive regulation of response to external stimulus | 3.51 | 2.18e-05 | 4.86e-04 | 15/211 | CCL5; CCL7; CCR1; CXCL10; DDX60; DHX58; FPR2; GBP5; HLA-E; IDO1; IL15; LGALS9; OPTN; STAP1; TLR3 |
| GO:0002237 | response to molecule of bacterial origin | 3.10 | 1.59e-04 | 2.88e-03 | 14/223 | B2M; CARD16; CARD17; CASP1; CCL5; CD274; CD80; CMPK2; CXCL10; CXCL11; IDO1; LGALS9; PDCD1LG2; STAP1 |
| GO:0002694 | regulation of leukocyte activation | 3.09 | 3.51e-06 | 1.10e-04 | 21/336 | CCL5; CD274; CD80; CTLA4; HLA-E; HLA-G; ICOS; IDO1; IFNB1; IL15; IL27; IFNL1; IRF1; LAG3; LGALS9; PDCD1LG2; PLSCR1; SOCS1; STAP1; TNFSF13B; ZNF683 |
| GO:0002521 | leukocyte differentiation | 3.09 | 6.14e-06 | 1.67e-04 | 20/320 | B2M; BATF2; CCR1; CD80; CTLA4; HLA-G; IFI16; IFNB1; IL15; IL27; IFNL1; IRF1; IRF7; LGALS9; RASSF2; RSAD2; SH3PXD2A; SOCS1; TLR3; ZNF683 |
| GO:0002764 | immune response-regulating signaling pathway | 3.06 | 2.05e-05 | 4.76e-04 | 18/290 | BTN3A1; BTN3A2; BTN3A3; CTLA4; DDX58; DDX60; DHX58; FPR2; GBP1; HLA-G; IFIH1; IRF1; IRF7; LGALS9; PLSCR1; RSAD2; STAP1; TLR3 |
| GO:0022407 | regulation of cell-cell adhesion | 3.01 | 4.47e-05 | 9.33e-04 | 17/279 | CCL5; CD274; CD80; CTLA4; HLA-E; HLA-G; ICOS; IDO1; IFNB1; IL15; IFNL1; IRF1; LAG3; LGALS9; PDCD1LG2; SOCS1; TNFSF13B |
| GO:0052547 | regulation of peptidase activity | 2.97 | 8.85e-05 | 1.71e-03 | 16/266 | AIM2; BST2; CARD16; CARD17; CASP1; FAS; IFI16; IFI27; IFI6; LAMP3; LGALS9; PSMB8; PSMB9; PSME2; SERPING1; TNFSF10 |
| GO:0050867 | positive regulation of cell activation | 2.93 | 4.73e-04 | 7.55e-03 | 13/219 | CCL5; CD274; CD80; CTLA4; HLA-E; HLA-G; ICOS; IL15; LGALS9; PDCD1LG2; SOCS1; STAP1; TNFSF13B |
| GO:0006959 | humoral immune response | 2.76 | 3.34e-03 | 4.46e-02 | 10/179 | BST2; C2; CFB; CXCL10; CXCL11; GNLY; HLA-E; IFNB1; PSMB10; SERPING1 |
| GO:0030099 | myeloid cell differentiation | 2.61 | 3.23e-03 | 4.38e-02 | 11/208 | B2M; BATF2; CCR1; IFI16; IRF7; ISG15; LGALS9; RASSF2; SH3PXD2A; STAT1; TLR3 |
| GO:0051090 | regulation of DNA-binding transcription factor activity | 2.59 | 3.48e-03 | 4.56e-02 | 11/210 | AIM2; CARD16; DDX58; EIF2AK2; IL18RAP; LGALS9; NLRC5; PARP10; TLR3; TRIM22; TRIM34 |
Factor 19 : 4 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0007272 | ensheathment of neurons | 3.63 | 9.43e-05 | 1.92e-02 | 12/74 | NCMAP; FAM126A; ID4; KLK6; KLK8; NRG1; NTRK2; NTRK3; RARB; SCN2A; SOX10; TLR2 |
| GO:0043588 | skin development | 2.88 | 8.21e-08 | 3.34e-05 | 31/241 | ALX4; CDH3; DACT2; DSC3; DSG1; DSG3; EDAR; FERMT1; ITGA2; ITGA3; ITGB4; KLK5; KRT14; KRT15; KRT16; KRT17; KRT5; KRT6A; KRT6B; KRT6C; KRT81; KRT83; KRT86; CERS3; LIPK; MET; PDGFA; PKP1; SOSTDC1; TGM5; TP63 |
| GO:0008544 | epidermis development | 2.87 | 1.23e-08 | 1.00e-05 | 35/273 | ALX4; BNC1; CDH3; COL17A1; COL7A1; DCT; DSC3; DSG1; DSG3; EDAR; FERMT1; KLK5; KLK7; KRT14; KRT15; KRT16; KRT17; KRT5; KRT6A; KRT6B; KRT6C; KRT81; KRT83; KRT86; LAMA3; LAMB3; LAMC2; CERS3; LIPK; NTF4; PDGFA; PKP1; SOSTDC1; TGM5; TP63 |
| GO:0043062 | extracellular structure organization | 2.11 | 6.03e-05 | 1.64e-02 | 31/329 | A2M; ADAMTSL2; COL11A1; COL17A1; COL4A6; COL7A1; FERMT1; GFAP; GPM6B; HAS3; HPSE2; ICAM5; ITGA10; ITGA2; ITGA3; ITGB4; ITGB6; KLK5; KLK7; LAMA3; LAMB3; LAMC2; LAMC3; LOXL4; MMP20; MMP7; MYH11; PDGFA; SERPINB5; SERPINF2; VIT |
Factor 20 : 60 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0001945 | lymph vessel development | 9.53 | 5.99e-07 | 3.48e-05 | 8/18 | EFNB2; FLT4; FOXC2; HEG1; SOX18; TMEM204; VEGFA; VEGFC |
| GO:0001570 | vasculogenesis | 7.42 | 7.01e-12 | 1.14e-09 | 18/52 | APLNR; CAV1; CD34; ACKR3; EGFL7; FOXF1; HEG1; HEY1; KDR; MYOCD; PDGFRB; RAMP2; RASIP1; SOX17; SOX18; TGFBR2; TIE1; VEGFA |
| GO:0035924 | cellular response to vascular endothelial growth factor stimulus | 6.95 | 5.64e-08 | 4.17e-06 | 12/37 | DLL4; FLT4; ADGRA2; KDR; JCAD; NRP1; PDGFRB; PGF; PTP4A3; RAMP2; VEGFA; VEGFC |
| GO:0003170 | heart valve development | 6.34 | 5.50e-08 | 4.17e-06 | 13/44 | ADAMTS9; DLL4; GJA5; HEY1; HEYL; JAG1; MEF2C; NOS3; PRDM1; SLIT2; SLIT3; TGFBR2; TNFRSF1B |
| GO:0003158 | endothelium development | 5.51 | 6.33e-10 | 7.37e-08 | 19/74 | APOLD1; CD34; CDH5; CLDN5; COL18A1; GJA4; GJA5; HEG1; HEY1; ID1; JAG1; KDR; NRP1; PDE2A; PECAM1; S1PR1; SOX17; SOX18; ZEB1 |
| GO:0060840 | artery development | 5.00 | 4.34e-07 | 2.72e-05 | 14/60 | ADAMTS9; DLL4; EFNB2; FOXF1; GJA5; ADGRF5; HEY1; JAG1; MYOCD; NRP1; PDGFRB; PRDM1; TBX2; VEGFA |
| GO:0010644 | cell communication by electrical coupling | 4.87 | 2.92e-03 | 3.78e-02 | 5/22 | CASQ2; CAV1; GJA5; GJC2; HRC |
| GO:0048010 | vascular endothelial growth factor receptor signaling pathway | 4.72 | 1.40e-05 | 5.71e-04 | 11/50 | ELMO1; FLT4; FYN; ITGB3; KDR; MMRN2; NRP1; PGF; TMEM204; VEGFA; VEGFC |
| GO:0009187 | cyclic nucleotide metabolic process | 4.55 | 6.74e-04 | 1.22e-02 | 7/33 | ADCY4; GUCY1A2; GUCY1B1; NPR1; PDE1A; PDE2A; PDE7B |
| GO:0048008 | platelet-derived growth factor receptor signaling pathway | 4.44 | 1.88e-03 | 2.59e-02 | 6/29 | MYOCD; NRP1; PDGFD; PDGFRB; PTGIR; VEGFA |
| GO:0060976 | coronary vasculature development | 4.44 | 1.88e-03 | 2.59e-02 | 6/29 | APLNR; MYOCD; NRP1; PDGFRB; PRDM1; VEGFA |
| GO:0061383 | trabecula morphogenesis | 4.41 | 8.15e-04 | 1.38e-02 | 7/34 | ADAMTS1; CCM2L; DLL4; HEG1; HEY1; S1PR1; TEK |
| GO:0001525 | angiogenesis | 4.41 | 0.00e+00 | 0.00e+00 | 71/345 | ADAMTS9; ANGPT2; APLNR; APOLD1; AQP1; ARHGAP24; CALCRL; CAV1; CD34; CDH13; CDH5; CLEC14A; COL15A1; COL18A1; COL4A1; COL4A2; CSPG4; ACKR3; DLL4; DYSF; ECSCR; EFNB2; EGFL7; ENPEP; ENPP2; EPAS1; ESM1; ETS1; FLT4; FOXC2; GJA5; ADGRA2; HEY1; HGF; HSPG2; ID1; ITGB3; JAG1; KDR; JCAD; MCAM; MEOX2; MMRN2; NOS3; NPR1; NRP1; PDGFRB; PECAM1; PGF; PTPRB; PTPRM; RAMP2; RAMP3; RASIP1; RHOJ; ROBO4; RSPO3; S1PR1; SCG2; SLIT2; SOX17; SOX18; STAB1; TAL1; TEK; TGFBR2; THSD7A; THY1; TIE1; VEGFA; VEGFC |
| GO:0090130 | tissue migration | 4.36 | 2.58e-14 | 1.05e-11 | 36/177 | ACTC1; ADAMTS9; ANGPT2; CDH13; CLEC14A; DLL4; EFNB2; ENPP2; ETS1; FLT4; FOXC2; FOXF1; ADGRA2; HBEGF; ID1; ITGB3; KDR; JCAD; MEF2C; MEOX2; MMRN2; NOS3; NRP1; PECAM1; PTP4A3; PTPRG; PTPRM; RHOJ; SCG2; SLIT2; SOX18; TEK; TGFBR2; VEGFA; VEGFC; ZEB2 |
| GO:0050918 | positive chemotaxis | 3.97 | 1.63e-04 | 3.68e-03 | 10/54 | ANGPT2; CDH13; HGF; KDR; NRP1; PGF; S1PR1; SCG2; VEGFA; VEGFC |
| GO:0001667 | ameboidal-type cell migration | 3.65 | 2.20e-13 | 5.96e-11 | 41/241 | ADAMTS9; ANGPT2; CDH13; CLEC14A; CYGB; DLL4; EDNRB; EFNB2; ENPP2; ETS1; FLT4; FOXC2; ADGRA2; HBEGF; ID1; ITGB3; KDR; JCAD; MEF2C; MEOX2; MMRN2; NOS3; NRP1; PECAM1; PTP4A3; PTPRG; PTPRM; RHOJ; SCG2; SEMA3G; SEMA5B; SEMA6B; SLIT2; SOX17; SOX18; TEK; TGFBR2; TNS1; VEGFA; VEGFC; ZEB2 |
| GO:1901342 | regulation of vasculature development | 3.62 | 4.44e-12 | 9.05e-10 | 37/219 | ADAMTS9; ANGPT2; APLNR; AQP1; CD34; CDH5; COL4A2; DLL4; ECSCR; EFNB2; ENPP2; ETS1; FOXC2; ADGRA2; HEY1; HGF; HSPG2; ID1; KDR; JCAD; MEOX2; MMRN2; MYOCD; NOS3; NPR1; NRP1; PDGFD; PGF; PTPRM; RAMP2; RHOJ; STAB1; TEK; TGFBR2; TIE1; VEGFA; VEGFC |
| GO:0003205 | cardiac chamber development | 3.45 | 1.85e-06 | 9.42e-05 | 19/118 | ADAMTS1; CCM2L; DLL4; FOXF1; GJA5; HEG1; HEY1; HEYL; JAG1; MEF2C; MYOCD; NOS3; NRP1; PRDM1; SLIT2; SLIT3; TBX2; TEK; TGFBR2 |
| GO:0003007 | heart morphogenesis | 3.25 | 4.39e-08 | 3.97e-06 | 27/178 | ACTC1; ADAMTS1; CCM2L; CLDN5; DLC1; DLL4; FAT4; FOXF1; GJA5; HEG1; HEY1; HEYL; JAG1; MEF2C; MYOM1; NOS3; NRP1; S1PR1; SLIT2; SLIT3; SOX17; SOX18; SPRY1; TBX2; TEK; TGFBR2; VEGFA |
| GO:0071559 | response to transforming growth factor beta | 3.24 | 1.55e-06 | 8.42e-05 | 21/139 | ADAMTSL2; CAV1; CAV2; CDH5; CLDN5; CLEC3B; COL4A2; FYN; ID1; ITGA8; LRRC32; NRROS; MEF2C; MYOCD; PDE2A; PDE3A; PDGFD; PRDM16; SMAD9; TGFBR2; ZEB1 |
| GO:0035051 | cardiocyte differentiation | 3.09 | 1.46e-04 | 3.68e-03 | 14/97 | ACTC1; ALPK3; EFNB2; JAG1; MEF2C; MYH11; MYOCD; MYOM1; PDGFRB; SOX17; SOX18; SPRY1; TBX2; VEGFA |
| GO:0014812 | muscle cell migration | 3.06 | 4.12e-03 | 4.73e-02 | 8/56 | ADAMTS1; ITGB3; MEF2C; MYOCD; NRP1; PDGFD; PDGFRB; SLIT2 |
| GO:0003014 | renal system process | 2.91 | 1.25e-03 | 1.95e-02 | 11/81 | ADCY4; AQP1; AVPR1A; CD34; EDNRB; F2R; GJA5; ADGRF5; HBB; MCAM; NPR1 |
| GO:0007498 | mesoderm development | 2.88 | 1.38e-03 | 2.12e-02 | 11/82 | BMX; ETS2; FOXC2; FOXF1; GDF3; ITGA8; ITGB3; SOX17; TAL1; TIE1; VEGFA |
| GO:0003013 | circulatory system process | 2.79 | 7.40e-11 | 1.00e-08 | 47/361 | ACTC1; ADRA1B; ARHGAP42; AVPR1A; BDKRB2; CASQ2; CAV1; CD34; CLIC2; DLL4; EDNRB; ENPEP; EPAS1; F2R; FLI1; FYN; GJA5; GUCY1B1; HBB; HBEGF; HEG1; HRC; HTR1B; ITGA1; KCNA5; KCNJ2; KCNJ8; KCNN2; MEOX2; MYL4; NOS2; NOS3; NPR1; P2RY1; PDE2A; PDE3A; PLN; PTP4A3; PTPRM; RAMP2; RAMP3; SCN4B; SLIT2; TBX2; TEK; VEGFC; VIP |
| GO:0050920 | regulation of chemotaxis | 2.75 | 3.41e-05 | 1.21e-03 | 20/156 | ANGPT2; CCL26; CDH13; DYSF; EFNB2; ADGRA2; KDR; MMP28; NRP1; PDGFD; PDGFRB; PGF; S1PR1; SCG2; SEMA3G; SEMA5B; SEMA6B; SLIT2; VEGFA; VEGFC |
| GO:0051056 | regulation of small GTPase mediated signal transduction | 2.75 | 3.54e-06 | 1.60e-04 | 25/195 | A2M; ADCYAP1R1; ARHGAP23; ARHGAP24; ARHGAP42; ARHGAP6; ARHGEF15; DLC1; F2RL3; F2R; FGD5; GPR20; HEG1; LPAR4; NGF; NRP1; P2RY8; PDGFRB; PLEKHG1; PREX2; RASIP1; RHOJ; SLIT2; SPRY1; SPRY4 |
| GO:0002576 | platelet degranulation | 2.74 | 2.04e-03 | 2.77e-02 | 11/86 | A2M; CLEC3B; HGF; ITGB3; ITIH3; MMRN1; PECAM1; SELP; VEGFA; VEGFC; VWF |
| GO:0048771 | tissue remodeling | 2.73 | 5.53e-04 | 1.04e-02 | 14/110 | CAV1; CSPG4; DLL4; EPAS1; F2R; FLT4; GJA5; HTR1B; ITGB3; JAG1; MEF2C; NOS3; RSPO3; S1PR1 |
| GO:0033002 | muscle cell proliferation | 2.72 | 1.50e-04 | 3.68e-03 | 17/134 | ADAMTS1; CALCRL; CDH13; HBEGF; HTR1B; MAPK11; MEF2C; MYOCD; NPR1; PDE1A; PDGFD; PDGFRB; PTGIR; S1PR1; TBX2; TGFBR2; VIP |
| GO:0090287 | regulation of cellular response to growth factor stimulus | 2.71 | 2.66e-05 | 1.03e-03 | 21/166 | ADAMTSL2; CAV1; CAV2; GDF3; ADGRA2; ITGA8; ITGB3; JCAD; NRROS; MMRN2; MYOCD; PRDM16; PTP4A3; SLIT2; SPRY1; SPRY4; TGFBR2; TMEM204; VEGFA; VEGFC; ZEB1 |
| GO:0019932 | second-messenger-mediated signaling | 2.59 | 3.04e-08 | 3.10e-06 | 39/323 | ADCY4; ADCYAP1R1; ADRA1B; ADRA1D; APLNR; AQP1; AVPR1A; CALCRL; CASQ2; CCR10; CDH13; CLIC2; ACKR3; EDNRB; ADGRL4; GNAI1; GNG2; GUCY1A2; GUCY1B1; HRC; KDR; ADGRL2; MCTP1; NOS2; NOS3; NPR1; NR5A2; PDE2A; PDE3A; PLN; PTGIR; RAMP2; RAMP3; RAPGEF4; RCAN2; SELE; SELP; VEGFA; VIP |
| GO:0007219 | Notch signaling pathway | 2.59 | 9.45e-04 | 1.57e-02 | 14/116 | CDH6; DLL4; EGFL7; FAT4; HEY1; HEYL; JAG1; KCNA5; MFNG; PEAR1; PLN; PTP4A3; TBX2; TGFBR2 |
| GO:2000147 | positive regulation of cell motility | 2.51 | 7.04e-08 | 4.77e-06 | 39/333 | ADAMTS1; VSIR; CAV1; CCL26; CDH13; COL18A1; ACKR3; DYSF; ENPP2; ETS1; F2R; FLT4; FOXC2; FOXF1; ADGRA2; HBEGF; HGF; ITGB3; KDR; JCAD; MCAM; NOS3; NRP1; PDGFD; PDGFRB; PGF; PLVAP; PODXL; RHOJ; S1PR1; SELP; SEMA3G; SEMA5B; SEMA6B; TEK; TGFBR2; THY1; VEGFA; VEGFC |
| GO:0034330 | cell junction organization | 2.46 | 1.61e-04 | 3.68e-03 | 20/174 | ARHGAP6; CAV1; CDH13; CDH5; CDH6; CDH8; CLDN5; DLC1; F2R; GJA4; GJA5; HEG1; KDR; LIMS2; NRP1; RAMP2; TEK; THY1; TNS1; VEGFA |
| GO:0035637 | multicellular organismal signaling | 2.46 | 1.09e-03 | 1.77e-02 | 15/131 | AVPR1A; CASQ2; CAV1; DMRT3; GJA5; HRC; KCNA5; KCNJ2; KCNN2; NPR1; PLN; S1PR1; SCN4A; SCN4B; SCN9A |
| GO:0031589 | cell-substrate adhesion | 2.45 | 1.26e-05 | 5.39e-04 | 27/236 | ADAMTS9; ANGPT2; ARHGAP6; CD34; CDH13; COL13A1; DLC1; EGFLAM; FOXF1; ID1; ITGA1; ITGA7; ITGA8; ITGB3; JAG1; KDR; LYVE1; MMRN1; MMRN2; NID1; NID2; NRP1; PECAM1; TEK; THY1; VEGFA; VWF |
| GO:0048863 | stem cell differentiation | 2.45 | 7.67e-04 | 1.36e-02 | 16/140 | A2M; EDNRB; FRZB; JAG1; LBH; MEF2C; MYOCD; NRP1; SEMA3G; SEMA5B; SEMA6B; SOX17; SOX18; TAL1; TBX2; ZEB2 |
| GO:0051271 | negative regulation of cellular component movement | 2.38 | 8.84e-05 | 2.48e-03 | 23/207 | ADAMTS9; ANGPT2; CD200; CYGB; DLC1; DLL4; IL33; JAG1; MCTP1; MEF2C; MEOX2; MMP28; MMRN2; MYOCD; NRP1; PTPRG; PTPRM; SEMA3G; SEMA5B; SEMA6B; SLIT2; THY1; TIE1 |
| GO:0040013 | negative regulation of locomotion | 2.36 | 1.03e-04 | 2.78e-03 | 23/209 | ADAMTS9; ANGPT2; CD200; CYGB; DLC1; DLL4; IL33; JAG1; MCTP1; MEF2C; MEOX2; MMP28; MMRN2; MYOCD; NRP1; PTPRG; PTPRM; SEMA3G; SEMA5B; SEMA6B; SLIT2; THY1; TIE1 |
| GO:0001655 | urogenital system development | 2.33 | 3.10e-05 | 1.15e-03 | 27/248 | ADAMTS1; ANGPT2; CD34; COL4A1; EFNB2; ENPEP; FAT4; FOXF1; HEYL; ID3; ITGA8; JAG1; KCNJ8; MEF2C; MYOCD; NID1; NRP1; PDGFD; PDGFRB; PECAM1; PGF; PODXL; SLIT2; SOX17; SPRY1; TEK; VEGFA |
| GO:0007265 | Ras protein signal transduction | 2.32 | 6.76e-05 | 2.12e-03 | 25/231 | ARHGAP42; ARHGAP6; ARHGEF15; CDH13; DLC1; ELMO1; F2RL3; F2R; FGD5; GPR20; GRAP; HEG1; LPAR4; NGF; NRP1; P2RY8; PDGFRB; PECAM1; PLEKHG1; PREX2; RASGRP2; RASIP1; RHOJ; SPRY1; SPRY4 |
| GO:0060537 | muscle tissue development | 2.30 | 4.12e-05 | 1.40e-03 | 27/252 | ACTC1; ADAMTS9; ALPK3; CCM2L; CAV1; CAV2; DLL4; EFNB2; GJA5; HEG1; HEYL; HLX; HOXD9; ITGA8; MAPK11; MEF2C; MEOX2; MYH11; MYOCD; MYOM1; PDGFRB; PLN; S1PR1; SOX17; TBX2; TGFBR2; VEGFA |
| GO:0006898 | receptor-mediated endocytosis | 2.29 | 4.22e-04 | 8.60e-03 | 20/187 | CALCRL; CAV1; CAV2; CFI; CUBN; ACKR3; EFNB2; ENPP2; HBA1; HBB; HSPG2; HTR1B; ITGB3; RAMP2; RAMP3; SELE; STAB1; SYT11; TGFBR2; VEGFA |
| GO:0043062 | extracellular structure organization | 2.28 | 3.37e-06 | 1.60e-04 | 35/329 | A2M; ACAN; ADAMTS4; ADAMTS9; ADAMTSL2; CD34; CETP; COL13A1; COL14A1; COL15A1; COL18A1; COL4A1; COL4A2; CSGALNACT1; EGFLAM; ETS1; FOXF1; HSPG2; ICAM2; ITGA1; ITGA7; ITGA8; ITGB3; JAM2; KDR; LAMA4; MYH11; NID1; NID2; OLFML2A; PECAM1; RAMP2; TNFRSF1B; TNXB; VWF |
| GO:0007187 | G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger | 2.25 | 5.56e-04 | 1.04e-02 | 20/191 | ADCY4; ADRA1B; ADRA1D; CALCRL; ADGRL4; GNAI1; GNG2; GRM2; GRM8; HTR1B; HTR1F; ADGRL2; P2RY1; PDE2A; PLN; PTGIR; RAMP2; RAMP3; S1PR1; VIP |
| GO:0046677 | response to antibiotic | 2.24 | 7.90e-04 | 1.37e-02 | 19/182 | ACTC1; ADCYAP1R1; AOC3; AQP1; CRHBP; ETS1; FYN; GNAI1; HBA1; HBB; HGF; HTR1B; ID1; ID3; KCNA5; MEF2C; PDGFD; PDGFRB; TRPC6 |
| GO:0050673 | epithelial cell proliferation | 2.23 | 7.11e-05 | 2.14e-03 | 27/260 | APLNR; CAV1; CAV2; CCL26; CD34; CDH13; DLL4; DYSF; EDNRB; EFNB2; EGFL7; FLT4; ID1; ITGB3; KDR; JCAD; LIMS2; MEF2C; NRP1; PGF; PTPRM; SCG2; TEK; VEGFA; VEGFC; VIP; WFDC1 |
| GO:0010959 | regulation of metal ion transport | 2.21 | 8.12e-05 | 2.36e-03 | 27/262 | ADCYAP1R1; APLNR; CASQ2; CAV1; CLIC2; DYSF; F2RL3; F2R; FYN; GEM; GJC2; HECW2; HRC; KCNA5; KCNJ2; KCNN2; LRRC55; NOS3; PDGFRB; PLN; RAMP3; REM1; SCN4B; TESC; THY1; TRPC6; VIP |
| GO:0001763 | morphogenesis of a branching structure | 2.20 | 3.22e-03 | 3.91e-02 | 15/146 | COL13A1; COL4A1; DLL4; FAT4; FOXF1; HGF; NRP1; PGF; PRDM1; RASIP1; RSPO3; SLIT2; SPRY1; TGFBR2; VEGFA |
| GO:0060485 | mesenchyme development | 2.17 | 1.17e-03 | 1.87e-02 | 19/188 | ACTC1; EDNRB; FOXF1; FRZB; HEY1; HEYL; HGF; JAG1; MEF2C; NOS3; NRP1; PDGFRB; SEMA3G; SEMA5B; SEMA6B; SPRY1; TBX2; TGFBR2; ZEB2 |
| GO:0090066 | regulation of anatomical structure size | 2.16 | 5.04e-05 | 1.64e-03 | 30/298 | ADRA1B; AQP1; ARHGAP42; AVPR1A; BDKRB2; CAV1; CCL26; EDNRB; F2R; GJA5; GMFG; HBB; HTR1B; ISLR2; ITGA1; KANK3; KCNA5; LRRC8C; NGF; NOS3; NPR1; NRP1; P2RY1; PTPRM; SEMA3G; SEMA5B; SEMA6B; SLIT2; VEGFA; VIP |
| GO:0042692 | muscle cell differentiation | 2.14 | 5.64e-04 | 1.04e-02 | 22/220 | ACTC1; ALPK3; AVPR1A; CASQ2; CAV2; DYSF; EDNRB; EFNB2; FOXF1; HEY1; ITGA8; MAPK11; MEF2C; MYH11; MYOCD; MYOM1; PDGFRB; RAMP2; TBX2; TMEM204; VEGFA; ZEB1 |
| GO:0043087 | regulation of GTPase activity | 2.14 | 3.40e-04 | 7.09e-03 | 24/241 | A2M; ARHGAP23; ARHGAP24; ARHGAP42; ARHGAP6; ARHGEF15; CAV2; CCL14; CCL26; DLC1; F2R; FGD5; GPR21; NRP1; PLXNA2; PREX2; PTPRN2; RABGAP1; RASGRP2; RASIP1; RGS5; S1PR1; SLIT2; SPRY1; THY1 |
| GO:0050817 | coagulation | 2.10 | 1.70e-03 | 2.47e-02 | 19/194 | A2M; CAV1; CD34; ENPP4; F2RL3; F2R; FYN; GNG2; HBB; ITGB3; MMRN1; NOS3; P2RY1; PEAR1; SELP; TFPI; THBD; TRPC6; VWF |
| GO:0043270 | positive regulation of ion transport | 2.06 | 2.70e-03 | 3.60e-02 | 18/187 | ABCB1; ADCYAP1R1; APLNR; AVPR1A; CAV1; F2RL3; F2R; GJC2; KCNJ2; LRRC55; P2RY1; PDGFRB; RAMP3; SCN4B; SLC6A1; TESC; THY1; TRPC6 |
| GO:0003012 | muscle system process | 2.03 | 1.56e-04 | 3.68e-03 | 30/317 | ACTC1; ADRA1B; ARHGAP42; BDKRB2; CALCRL; CASQ2; CAV1; CLIC2; DYSF; EDNRB; F2R; GJA5; HRC; ITGA1; KCNA5; KCNJ2; KCNJ8; KCNN2; MEF2C; MYH11; MYL4; MYOCD; MYOM1; NOS3; P2RY1; PLN; SCN4A; SCN4B; SGCA; TNFRSF1B |
| GO:0050878 | regulation of body fluid levels | 2.03 | 2.08e-04 | 4.46e-03 | 29/307 | A2M; ADCY4; APLNR; AQP1; CAV1; CD34; EDNRB; ENPP4; F2RL3; F2R; FLI1; FYN; GJA5; GNG2; HBB; HEG1; ITGB3; MMRN1; NOS3; NPR1; P2RY1; PEAR1; SELP; TFPI; THBD; TRPC6; VEGFA; VIP; VWF |
| GO:0048880 | sensory system development | 2.02 | 1.62e-03 | 2.40e-02 | 21/223 | ARHGEF15; COL4A1; DLL4; GDF3; JAG1; MAF; MFRP; MYOM1; NES; NRP1; PDGFRB; PRDM1; PTPRM; RHOJ; TBX2; TGFBR2; THY1; UNC45B; VEGFA; ZEB1; ZEB2 |
| GO:0034765 | regulation of ion transmembrane transport | 2.01 | 1.39e-04 | 3.65e-03 | 31/330 | ABCB1; APLNR; CASQ2; CAV1; CLIC2; CLIC5; CRHBP; DYSF; F2RL3; F2R; FYN; GEM; GJA5; GJC2; HECW2; HRC; KCNA5; KCNJ2; KCNJ8; KCNN2; LRRC55; MEF2C; PLN; RAMP3; REM1; SCN4A; SCN4B; SCN9A; TESC; THY1; TRPC6 |
| Factor | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
| Signif_GO_terms | 0 | 0 | 12 | 49 | 106 | 0 | 4 | 27 | 11 | 0 | 13 | 0 | 67 | 59 | 6 | 94 | 2 | 63 | 4 | 60 |
5 DEG Interpretation
In GSFA, differential expression analysis can be performed based on the LFSR method. Here we evaluate the specific downstream genes affected by the perturbations detected by GSFA.
We also performed several other differential expression methods for comparison, including Welch's t-test.
5.1 Number of DEGs detected by different methods
| KO | TP53_LOF | TP53_mis | CDH1_LOF | CDH1_mis | GATA3_LOF |
| Num_genes | 772 | 1610 | 1633 | 0 | 377 |
| KO | GATA3_mis | MAP3K1_LOF | MAP3K1_mis | PIK3CA_LOF | PIK3CA_mis |
| Num_genes | 0 | 0 | 0 | 0 | 1141 |
Number of DEGs detected under each perturbation using different methods:
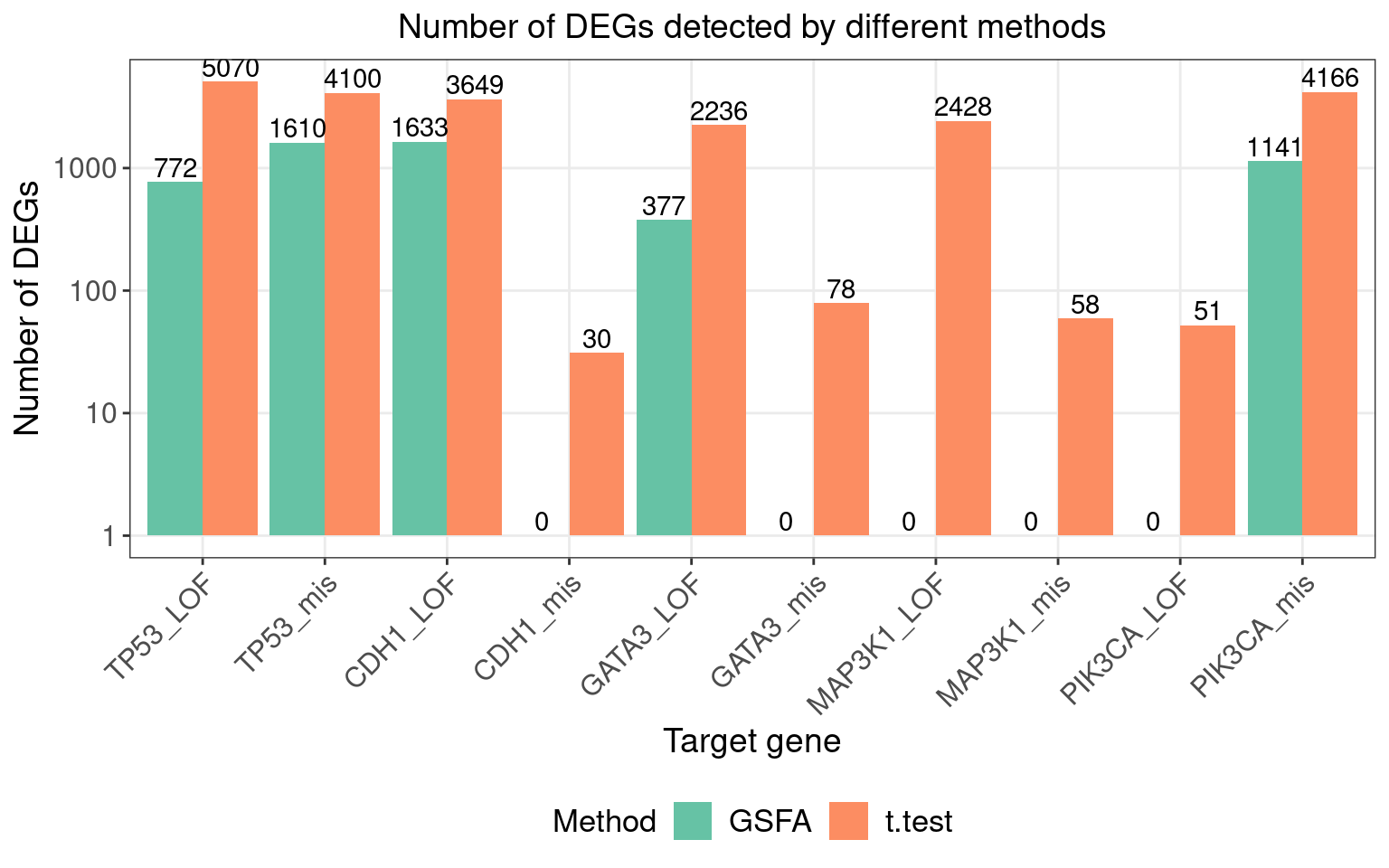
5.2 Perturbation effects on marker genes
To better understand the functions of these mutations, we examined their effects on marker genes for cell proliferation and T cell activation/resting states.
5.2.1 GSFA
Here are the summarized effects of perturbations on marker genes estimated by GSFA.
As we can see, TP53 loss of function/missense mutations have positive effects on cell proliferation, while CDH1 loss of function and PIK2CA missense mutations have negative effects on cell proliferation.
CDH1 LOF is also positively associated with T cell markers (for both activation and resting states).
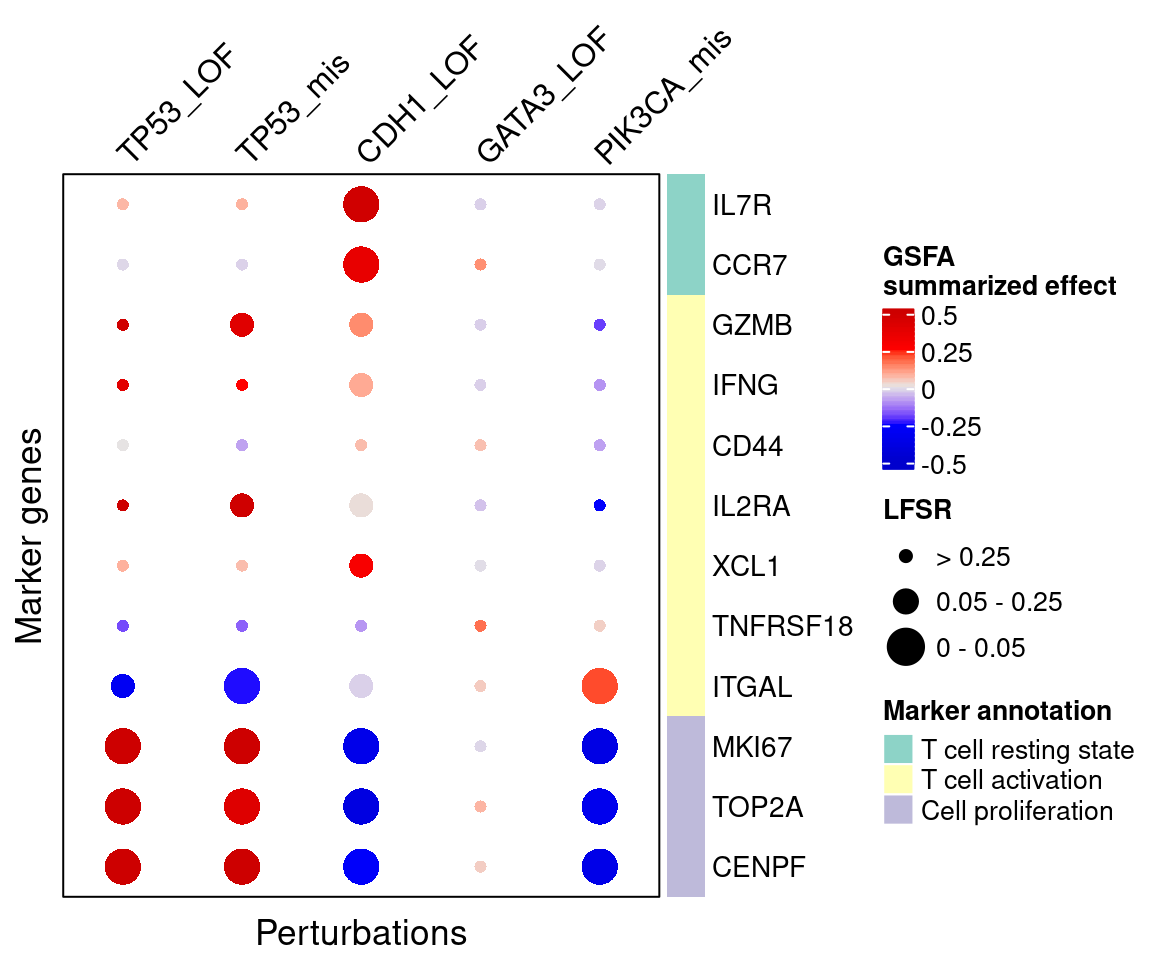
5.3 GO enrichment in DEGs
We further examine these DEGs for enrichment of relevant biological processes through GO enrichment analysis.
Foreground genes: Genes w/ GSFA LFSR < 0.05 under each perturbation;
Background genes: all 10k genes used in GSFA;
Statistical test: hypergeometric test (over-representation test);
Gene sets: Gene ontology "Biological Process" (non-redundant).
GO terms that passed over-representation test fold change \(\geq\) 2 and q value \(<\) 0.05:
TP53_LOF : 62 significant GO terms| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0051383 | kinetochore organization | 12.00 | 5.80e-11 | 1.75e-09 | 10/11 | CDT1; CENPA; CENPE; CENPF; CENPK; CENPN; CENPW; NDC80; NUF2; SMC4 |
| GO:0061641 | CENP-A containing chromatin organization | 10.20 | 1.31e-09 | 3.33e-08 | 10/13 | KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; HJURP; CENPU; OIP5 |
| GO:0034502 | protein localization to chromosome | 9.62 | 4.51e-14 | 1.67e-12 | 16/22 | AURKB; BRCA2; BUB1B; KNL1; CDCA5; CDK1; CDT1; CENPA; ESCO2; EZH2; HASPIN; MTBP; NDC80; PLK1; RAD21; TTK |
| GO:0050000 | chromosome localization | 9.31 | 4.44e-16 | 1.81e-14 | 19/27 | CCNB1; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; CHMP4C; DLGAP5; FAM83D; KIF14; KIF18A; KIF2C; KIFC1; NDC80; NUF2; PSRC1; RRS1; SPAG5 |
| GO:0036297 | interstrand cross-link repair | 8.82 | 3.83e-07 | 8.20e-06 | 8/12 | EME1; FANCA; FANCB; FANCE; FANCI; RAD51AP1; RAD51; UBE2T |
| GO:0006260 | DNA replication | 7.99 | 0.00e+00 | 0.00e+00 | 58/96 | ATAD5; BARD1; BLM; BRCA1; BRCA2; BRIP1; TICRR; RMI2; MCMDC2; CCNA2; CCNE1; CCNE2; CDC45; CDC6; CDC7; CDK1; CDT1; CHAF1B; CHEK1; CHTF18; CLSPN; DBF4; DDX11; DNA2; DSCC1; DTL; E2F7; E2F8; EME1; ESCO2; EXO1; FBXO5; FEN1; GINS1; GINS2; GINS4; GMNN; PCLAF; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; TONSL; ORC1; PIF1; POLD1; POLE2; POLQ; RAD51; RECQL4; RFC4; RNASEH2A; RRM2; ALYREF; TOP1MT; WDHD1 |
| GO:0007059 | chromosome segregation | 7.83 | 0.00e+00 | 0.00e+00 | 74/125 | AURKB; BIRC5; BLM; BRCA1; BRIP1; BUB1B; BUB1; KNSTRN; RMI2; MCMDC2; KNL1; CCNB1; CCNE1; CCNE2; CDC20; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CENPN; CENPW; CEP55; CHMP4C; DDX11; DLGAP5; DSCC1; ECT2; EME1; ESCO2; ESPL1; FAM83D; FBXO5; FEN1; HASPIN; HJURP; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; MAD2L1; MKI67; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NUF2; NUSAP1; OIP5; PLK1; PRC1; PSRC1; PTTG1; RACGAP1; RAD21; RRS1; SGO1; SGO2; SKA1; SKA2; SKA3; SMC4; SPAG5; TACC3; NDC1; TOP2A; TRIP13; TTK; ZWINT |
| GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 7.74 | 0.00e+00 | 0.00e+00 | 31/53 | AURKA; AURKB; SAPCD2; CCNB1; CDC20; CENPA; CENPE; CHMP4C; ESPL1; GPSM2; CEP126; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; NUSAP1; PLK1; POC1A; PRC1; PSRC1; RACGAP1; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0045023 | G0 to G1 transition | 7.12 | 1.56e-05 | 2.76e-04 | 7/13 | BRCA1; CDC7; CHEK1; E2F1; EZH2; RAD51; RRM2 |
| GO:0000075 | cell cycle checkpoint | 7.07 | 0.00e+00 | 0.00e+00 | 39/73 | AURKA; AURKB; BLM; BRCA1; BRIP1; BUB1B; BUB1; TICRR; CCNB1; CDC20; CDC25C; CDC45; CDC6; CDK1; CDT1; CENPF; CHEK1; CHMP4C; CLSPN; DNA2; DTL; E2F1; E2F7; E2F8; EME1; GTSE1; H2AFX; KNTC1; MAD2L1; NDC80; NEK11; ORC1; PLK1; PRKDC; TOP2A; TRIP13; TTK; WDR76; ZWINT |
| GO:0007051 | spindle organization | 6.81 | 0.00e+00 | 0.00e+00 | 34/66 | ASPM; AURKA; AURKB; KNSTRN; AUNIP; CCNB1; CDC20; CENPE; CHMP4C; ESPL1; FBXO5; GPSM2; WASHC5; CEP126; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; PLK1; POC1A; PRC1; PSRC1; RACGAP1; SPAG5; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0032200 | telomere organization | 6.61 | 1.77e-10 | 4.98e-09 | 16/32 | AURKB; BLM; BRCA2; CCNE1; CCNE2; DNA2; EXO1; FEN1; MYC; NEK2; PIF1; POLD1; PRKDC; RAD51; RECQL4; RFC4 |
| GO:0006301 | postreplication repair | 6.61 | 1.13e-04 | 1.88e-03 | 6/12 | BRCA1; DTL; PCLAF; POLD1; POLE2; RFC4 |
| GO:0006302 | double-strand break repair | 6.52 | 0.00e+00 | 0.00e+00 | 35/71 | BARD1; BLM; BRCA1; BRCA2; BRIP1; PARPBP; RMI2; AUNIP; MCMDC2; CDC45; CDC7; CDCA5; CHEK1; DDX11; DEK; DNA2; EME1; ESCO2; EXO1; FANCB; FEN1; FOXM1; GINS2; GINS4; H2AFX; TONSL; POLQ; PRKDC; RAD21; RAD51AP1; RAD51; RAD54B; RAD54L; RECQL4; TRIP13 |
| GO:0000910 | cytokinesis | 6.28 | 1.11e-16 | 5.02e-15 | 28/59 | ANLN; AURKA; AURKB; BRCA2; CDC25B; CDC6; CDT1; CENPA; CEP55; CHMP4C; CIT; CKAP2; CSPP1; E2F7; E2F8; ECT2; ESPL1; WASHC5; KIF14; KIF20A; KIF23; KIF4A; NUSAP1; PLK1; PRC1; RACGAP1; SEPT3; STMN1 |
| GO:0000726 | non-recombinational repair | 6.23 | 1.34e-05 | 2.47e-04 | 8/17 | BARD1; BLM; BRCA1; AUNIP; DEK; H2AFX; POLQ; PRKDC |
| GO:0071166 | ribonucleoprotein complex localization | 6.11 | 1.97e-04 | 3.02e-03 | 6/13 | DDX39A; HHEX; NUP210; RRS1; ALYREF; NDC1 |
| GO:0044839 | cell cycle G2/M phase transition | 6.06 | 0.00e+00 | 0.00e+00 | 38/83 | ATAD5; AURKA; AURKB; BLM; BRCA1; TICRR; CCNA2; CCNB1; CCNB2; CDC25A; CDC25B; CDC25C; CDC6; CDC7; CDK1; CENPF; CHEK1; CHMP4C; CIT; CLSPN; DTL; FBXO5; FOXM1; GTSE1; HMMR; KIF14; MELK; NEK10; NEK2; ORC1; PCM1; PKMYT1; PLK1; PLK4; RAD21; RECQL4; SKP2; TPX2 |
| GO:0071103 | DNA conformation change | 5.94 | 0.00e+00 | 0.00e+00 | 44/98 | ASF1B; BLM; BRIP1; KNL1; CCNB1; CDC45; CDCA5; CDK1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; DDX11; DNA2; GINS1; GINS2; GINS4; H2AFX; HELLS; HHEX; HJURP; HMGB2; MCM2; MCM4; MCM6; MCM7; CENPU; NCAPD2; NCAPG2; NCAPG; NCAPH; NUSAP1; OIP5; PIF1; RAD51; RAD54B; RAD54L; RECQL4; SMC4; TOP1MT; TOP2A |
| GO:0031023 | microtubule organizing center organization | 5.85 | 5.24e-11 | 1.64e-09 | 19/43 | AURKA; BRCA1; BRCA2; CCNF; CDK1; CHEK1; CHMP4C; CNTLN; PCLAF; KIF11; NEK2; PCM1; PLK1; PLK4; POC1A; SGO1; STIL; TRIM37; WDR62 |
| GO:0051653 | spindle localization | 5.79 | 8.50e-05 | 1.44e-03 | 7/16 | ASPM; SAPCD2; CENPA; ESPL1; GPSM2; NDC80; NUSAP1 |
| GO:0001578 | microtubule bundle formation | 5.70 | 8.89e-14 | 3.15e-12 | 25/58 | ARMC4; CFAP43; DRC1; CFAP206; CFAP157; CCDC103; CCDC114; CCDC65; DNAH7; DNAI2; DNAJB13; HYDIN; KIF20A; LRGUK; LRRC49; DNAAF1; LRRC6; NCKAP5; PLK1; PRC1; PSRC1; RSPH1; RSPH4A; TEKT2; TPPP3 |
| GO:0006284 | base-excision repair | 5.67 | 3.22e-04 | 4.86e-03 | 6/14 | DNA2; FEN1; NEIL3; POLD1; POLQ; RECQL4 |
| GO:0044772 | mitotic cell cycle phase transition | 5.65 | 0.00e+00 | 0.00e+00 | 82/192 | ANLN; ATAD5; AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; TICRR; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN2C; CDKN2D; CDKN3; CDT1; CENPE; CENPF; CHMP4C; CIT; CKS1B; CKS2; CLSPN; DBF4; DLGAP5; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FBXO5; FOXM1; GMNN; GTSE1; HMMR; IQGAP3; KIF14; KNTC1; MAD2L1; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MELK; MTBP; MYC; NDC80; NEK11; NEK2; ORC1; PCM1; PKMYT1; PLK1; PLK4; POLE2; PRKDC; RAD21; RBL1; RECQL4; RRM2; SKP2; TACC3; TPX2; TRIP13; TTK; TYMS; UBE2C; UBE2S |
| GO:0021591 | ventricular system development | 5.51 | 1.23e-03 | 1.67e-02 | 5/12 | ARMC4; AK8; CENPF; HYDIN; NME5 |
| GO:0001539 | cilium or flagellum-dependent cell motility | 5.41 | 1.54e-05 | 2.76e-04 | 9/22 | DRC1; CCDC65; DNAH6; DNAH7; SPEF1; TEKT1; TEKT2; TEKT3; CFAP57 |
| GO:0048285 | organelle fission | 5.27 | 0.00e+00 | 0.00e+00 | 86/216 | ANLN; ASPM; AURKA; AURKB; BLM; BRCA2; BRIP1; BUB1B; BUB1; KNSTRN; MCMDC2; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CCNO; CDC20; CDC25B; CDC25C; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; CHEK1; CHMP4C; CKS2; DLGAP5; DSCC1; EME1; ESPL1; MTFR2; FANCA; FBXO5; HASPIN; WASHC5; KIF11; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; KNTC1; MAD2L1; MKI67; MND1; MTBP; MTFR1; MYBL1; MYBL2; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NUF2; NUSAP1; PKMYT1; PLK1; PRC1; PSRC1; PTTG1; RACGAP1; RAD21; RAD51; RAD54B; RRS1; SGO1; SGO2; SMC4; SPAG5; TACC3; NDC1; TOP2A; TPX2; TRIP13; TTK; UBE2C; UBE2S; ZWINT |
| GO:1901987 | regulation of cell cycle phase transition | 5.15 | 0.00e+00 | 0.00e+00 | 58/149 | ANLN; ATAD5; AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; TICRR; CCNB1; CCNE1; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN2D; CDT1; CENPE; CENPF; CHEK1; CHMP4C; CLSPN; DLGAP5; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FAM83D; FBXO5; GTSE1; HMMR; KIF14; KNTC1; MAD2L1; MTBP; NDC80; NEK10; NEK11; NEK2; ORC1; PCM1; PLK1; PLK4; PRKDC; RAD21; RBL1; RECQL4; TPX2; TRIP13; TTK; UBE2C |
| GO:0010948 | negative regulation of cell cycle process | 5.12 | 0.00e+00 | 0.00e+00 | 43/111 | AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; TICRR; CCNB1; CCNF; CDC20; CDC25C; CDC6; CDC7; CDK1; CDKN2D; CDT1; CENPF; CHEK1; CHMP4C; CLSPN; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FBXO5; GTSE1; MAD2L1; MTBP; NDC80; NEK2; ORC1; PLK1; PRKDC; PTTG1; RAD21; RAD51; RBL1; RRM2; TRIM37; TRIP13; TTK |
| GO:0044843 | cell cycle G1/S phase transition | 4.98 | 0.00e+00 | 0.00e+00 | 41/109 | AURKA; CCNA2; CCNB1; CCNE1; CCNE2; CDC25A; CDC25C; CDC45; CDC6; CDC7; CDK1; CDKN2C; CDKN2D; CDKN3; CDT1; DBF4; E2F1; E2F7; E2F8; EZH2; FAM83D; FBXO5; GMNN; GTSE1; IQGAP3; KIF14; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MTBP; MYC; ORC1; POLE2; PRKDC; RBL1; RRM2; SKP2; TYMS |
| GO:0007018 | microtubule-based movement | 4.96 | 0.00e+00 | 0.00e+00 | 48/128 | ARMC4; DRC1; MAATS1; CFAP206; CCDC103; CCDC114; CFAP53; CCDC65; CENPE; CNIH2; DLGAP5; DNAH12; DNAH6; DNAH7; DNAH9; DNAI2; HYDIN; KIF11; KIF14; KIF15; KIF17; KIF18A; KIF18B; KIF20A; KIF21A; KIF23; KIF24; KIF2C; KIF4A; KIF6; KIFC1; KIFC2; DNAAF1; LRRC6; MAK; NME5; CFAP221; PCM1; RACGAP1; ROPN1L; RSPH4A; SPA17; TEKT1; TEKT2; TEKT3; WDR34; WDR63; WDR78 |
| GO:1904029 | regulation of cyclin-dependent protein kinase activity | 4.81 | 1.09e-09 | 2.87e-08 | 20/55 | BLM; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CCNO; CDC25A; CDC25C; CDC6; CDKN2C; CDKN2D; CDKN3; CKS1B; CKS2; HHEX; PKMYT1; PLK1; PSRC1 |
| GO:0044380 | protein localization to cytoskeleton | 4.81 | 1.25e-04 | 2.04e-03 | 8/22 | ANLN; AURKA; FAM83D; FAM83H; PCM1; SPAG5; STIL; TTK |
| GO:0045930 | negative regulation of mitotic cell cycle | 4.80 | 0.00e+00 | 0.00e+00 | 41/113 | AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; TICRR; CCNB1; CDC20; CDC25C; CDC6; CDK1; CDT1; CENPF; CHEK1; CHMP4C; CLSPN; E2F1; E2F7; E2F8; EME1; EZH2; FBXO5; GTSE1; KNTC1; MAD2L1; MTBP; NDC80; NEK11; NKX3-1; ORC1; PLK1; PRKDC; PTTG1; RAD21; RBL1; SCRIB; TOP2A; TRIP13; TTK; ZWINT |
| GO:0006310 | DNA recombination | 4.76 | 2.22e-16 | 9.51e-15 | 36/100 | ATAD5; BLM; BRCA1; BRCA2; BRIP1; PARPBP; RMI2; AUNIP; MCMDC2; CDC45; CDC7; CHEK1; EME1; EXO1; FANCB; FEN1; GINS2; GINS4; H2AFX; HMGB2; KPNA2; MND1; TONSL; PIF1; POLQ; PRKDC; RAD21; RAD51AP1; RAD51; RAD54B; RAD54L; RECQL4; TFRC; ALYREF; TOP2A; TRIP13 |
| GO:0033044 | regulation of chromosome organization | 4.58 | 1.22e-15 | 4.73e-14 | 36/104 | ATAD2; AURKB; BRCA1; BUB1B; BUB1; RMI2; CCNB1; CDC20; CDC45; CDC6; CDCA5; CDT1; CENPE; CENPF; CHEK1; DDX11; DLGAP5; DNMT3B; ESPL1; FBXO5; FEN1; MAD2L1; MCM2; MKI67; MYC; NDC80; NEK2; PHF19; PIF1; PLK1; PTTG1; RAD21; TACC3; TOP2A; TRIP13; TTK |
| GO:0042770 | signal transduction in response to DNA damage | 4.58 | 1.86e-08 | 4.33e-07 | 18/52 | ATAD5; AURKA; BRCA1; BRCA2; CCNB1; CDC25C; CDK1; CHEK1; DTL; E2F1; E2F7; E2F8; FOXM1; GTSE1; NDRG1; PLK1; PMAIP1; PRKDC |
| GO:0051321 | meiotic cell cycle | 4.58 | 0.00e+00 | 0.00e+00 | 45/130 | ASPM; AURKA; BLM; BRCA2; BRIP1; BUB1B; BUB1; MCMDC2; CCNE1; CCNE2; CDC20; CDC25A; CDC25B; CDC25C; CKS2; EME1; ESPL1; EXO1; FANCA; FBXO5; H2AFX; WASHC5; KIF18A; MND1; MYBL1; NCAPD2; NCAPH; NEK2; NUF2; PKMYT1; PLK1; PRKACB; PTTG1; RAD21; RAD51; RAD54B; RAD54L; RSPH1; SGO1; SGO2; SMC4; NDC1; TOP2A; TRIP13; TTK |
| GO:0015949 | nucleobase-containing small molecule interconversion | 4.41 | 3.85e-03 | 4.74e-02 | 5/15 | AK8; CTPS1; GLRX; NME2; TYMS |
| GO:0071824 | protein-DNA complex subunit organization | 4.03 | 6.81e-10 | 1.85e-08 | 25/82 | ANP32E; ASF1B; MCMDC2; KNL1; CDC45; CDT1; CENPA; CENPE; CENPF; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; GMNN; H2AFX; HELLS; HJURP; HMGB2; MCM2; CENPU; MYC; OIP5; RAD51 |
| GO:0071897 | DNA biosynthetic process | 3.97 | 2.27e-07 | 5.00e-06 | 18/60 | AURKB; BLM; CDKN2D; CENPF; CHTF18; DSCC1; DTL; PCLAF; LIN9; MYC; NEK2; PIF1; POLD1; POLE2; POLQ; RFC4; TK1; TYMS |
| GO:0006338 | chromatin remodeling | 3.97 | 2.34e-06 | 4.75e-05 | 15/50 | ANP32E; KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHEK1; HELLS; HJURP; HMGB2; CENPU; MYC; OIP5 |
| GO:2001020 | regulation of response to DNA damage stimulus | 3.90 | 6.82e-09 | 1.68e-07 | 23/78 | ATAD5; BLM; BRCA1; DDIAS; PARPBP; RMI2; AUNIP; CDKN2D; CHEK1; DDX11; DEK; ELL3; FANCB; FBXO5; FOXM1; H2AFX; MYC; NKX3-1; PMAIP1; POLQ; RAD51AP1; RAD51; WDR76 |
| GO:0045787 | positive regulation of cell cycle | 3.81 | 0.00e+00 | 0.00e+00 | 55/191 | ATAD5; AURKA; AURKB; BRCA1; BRCA2; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CCNO; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDT1; CHEK1; CIT; CKS1B; CKS2; CSPP1; DDX11; DLGAP5; DTL; E2F1; E2F7; E2F8; ECT2; ESPL1; EZH2; FAM83D; FBXO5; FEN1; GPSM2; GTSE1; KIF14; KIF23; MAD2L1; MTBP; NDC80; NKX3-1; NUSAP1; PLK4; POC1A; PSRC1; RACGAP1; RAD21; RECQL4; SPAG5; UBE2C |
| GO:0032886 | regulation of microtubule-based process | 3.74 | 1.03e-10 | 3.00e-09 | 30/106 | ARMC4; AURKA; BRCA1; DRC1; CCDC65; CCNF; CHEK1; CHMP4C; CKAP2; CNIH2; GPSM2; KIF11; KIF18A; DNAAF1; NEK2; PLK1; PLK4; POC1A; PRKAA2; PSRC1; SKA1; SKA2; SKA3; SPAG5; SPEF1; STIL; STMN1; TACC3; TPX2; TRIM37 |
| GO:0006997 | nucleus organization | 3.61 | 5.61e-04 | 8.16e-03 | 9/33 | CCNB1; CCNB2; CDK1; CEP55; CHMP4C; GOLM1; HMGB2; PLK1; NDC1 |
| GO:0051052 | regulation of DNA metabolic process | 3.60 | 2.95e-13 | 9.61e-12 | 40/147 | ATAD5; AURKB; BLM; BRCA1; BRCA2; PARPBP; TICRR; RMI2; AUNIP; CCNA2; CDC6; CDC7; CDK1; CDT1; CHEK1; CHTF18; DBF4; DDX11; DEK; DNA2; DSCC1; E2F7; E2F8; ESCO2; FANCB; FBXO5; FOXM1; GMNN; H2AFX; KPNA2; MYC; NEK2; PIF1; POLQ; RAD51AP1; RAD51; RECQL4; RFC4; TFRC; ALYREF |
| GO:0031109 | microtubule polymerization or depolymerization | 3.54 | 1.10e-05 | 2.19e-04 | 15/56 | CKAP2; FBXO5; KIF14; KIF18A; KIF18B; KIF24; KIF2C; NCKAP5; PSRC1; SKA1; SKA2; SKA3; SPEF1; STMN1; TPPP3 |
| GO:0044782 | cilium organization | 3.54 | 1.39e-13 | 4.71e-12 | 42/157 | ARMC4; CFAP43; CFAP126; DRC1; CATIP; CFAP206; CFAP157; CCDC103; CCDC114; CFAP53; CCDC13; CCDC65; CCNO; CDK1; DNAH7; DNAI2; DNAJB13; HYDIN; IQUB; CEP126; KIF17; KIF24; LRGUK; LRRC49; DNAAF1; LRRC6; MAK; NEK2; NME5; NPHP1; CFAP221; PCM1; PLK1; PLK4; POC1A; RSPH1; RSPH4A; SEPT3; TEKT1; TEKT2; TEKT3; WDR34 |
| GO:0072331 | signal transduction by p53 class mediator | 3.42 | 1.93e-07 | 4.37e-06 | 22/85 | ATAD5; AURKA; AURKB; BOP1; BRCA1; BRCA2; CCNB1; CDC25C; CDK1; CHEK1; E2F1; E2F2; E2F7; E2F8; ELL3; FOXM1; GTSE1; NDRG1; PMAIP1; PYCARD; RRS1; TPX2 |
| GO:0071826 | ribonucleoprotein complex subunit organization | 3.40 | 8.97e-04 | 1.28e-02 | 9/35 | BOP1; TICRR; AGO2; EIF3H; MTERF3; PIH1D2; PRKAA2; PSIP1; RRS1 |
| GO:0006333 | chromatin assembly or disassembly | 3.36 | 1.18e-05 | 2.26e-04 | 16/63 | ASF1B; KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; H2AFX; HELLS; HJURP; HMGB2; MCM2; CENPU; OIP5 |
| GO:0051302 | regulation of cell division | 3.17 | 4.63e-07 | 9.67e-06 | 23/96 | ASPM; AURKA; AURKB; BLM; BRCA2; CDC25B; CDC6; CHMP4C; CIT; CSPP1; E2F7; E2F8; ECT2; KIF14; KIF18B; KIF20A; KIF23; MYC; NKX3-1; PLK1; PRC1; RACGAP1; RBL1 |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 3.09 | 1.19e-05 | 2.26e-04 | 18/77 | ATAD5; BOP1; DNA2; AGO2; EME1; EXO1; EXOSC4; FEN1; HMGB2; RIDA; ISG20; LACTB2; REXO5; POLD1; POP1; RFC4; RNASEH2A; RRS1 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 3.05 | 3.87e-04 | 5.72e-03 | 12/52 | BOP1; TICRR; DCAF13; AGO2; EIF3H; EXOSC4; ISG20; MTERF3; PIH1D2; PRKAA2; PSIP1; RRS1 |
| GO:0031503 | protein-containing complex localization | 2.94 | 7.02e-05 | 1.22e-03 | 16/72 | BIRC5; CNIH2; DDX39A; GPC4; HASPIN; HHEX; KIF17; MAK; NUP210; PCM1; RRS1; SCRIB; STX1B; ALYREF; NDC1; WDR34 |
| GO:0010639 | negative regulation of organelle organization | 2.84 | 1.02e-08 | 2.43e-07 | 35/163 | ATAD2; AURKB; BRCA1; BUB1B; BUB1; ARPIN; CCNB1; CCNF; CDC20; CDT1; CENPF; CHEK1; CKAP2; DLC1; DNMT3B; ESPL1; FBXO5; MAD2L1; MAK; MCM2; MTBP; NDC80; NEK2; PIF1; PLEKHH2; PLK1; PTTG1; RAD21; SPEF1; STMN1; TMSB15A; TMSB15B; TOP2A; TRIM37; TRIP13; TTK |
| GO:0022406 | membrane docking | 2.80 | 1.45e-03 | 1.94e-02 | 11/52 | CDK1; KIF24; NEK2; NPHP1; PCM1; PLK1; PLK4; SNPH; STX1B; SYTL2; TSNARE1 |
| GO:1903320 | regulation of protein modification by small protein conjugation or removal | 2.46 | 1.97e-03 | 2.51e-02 | 13/70 | BRCA1; CDC20B; CDC20; FANCI; FBXO5; MAD2L1; MTBP; PLK1; RIPK2; SKP2; UBE2C; UBE2S; VPS28 |
| GO:0007050 | cell cycle arrest | 2.43 | 1.62e-04 | 2.53e-03 | 20/109 | AURKA; BARD1; BRCA1; DDIAS; CCNB1; CDC25C; CDK1; CDKN2C; CDKN2D; CDKN3; E2F1; E2F7; E2F8; FOXM1; GTSE1; MTBP; MYC; NKX3-1; PRKAA2; PRR11 |
| GO:0007292 | female gamete generation | 2.34 | 2.14e-03 | 2.68e-02 | 14/79 | ASPM; AURKA; BRCA2; MCMDC2; CCNB1; CDC25B; FBXO5; WASHC5; NCAPH; PLK1; TOP2A; TRIP13; TTK; YBX2 |
| GO:0010498 | proteasomal protein catabolic process | 2.15 | 1.14e-03 | 1.57e-02 | 19/117 | AURKA; AURKB; BUB1B; CCNB1; CCNF; CDC20; CDK1; FBXL6; FBXO5; KIF14; MAD2L1; PBK; PLK1; PMAIP1; PTTG1; SHARPIN; SKP2; UBE2C; UBE2S |
TP53_mis : 44 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0051383 | kinetochore organization | 5.58 | 1.19e-07 | 4.02e-06 | 10/11 | CDT1; CENPA; CENPE; CENPF; CENPK; CENPN; CENPW; NDC80; NUF2; SMC4 |
| GO:0034502 | protein localization to chromosome | 5.02 | 2.21e-11 | 1.28e-09 | 18/22 | AURKB; BRCA2; BUB1B; KNL1; CDCA5; CDK1; CDT1; CENPA; ESCO2; ESR1; EZH2; HASPIN; MTBP; NDC80; PARP3; PLK1; RAD21; TTK |
| GO:0061641 | CENP-A containing chromatin organization | 4.72 | 2.26e-06 | 6.58e-05 | 10/13 | KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; HJURP; CENPU; OIP5 |
| GO:0050000 | chromosome localization | 4.09 | 6.21e-09 | 2.66e-07 | 18/27 | CCNB1; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; DLGAP5; FAM83D; KIF14; KIF18A; KIF2C; KIFC1; NDC80; NUF2; PSRC1; RRS1; SPAG5 |
| GO:0036297 | interstrand cross-link repair | 4.09 | 1.29e-04 | 3.01e-03 | 8/12 | EME1; FANCA; FANCB; FANCE; FANCI; RAD51AP1; RAD51; UBE2T |
| GO:0007059 | chromosome segregation | 3.83 | 0.00e+00 | 0.00e+00 | 78/125 | AURKB; BIRC5; BLM; BRCA1; BRIP1; BUB1B; BUB1; KNSTRN; RMI2; M1AP; MCMDC2; C9orf84; KNL1; CCNB1; CCNE1; CCNE2; CDC20; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CENPN; CENPW; CEP55; DDX11; DLGAP5; DSCC1; ECT2; EME1; ESCO2; ESPL1; FAM83D; FBXO5; FEN1; HASPIN; HJURP; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; MAD2L1; MKI67; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NUF2; NUSAP1; OIP5; PLK1; PRC1; PSRC1; PTTG1; RACGAP1; RAD21; RGS14; RRS1; SGO1; SGO2; SKA1; SKA2; SKA3; SMC1B; SMC4; SPAG5; SYCP1; TACC3; NDC1; TOP2A; TRIP13; TTK; ZWINT |
| GO:0006260 | DNA replication | 3.77 | 0.00e+00 | 0.00e+00 | 59/96 | ATAD5; BARD1; BLM; BRCA1; BRCA2; BRIP1; TICRR; RMI2; MCMDC2; CCNA2; CCNE1; CCNE2; CDC45; CDC6; CDC7; CDK1; CDT1; CHAF1B; CHEK1; CHTF18; CLSPN; DBF4; DDX11; DNA2; DSCC1; DTL; E2F7; E2F8; EGFR; EME1; ESCO2; EXO1; FBXO5; FEN1; GINS1; GINS2; GINS4; GMNN; PCLAF; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; TONSL; NT5M; ORC1; PIF1; POLD1; POLE2; POLQ; RAD51; RECQL4; RFC4; RNASEH2A; RRM2; ALYREF; WDHD1 |
| GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 3.59 | 3.09e-12 | 2.51e-10 | 31/53 | AURKA; AURKB; SAPCD2; CCNB1; CDC20; CENPA; CENPE; ESPL1; GPSM2; CEP126; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; NUSAP1; PARP3; PLK1; POC1A; PRC1; PSRC1; RACGAP1; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0000075 | cell cycle checkpoint | 3.36 | 4.26e-14 | 4.96e-12 | 40/73 | AURKA; AURKB; BLM; BRCA1; BRIP1; BUB1B; BUB1; TICRR; CCNB1; CCND1; CDC20; CDC25C; CDC45; CDC6; CDK1; CDT1; CENPF; CHEK1; CLSPN; DNA2; DTL; E2F1; E2F7; E2F8; EME1; GTSE1; H2AFX; KNTC1; MAD2L1; NDC80; NEK11; ORC1; PLK1; PRKDC; SOX11; TOP2A; TRIP13; TTK; WDR76; ZWINT |
| GO:0001539 | cilium or flagellum-dependent cell motility | 3.35 | 4.32e-05 | 1.13e-03 | 12/22 | DRC1; CCDC39; CCDC65; DNAH11; DNAH6; DNAH7; SPATA4; SPEF1; TEKT1; TEKT2; TEKT3; CFAP57 |
| GO:0045023 | G0 to G1 transition | 3.31 | 2.07e-03 | 3.44e-02 | 7/13 | BRCA1; CDC7; CHEK1; E2F1; EZH2; RAD51; RRM2 |
| GO:0015949 | nucleobase-containing small molecule interconversion | 3.27 | 1.06e-03 | 1.92e-02 | 8/15 | AK4; AK8; CTPS1; GSR; GUK1; NME1; NME2; TYMS |
| GO:0032200 | telomere organization | 3.26 | 1.75e-06 | 5.27e-05 | 17/32 | AURKB; BLM; BRCA2; CCNE1; CCNE2; DNA2; EXO1; FEN1; HMBOX1; NEK2; PARP3; PIF1; POLD1; PRKDC; RAD51; RECQL4; RFC4 |
| GO:0007051 | spindle organization | 3.26 | 6.05e-12 | 4.10e-10 | 35/66 | ASPM; AURKA; AURKB; KNSTRN; AUNIP; CCNB1; CDC20; CENPE; ESPL1; FBXO5; GPSM2; WASHC5; CEP126; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; PARP3; PLK1; POC1A; PRC1; PSRC1; RACGAP1; RGS14; SPAG5; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0071103 | DNA conformation change | 3.13 | 1.33e-15 | 2.17e-13 | 50/98 | ASF1B; BLM; BRIP1; M1AP; KNL1; CCNB1; CDC45; CDCA5; CDK1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; CHD5; DDX11; DNA2; GINS1; GINS2; GINS4; GPER1; H2AFX; HELLS; HHEX; HJURP; HMGB2; HMGB3; MCM2; MCM4; MCM6; MCM7; CENPU; NAP1L5; NCAPD2; NCAPG2; NCAPG; NCAPH; NUSAP1; OIP5; PIF1; RAD51; RAD54B; RAD54L; RECQL4; SMC4; SYCP1; SYCP3; TOP2A |
| GO:0006302 | double-strand break repair | 3.11 | 1.70e-11 | 1.06e-09 | 36/71 | BARD1; BLM; BRCA1; BRCA2; BRIP1; PARPBP; RMI2; AUNIP; MCMDC2; CDC45; CDC7; CDCA5; CHEK1; DDX11; DEK; DNA2; EME1; ESCO2; EXO1; FANCB; FEN1; FOXM1; GINS2; GINS4; H2AFX; TONSL; PARP3; POLQ; PRKDC; RAD21; RAD51AP1; RAD51; RAD54B; RAD54L; RECQL4; TRIP13 |
| GO:0051653 | spindle localization | 3.07 | 1.82e-03 | 3.09e-02 | 8/16 | ASPM; SAPCD2; CENPA; ESPL1; GPSM2; LLGL2; NDC80; NUSAP1 |
| GO:0044839 | cell cycle G2/M phase transition | 3.03 | 1.99e-12 | 1.89e-10 | 41/83 | ATAD5; AURKA; AURKB; BLM; BRCA1; BTRC; TICRR; CCNA2; CCNB1; CCNB2; CCNB3; CCND1; CDC25A; CDC25B; CDC25C; CDC6; CDC7; CDK1; CENPF; CHEK1; CIT; CLSPN; DTL; FBXO5; FOXM1; GTSE1; HMMR; KIF14; MELK; NEK10; NEK2; ORC1; PCM1; PKMYT1; PLK1; PLK4; RAD21; RECQL4; SKP2; STOX1; TPX2 |
| GO:0001578 | microtubule bundle formation | 2.96 | 1.27e-08 | 4.91e-07 | 28/58 | ARMC4; CFAP43; DRC1; CFAP206; CFAP157; CCDC103; CCDC114; CCDC39; CCDC65; DNAH7; DNAI2; DNAJB13; DNALI1; HYDIN; KIF20A; LRGUK; LRRC49; DNAAF1; LRRC6; NCKAP5; NEURL1; PLK1; PRC1; PSRC1; RSPH1; RSPH4A; TEKT2; ZMYND10 |
| GO:0000726 | non-recombinational repair | 2.89 | 2.95e-03 | 4.45e-02 | 8/17 | BARD1; BLM; BRCA1; AUNIP; DEK; H2AFX; POLQ; PRKDC |
| GO:0048285 | organelle fission | 2.84 | 0.00e+00 | 0.00e+00 | 100/216 | ANLN; ASPM; AURKA; AURKB; BLM; BMP7; BNIP3; BRCA2; BRIP1; BUB1B; BUB1; KNSTRN; M1AP; MCMDC2; C9orf84; CCNA2; CCNB1; CCNB2; CCNB3; CCND1; CCNE1; CCNE2; CCNF; CCNI2; CDC20; CDC25B; CDC25C; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; CHEK1; CKS2; DDHD2; DLGAP5; DMRTC2; DSCC1; EME1; EPGN; ESPL1; MTFR2; FANCA; FBXO5; GDAP1; HASPIN; IL1A; WASHC5; KIF11; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; KNTC1; MAD2L1; MKI67; MND1; MTBP; MTFR1; MYBL1; MYBL2; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NUF2; NUSAP1; PKMYT1; PLK1; PPARGC1A; PRC1; PSRC1; PTTG1; RACGAP1; RAD21; RAD51; RAD54B; RRS1; SGO1; SGO2; SMC4; SPAG5; SYCP1; SYCP3; TACC3; NDC1; TOP2A; TPX2; TRIP13; TTK; UBE2C; UBE2S; WNT4; ZWINT |
| GO:0000910 | cytokinesis | 2.81 | 9.50e-08 | 3.36e-06 | 27/59 | ANLN; AURKA; AURKB; BRCA2; CDC25B; CDC6; CDT1; CENPA; CEP55; CIT; CKAP2; E2F7; E2F8; ECT2; ESPL1; WASHC5; KIF14; KIF20A; KIF23; KIF4A; NUSAP1; PLK1; PRC1; PRKCE; RACGAP1; SEPT3; STMN1 |
| GO:0044772 | mitotic cell cycle phase transition | 2.78 | 0.00e+00 | 0.00e+00 | 87/192 | ANLN; ATAD5; AURKA; AURKB; BCL2; BLM; BRCA1; BTRC; BUB1B; BUB1; TICRR; CCNA2; CCNB1; CCNB2; CCNB3; CCND1; CCNE1; CCNE2; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN2C; CDKN2D; CDKN3; CDT1; CENPE; CENPF; CIT; CKS1B; CKS2; CLSPN; DBF4; DLGAP5; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FBXO5; FOXM1; GMNN; GTSE1; HMMR; IQGAP3; KIF14; KNTC1; MAD2L1; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MELK; MTBP; NDC80; NEK11; NEK2; ORC1; PCM1; PKMYT1; PLK1; PLK4; POLE2; PRKDC; RAD21; RBL1; RECQL4; RHOU; RRM2; SKP2; STOX1; TACC3; TPX2; TRIP13; TTK; TYMS; UBE2C; UBE2S; USP26 |
| GO:1901987 | regulation of cell cycle phase transition | 2.64 | 6.33e-15 | 8.59e-13 | 64/149 | ANLN; ATAD5; AURKA; AURKB; BCL2; BLM; BRCA1; BTRC; BUB1B; BUB1; C10orf99; TICRR; CCNB1; CCNB3; CCND1; CCNE1; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN2D; CDT1; CENPE; CENPF; CHEK1; CLSPN; DLGAP5; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FAM83D; FBXO5; GTSE1; HMMR; KIF14; KNTC1; MAD2L1; MTBP; NDC80; NEK10; NEK11; NEK2; ORC1; PCM1; PLK1; PLK4; PRKDC; RAD21; RBL1; RECQL4; STOX1; SUSD2; TPX2; TRIP13; TTK; UBE2C |
| GO:0010948 | negative regulation of cell cycle process | 2.60 | 4.94e-11 | 2.68e-09 | 47/111 | AURKA; AURKB; BCL2; BLM; BMP7; BRCA1; BUB1B; BUB1; C10orf99; TICRR; CCNB1; CCND1; CCNF; CDC20; CDC25C; CDC6; CDC7; CDK1; CDKN2D; CDT1; CENPF; CHEK1; CLSPN; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FBXO5; GPER1; GTSE1; MAD2L1; MTBP; NDC80; NEK2; ORC1; PLK1; PRKDC; PTTG1; RAD21; RAD51; RBL1; RRM2; SUSD2; TRIP13; TTK |
| GO:0044843 | cell cycle G1/S phase transition | 2.59 | 9.11e-11 | 4.64e-09 | 46/109 | AURKA; BCL2; C10orf99; CCNA2; CCNB1; CCND1; CCNE1; CCNE2; CDC25A; CDC25C; CDC45; CDC6; CDC7; CDK1; CDKN2C; CDKN2D; CDKN3; CDT1; DBF4; E2F1; E2F7; E2F8; EZH2; FAM83D; FBXO5; GMNN; GTSE1; IQGAP3; KIF14; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MTBP; ORC1; POLE2; PRKDC; RBL1; RHOU; RRM2; SKP2; SUSD2; TYMS; USP26 |
| GO:0007018 | microtubule-based movement | 2.59 | 2.09e-12 | 1.89e-10 | 54/128 | ARMC4; DRC1; MAATS1; CFAP206; CABYR; CCDC103; CCDC114; CFAP53; CCDC39; CCDC65; CENPE; CNIH2; DLGAP5; DNAH11; DNAH12; DNAH6; DNAH7; DNAH9; DNAI2; HIF1A; HYDIN; KIF11; KIF14; KIF15; KIF18A; KIF18B; KIF20A; KIF21A; KIF21B; KIF23; KIF24; KIF2C; KIF4A; KIF6; KIFC1; KIFC2; DNAAF1; LRRC6; MAK; NME5; CFAP221; PCM1; RACGAP1; RASGRP1; ROPN1L; RSPH4A; SPA17; TEKT1; TEKT2; TEKT3; WDR34; WDR63; WDR66; WDR78 |
| GO:0031023 | microtubule organizing center organization | 2.57 | 5.85e-05 | 1.44e-03 | 18/43 | AURKA; BRCA1; BRCA2; CALML5; CCNF; CDK1; CHEK1; PCLAF; KIF11; NEK2; PARD6B; PCM1; PLK1; PLK4; POC1A; SGO1; STIL; WDR62 |
| GO:1904029 | regulation of cyclin-dependent protein kinase activity | 2.57 | 5.74e-06 | 1.61e-04 | 23/55 | BLM; CCNA2; CCNB1; CCNB2; CCNB3; CCND1; CCNE1; CCNE2; CCNF; CCNI2; CDC25A; CDC25C; CDC6; CDKN2C; CDKN2D; CDKN3; CKS1B; CKS2; HHEX; PKMYT1; PLK1; PSRC1; STOX1 |
| GO:0051321 | meiotic cell cycle | 2.55 | 4.40e-12 | 3.26e-10 | 54/130 | ASPM; AURKA; BLM; BRCA2; BRIP1; BUB1B; BUB1; M1AP; MCMDC2; C9orf84; CCNB3; CCNE1; CCNE2; CDC20; CDC25A; CDC25B; CDC25C; CKS2; DMRTC2; EME1; ESPL1; EXO1; FAM9A; FANCA; FBXO5; H2AFX; WASHC5; KIF18A; MND1; MYBL1; NCAPD2; NCAPH; NEK2; NUF2; PKMYT1; PLK1; PTTG1; RAD21; RAD51; RAD54B; RAD54L; RSPH1; SGO1; SGO2; SMC1B; SMC4; SYCP1; SYCP3; TDRD1; NDC1; TOP2A; TRIP13; TTK; WNT4 |
| GO:0006338 | chromatin remodeling | 2.46 | 5.03e-05 | 1.28e-03 | 20/50 | KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHD5; CHEK1; ESR1; HELLS; HJURP; HMGB2; HMGB3; CENPU; OIP5; PABPC1L; PADI2; PAX7; SYCP3 |
| GO:0018149 | peptide cross-linking | 2.42 | 5.16e-04 | 1.05e-02 | 15/38 | CSTA; IVL; KRT2; PI3; SPRR1A; SPRR1B; SPRR2A; SPRR2B; SPRR2D; SPRR2E; SPRR2F; SPRR3; SPRR4; TGM2; TGM4 |
| GO:0033044 | regulation of chromosome organization | 2.42 | 1.08e-08 | 4.41e-07 | 41/104 | ATAD2; AURKB; BRCA1; BUB1B; BUB1; RMI2; CAMK2D; CCNB1; CDC20; CDC45; CDC6; CDCA5; CDT1; CENPE; CENPF; CHEK1; DDX11; DLGAP5; DMRTC2; DNMT3B; ESPL1; FBXO5; FEN1; HMBOX1; MAD2L1; MCM2; MKI67; NDC80; NEK2; PADI2; PAX7; PHF19; PIF1; PLK1; PPARGC1A; PTTG1; RAD21; TACC3; TOP2A; TRIP13; TTK |
| GO:0006997 | nucleus organization | 2.42 | 1.24e-03 | 2.17e-02 | 13/33 | CCNB1; CCNB2; CDK1; CEP55; CHD5; DMRTC2; GPER1; HMGB2; PLK1; REEP4; SYCP1; SYCP3; NDC1 |
| GO:0006310 | DNA recombination | 2.39 | 3.52e-08 | 1.30e-06 | 39/100 | ATAD5; BLM; BRCA1; BRCA2; BRIP1; PARPBP; RMI2; AUNIP; MCMDC2; C9orf84; CDC45; CDC7; CHEK1; EME1; EXO1; FANCB; FEN1; GINS2; GINS4; H2AFX; HMGB2; HMGB3; KPNA2; MND1; TONSL; PIF1; POLQ; PRKDC; RAD21; RAD51AP1; RAD51; RAD54B; RAD54L; RECQL4; SYCP1; TFRC; ALYREF; TOP2A; TRIP13 |
| GO:0045930 | negative regulation of mitotic cell cycle | 2.39 | 4.95e-09 | 2.24e-07 | 44/113 | AURKA; AURKB; BCL2; BLM; BMP7; BRCA1; BUB1B; BUB1; TICRR; CCNB1; CCND1; CDC20; CDC25C; CDC6; CDK1; CDT1; CENPF; CHEK1; CLSPN; E2F1; E2F7; E2F8; EGFR; EME1; EZH2; BRINP3; FBXO5; GTSE1; KNTC1; MAD2L1; MTBP; NDC80; NEK11; ORC1; PLK1; PRKDC; PTTG1; RAD21; RBL1; SCRIB; TOP2A; TRIP13; TTK; ZWINT |
| GO:0042770 | signal transduction in response to DNA damage | 2.24 | 3.15e-04 | 6.76e-03 | 19/52 | ATAD5; AURKA; BRCA1; BRCA2; CCNB1; CDC25C; CDK1; CHEK1; DTL; E2F1; E2F7; E2F8; FOXM1; GTSE1; NDRG1; PLK1; PMAIP1; PRKDC; SNAI1 |
| GO:0045787 | positive regulation of cell cycle | 2.15 | 1.12e-10 | 5.37e-09 | 67/191 | ASCL1; ATAD5; AURKA; AURKB; BRCA1; BRCA2; CCNA2; CCNB1; CCNB2; CCNB3; CCND1; CCNE1; CCNE2; CCNF; CCNI2; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDT1; CHEK1; CIT; CKS1B; CKS2; DDX11; DLGAP5; DTL; E2F1; E2F7; E2F8; ECT2; EPGN; ESPL1; EZH2; FAM83D; FBXO5; FEN1; FGFR2; FOSL1; GATA6; GPER1; GPSM2; GTSE1; IL1A; KIF14; KIF23; MAD2L1; MTBP; MYOG; NDC80; NUSAP1; PLK4; POC1A; PRKCE; PSRC1; RACGAP1; RAD21; RECQL4; SLC6A4; SPAG5; STOX1; UBE2C; WNT4 |
| GO:0071897 | DNA biosynthetic process | 2.15 | 3.09e-04 | 6.76e-03 | 21/60 | AURKB; BLM; CDKN2D; CENPF; CHTF18; DSCC1; DTL; FGFR4; HMBOX1; PCLAF; LIN9; NEK2; PARP3; PIF1; POLD1; POLE2; POLQ; RFC4; SYCP1; TK1; TYMS |
| GO:0018210 | peptidyl-threonine modification | 2.05 | 2.79e-03 | 4.29e-02 | 16/48 | AZU1; BCL2; BMP7; CAMK2D; CDC7; CDK1; CHEK1; DDIT4; DMTN; GALNT3; HASPIN; PLK1; RIPK2; STOX1; TGFBR1; TTK |
| GO:0044782 | cilium organization | 2.03 | 1.24e-07 | 4.02e-06 | 52/157 | ABLIM1; ARMC4; B9D1; CFAP43; CFAP126; DRC1; CATIP; C5orf30; CFAP206; CFAP157; CCDC103; CCDC114; CFAP53; CCDC13; CCDC39; CCDC65; CDK1; DCDC2; DNAH7; DNAI2; DNAJB13; DNALI1; FOXJ1; FUZ; HYDIN; IQUB; CEP126; KIF24; LRGUK; LRRC49; DNAAF1; LRRC6; MAK; NEK2; NEURL1; NME5; NOTO; NPHP1; CFAP221; PCM1; PLK1; PLK4; POC1A; RSPH1; RSPH4A; SEPT3; TEKT1; TEKT2; TEKT3; UBXN10; WDR34; ZMYND10 |
| GO:0072331 | signal transduction by p53 class mediator | 2.02 | 1.12e-04 | 2.67e-03 | 28/85 | ATAD5; AURKA; AURKB; BCL2; BOP1; BRCA1; BRCA2; CCNB1; CDC25C; CDK1; CHD5; CHEK1; DDIT4; E2F1; E2F2; E2F7; E2F8; ELL3; FOXM1; GTSE1; NDRG1; NUPR1; PERP; PMAIP1; PYCARD; RRS1; SNAI1; TPX2 |
| GO:0071824 | protein-DNA complex subunit organization | 2.02 | 1.49e-04 | 3.36e-03 | 27/82 | ASF1B; MCMDC2; KNL1; CDC45; CDT1; CENPA; CENPE; CENPF; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; CHD5; ESR1; GMNN; H2AFX; HELLS; HJURP; HMGB2; MCM2; CENPU; NAP1L5; OIP5; RAD51; SYCP3 |
| GO:0051052 | regulation of DNA metabolic process | 2.00 | 6.08e-07 | 1.90e-05 | 48/147 | ANXA3; ATAD5; AURKB; BLM; BRCA1; BRCA2; PARPBP; TICRR; RMI2; AUNIP; CCNA2; CDC6; CDC7; CDK1; CDT1; CHEK1; CHTF18; DBF4; DDX11; DEK; DNA2; DSCC1; E2F7; E2F8; EGFR; ESCO2; FANCB; FBXO5; FGFR4; FOXM1; GMNN; GPER1; H2AFX; HMBOX1; KPNA2; NEK2; PARP3; PIF1; POLQ; PPARGC1A; RAD51AP1; RAD51; RECQL4; RFC4; STOX1; TFRC; ALYREF; ZNF93 |
CDH1_LOF : 31 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0051383 | kinetochore organization | 4.95 | 3.88e-07 | 1.31e-05 | 10/11 | CDT1; CENPA; CENPE; CENPF; CENPK; CENPN; CENPW; NDC80; NUF2; SMC4 |
| GO:0061641 | CENP-A containing chromatin organization | 4.19 | 7.02e-06 | 1.63e-04 | 10/13 | KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; HJURP; CENPU; OIP5 |
| GO:0050000 | chromosome localization | 3.83 | 4.53e-09 | 2.17e-07 | 19/27 | ANKRD53; CCNB1; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; DLGAP5; FAM83D; GEM; KIF14; KIF18A; KIF2C; KIFC1; NDC80; NUF2; PSRC1; SPAG5 |
| GO:0031579 | membrane raft organization | 3.63 | 3.09e-04 | 5.48e-03 | 8/12 | CAV1; CAV2; CD2; DOCK2; GSN; MAL; PTPRC; RFTN1 |
| GO:0034502 | protein localization to chromosome | 3.47 | 3.38e-06 | 8.59e-05 | 14/22 | AURKB; BUB1B; KNL1; CDCA5; CDK1; CDT1; CENPA; ESCO2; EZH2; HASPIN; MTBP; NDC80; PLK1; TTK |
| GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 3.39 | 1.60e-12 | 2.61e-10 | 33/53 | ANKRD53; AURKA; AURKB; SAPCD2; CCNB1; CDC20; CENPA; CENPE; ESPL1; GNAI1; GPSM2; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; NUSAP1; PLK1; POC1A; PRC1; PSRC1; RACGAP1; SPRY1; SPRY2; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0007059 | chromosome segregation | 3.36 | 0.00e+00 | 0.00e+00 | 77/125 | ANKRD53; AURKB; BIRC5; BLM; BRCA1; BRIP1; BUB1B; BUB1; KNSTRN; RMI2; C9orf84; KNL1; CCNB1; CCNE1; CCNE2; CDC20; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CENPN; CENPW; CEP55; DDX11; DLGAP5; DSCC1; DUSP1; ECT2; EME1; ESCO2; ESPL1; FAM83D; FBXO5; FEN1; GEM; HASPIN; HJURP; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; MAD2L1; MKI67; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NUF2; NUSAP1; OIP5; PLK1; PRC1; PSRC1; PTTG1; RACGAP1; RAD51C; SGO1; SGO2; SKA1; SKA2; SKA3; SMC4; SPAG5; SPATA22; SYCE2; TACC3; TOP2A; TRIP13; TTK; USP44; ZWINT |
| GO:0006260 | DNA replication | 3.18 | 0.00e+00 | 0.00e+00 | 56/96 | ATAD5; BLM; BRCA1; BRIP1; TICRR; RMI2; CCNA2; CCNE1; CCNE2; CDC45; CDC6; CDC7; CDK1; CDT1; CHAF1B; CHEK1; CLSPN; DBF4; DDX11; DNA2; DSCC1; DTL; E2F7; E2F8; EME1; ESCO2; EXO1; FBXO5; FEN1; GINS1; GINS2; GINS4; GLI1; GMNN; JUN; PCLAF; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; NFIB; TONSL; NPM2; ORC1; PIF1; POLE2; POLQ; RAD51; RECQL4; RFC4; RNASEH2A; RRM2; ALYREF; WDHD1 |
| GO:0036297 | interstrand cross-link repair | 3.18 | 2.31e-03 | 2.58e-02 | 7/12 | EME1; FANCA; FANCB; FANCI; RAD51AP1; RAD51; UBE2T |
| GO:0051653 | spindle localization | 3.06 | 7.60e-04 | 1.08e-02 | 9/16 | ASPM; SAPCD2; CENPA; ESPL1; GPSM2; NDC80; NUSAP1; SPRY1; SPRY2 |
| GO:0045023 | G0 to G1 transition | 2.93 | 4.22e-03 | 4.34e-02 | 7/13 | BRCA1; CDC7; CHEK1; E2F1; EZH2; RAD51; RRM2 |
| GO:0000075 | cell cycle checkpoint | 2.84 | 6.85e-11 | 7.97e-09 | 38/73 | AURKA; AURKB; BLM; BRCA1; BRIP1; BUB1B; BUB1; TICRR; CCNB1; CDC20; CDC25C; CDC45; CDC6; CDK1; CDT1; CENPF; CHEK1; CLSPN; DNA2; DTL; DUSP1; E2F1; E2F7; E2F8; EME1; GTSE1; H2AFX; KNTC1; MAD2L1; NDC80; ORC1; PLK1; TOP2A; TRIP13; TTK; USP44; WDR76; ZWINT |
| GO:0007051 | spindle organization | 2.81 | 1.00e-09 | 8.14e-08 | 34/66 | ANKRD53; ASPM; AURKA; AURKB; KNSTRN; AUNIP; CCDC69; CCNB1; CDC20; CENPE; ESPL1; FBXO5; GNAI1; GPSM2; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; PLK1; POC1A; PRC1; PSRC1; RACGAP1; SPAG5; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0000910 | cytokinesis | 2.77 | 1.45e-08 | 6.19e-07 | 30/59 | ANKRD53; ANLN; AURKA; AURKB; CDC25B; CDC6; CDT1; CENPA; CEP55; CIT; CKAP2; CXCR5; E2F7; E2F8; ECT2; ESPL1; KIF14; KIF20A; KIF23; KIF4A; KLHL13; NUSAP1; PLK1; PRC1; RACGAP1; ROPN1B; SEPT3; SEPT4; STMN1; SVIL |
| GO:0044772 | mitotic cell cycle phase transition | 2.55 | 0.00e+00 | 0.00e+00 | 90/192 | ABCB1; ADAMTS1; ANLN; ATAD5; AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; TICRR; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN2D; CDKN3; CDT1; CENPE; CENPF; CIT; CKS1B; CKS2; CLSPN; DBF4; DLGAP5; DTL; DUSP1; E2F1; E2F7; E2F8; ESPL1; EZH2; FAM107A; FBXO5; FHL1; FOXM1; GFI1; GMNN; GTSE1; HMMR; HYAL1; ID4; IQGAP3; KCNA5; KIF14; KLF4; KNTC1; LZTS1; MAD2L1; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MELK; MTBP; MYC; NDC80; NEK2; NES; NPM2; ORC1; PKMYT1; PLK1; PLK4; POLE2; RAD51C; RECQL4; RRM2; TACC3; TPX2; TRIP13; TTK; TYMS; UBE2C; UBE2S; USP44; ZFP36L2 |
| GO:0032200 | telomere organization | 2.55 | 2.04e-04 | 3.78e-03 | 15/32 | AURKB; BLM; CCNE1; CCNE2; DNA2; EXO1; FEN1; MYC; NEK2; PIF1; PRKCQ; RAD51C; RAD51; RECQL4; RFC4 |
| GO:0071897 | DNA biosynthetic process | 2.54 | 4.45e-07 | 1.45e-05 | 28/60 | ADIPOQ; AURKB; BLM; CDKN2D; CENPF; DSCC1; DTL; DUSP1; FGF2; HGF; PCLAF; KLF4; LIN9; MYC; NEK2; PAK3; PDGFA; PDGFRB; PIF1; POLE2; POLQ; PPARG; PRKCQ; RFC4; RGN; SPATA22; TK1; TYMS |
| GO:0006302 | double-strand break repair | 2.46 | 1.85e-07 | 6.54e-06 | 32/71 | BLM; BRCA1; BRIP1; PARPBP; RMI2; AUNIP; CDC45; CDC7; CDCA5; CHEK1; DDX11; DEK; DNA2; EME1; ESCO2; EXO1; FANCB; FEN1; FOXM1; GINS2; GINS4; H2AFX; TONSL; POLQ; RAD51AP1; RAD51C; RAD51; RAD54B; RAD54L; RECQL4; TRIP13; WAS |
| GO:0044839 | cell cycle G2/M phase transition | 2.43 | 2.93e-08 | 1.08e-06 | 37/83 | ABCB1; ATAD5; AURKA; AURKB; BLM; BRCA1; TICRR; CCNA2; CCNB1; CCNB2; CDC25A; CDC25B; CDC25C; CDC6; CDC7; CDK1; CENPF; CHEK1; CIT; CLSPN; DTL; FBXO5; FHL1; FOXM1; GTSE1; HMMR; KIF14; MELK; NEK2; NES; ORC1; PKMYT1; PLK1; PLK4; RAD51C; RECQL4; TPX2 |
| GO:0071103 | DNA conformation change | 2.39 | 4.02e-09 | 2.05e-07 | 43/98 | ASF1B; BLM; BRIP1; KNL1; CCNB1; CDC45; CDCA5; CDK1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; DDX11; DNA2; GINS1; GINS2; GINS4; H2AFX; HELLS; HJURP; HMGB2; MCM2; MCM4; MCM6; MCM7; CENPU; NAP1L2; NCAPD2; NCAPG2; NCAPG; NCAPH; NUSAP1; OIP5; PIF1; RAD51; RAD54B; RAD54L; RECQL4; SMC4; TOP2A |
| GO:0048285 | organelle fission | 2.37 | 0.00e+00 | 0.00e+00 | 94/216 | ANKRD53; ANLN; ASPM; AURKA; AURKB; BLM; BRIP1; BUB1B; BUB1; KNSTRN; C9orf84; CAV2; CCDC8; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CDC20; CDC25B; CDC25C; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; CHEK1; CKS2; CNTD2; DLGAP5; DSCC1; DUSP1; EDN1; EDN3; EME1; ESPL1; MTFR2; FANCA; FBXO5; HASPIN; IGF1; IL1A; KIF11; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; KNTC1; MAD2L1; MKI67; MND1; MTBP; MYBL1; MYBL2; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NPM2; NUF2; NUSAP1; PDE3A; PDGFRB; PKMYT1; PLK1; PRC1; PSRC1; PTTG1; RACGAP1; RAD51C; RAD51; RAD54B; SGO1; SGO2; SMC4; SPAG5; SPATA22; SYCE2; TACC3; TOP2A; TPX2; TRIP13; TTK; UBE2C; UBE2S; USP44; ZWINT |
| GO:0010948 | negative regulation of cell cycle process | 2.36 | 9.15e-10 | 8.14e-08 | 48/111 | AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; C10orf99; TICRR; CCNB1; CCNF; CDC20; CDC25C; CDC6; CDC7; CDK1; CDKN2D; CDT1; CENPF; CHEK1; CLSPN; DTL; DUSP1; E2F1; E2F7; E2F8; ESPL1; EVI2B; EZH2; FAM107A; FBXO5; FHL1; GTSE1; KLF4; MAD2L1; MLXIPL; MTBP; NDC80; NEK2; ORC1; PLK1; PTTG1; RAD51; RRM2; TRIM37; TRIP13; TTK; USP44; ZFP36L2 |
| GO:0044843 | cell cycle G1/S phase transition | 2.35 | 1.54e-09 | 9.64e-08 | 47/109 | ADAMTS1; AURKA; C10orf99; CCNA2; CCNB1; CCNE1; CCNE2; CDC25A; CDC25C; CDC45; CDC6; CDC7; CDK1; CDKN2D; CDKN3; CDT1; DBF4; E2F1; E2F7; E2F8; EZH2; FAM107A; FAM83D; FBXO5; FHL1; GFI1; GLI1; GMNN; GTSE1; HYAL1; ID4; IQGAP3; KCNA5; KIF14; KLF4; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MTBP; MYC; ORC1; POLE2; RRM2; TYMS |
| GO:1901987 | regulation of cell cycle phase transition | 2.34 | 2.01e-12 | 2.72e-10 | 64/149 | ADAMTS1; ANLN; ATAD5; AURKA; AURKB; BLM; BRCA1; BUB1B; BUB1; C10orf99; TICRR; CCNB1; CCNE1; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN2D; CDT1; CENPE; CENPF; CHEK1; CLSPN; DLGAP5; DTL; DUSP1; E2F1; E2F7; E2F8; ESPL1; EZH2; FAM107A; FAM83D; FBXO5; FHL1; GLI1; GTSE1; HMMR; HYAL1; KCNA5; KIF14; KLF4; KNTC1; MAD2L1; MTBP; NDC80; NEK2; NPM2; ORC1; PLK1; PLK4; RAD51C; RECQL4; TPX2; TRIP13; TTK; UBE2C; USP44; ZFP36L2 |
| GO:0033044 | regulation of chromosome organization | 2.30 | 1.04e-08 | 4.72e-07 | 44/104 | ATAD2; AURKB; BRCA1; BUB1B; BUB1; RMI2; CCNB1; CDC20; CDC45; CDC6; CDCA5; CDT1; CENPE; CENPF; CHEK1; DDX11; DLGAP5; DNMT3B; DUSP1; ESPL1; FBXO5; FEN1; GFI1; MAD2L1; MCM2; MKI67; MYC; MYOCD; NAP1L2; NDC80; NEK2; PAX5; PHF19; PIF1; PLK1; PRKCQ; PTTG1; SNCA; TACC3; TAL1; TOP2A; TRIP13; TTK; USP44 |
| GO:0018149 | peptide cross-linking | 2.29 | 5.72e-04 | 8.78e-03 | 16/38 | CSTA; EGFLAM; EPB42; IVL; SPOCK2; SPOCK3; SPRR1A; SPRR1B; SPRR2A; SPRR2B; SPRR2D; SPRR2E; SPRR2F; SPRR3; SPRR4; TGM5 |
| GO:0006310 | DNA recombination | 2.29 | 2.91e-08 | 1.08e-06 | 42/100 | ATAD5; BCL11B; BLM; BRCA1; BRIP1; PARPBP; RMI2; AUNIP; C9orf84; CCR6; CD40LG; CD40; CDC45; CDC7; CHEK1; EME1; EXO1; FANCB; FEN1; GINS2; GINS4; H2AFX; HMGB2; IL7R; KPNA2; MND1; TONSL; PIF1; POLQ; PTPRC; RAD51AP1; RAD51C; RAD51; RAD54B; RAD54L; RECQL4; TBX21; TCF7; ALYREF; TOP2A; TRIP13; WAS |
| GO:0051052 | regulation of DNA metabolic process | 2.15 | 1.24e-09 | 9.15e-08 | 58/147 | ADIPOQ; ANXA3; ATAD5; AURKB; BLM; BRCA1; PARPBP; TICRR; RMI2; AUNIP; CCNA2; CD40; CDC6; CDC7; CDK1; CDT1; CHEK1; CIDEA; DBF4; DDX11; DEK; DNA2; DSCC1; DUSP1; E2F7; E2F8; ESCO2; FANCB; FBXO5; FGF2; FOXM1; GLI1; GMNN; H2AFX; HGF; IL7R; JUN; KLF4; KPNA2; MYC; NEK2; NPM2; PAK3; PDGFA; PDGFRB; PIF1; POLQ; PPARG; PRKCQ; PTPRC; RAD51AP1; RAD51; RECQL4; RFC4; RGN; TBX21; ALYREF; WAS |
| GO:0051302 | regulation of cell division | 2.10 | 2.43e-06 | 6.83e-05 | 37/96 | ANKRD53; ASPM; AURKA; AURKB; BLM; C10orf99; CDC25B; CDC6; CIT; CXCR5; E2F7; E2F8; ECT2; EVI2B; FGF1; FGF2; FGF7; VEGFD; IL1A; KIF14; KIF18B; KIF20A; KIF23; KLHL13; MYC; NAP1L2; PDGFA; PDGFD; PLK1; PRC1; PROK1; PTN; RACGAP1; SOX17; SVIL; TAL1; THBS4 |
| GO:1904029 | regulation of cyclin-dependent protein kinase activity | 2.08 | 4.21e-04 | 6.98e-03 | 21/55 | BLM; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CDC25A; CDC25C; CDC6; CDKN2D; CDKN3; CEBPA; CKS1B; CKS2; CNTD2; DIRAS3; MYOCD; PKMYT1; PLK1; PSRC1 |
| GO:0031023 | microtubule organizing center organization | 2.03 | 2.72e-03 | 2.95e-02 | 16/43 | AURKA; BRCA1; CCNF; CDK1; CHEK1; PCLAF; KIF11; NEK2; PKHD1; PLK1; PLK4; POC1A; SGO1; STIL; TRIM37; WDR62 |
GATA3_LOF : 24 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0140029 | exocytic process | 6.37 | 2.06e-07 | 2.79e-05 | 12/47 | CPLX1; CPLX2; KCNB1; NAPB; PPFIA3; RAB3A; RAB3C; SNAP25; STX1A; STXBP1; SYT7; UNC13A |
| GO:0048284 | organelle fusion | 5.67 | 6.83e-06 | 5.56e-04 | 10/44 | CPLX1; CPLX2; GDAP1; RAB39A; RAB3A; SNAP25; STX1A; STXBP1; SYT4; SYT7 |
| GO:0017156 | calcium ion regulated exocytosis | 5.08 | 4.77e-10 | 1.94e-07 | 21/103 | CACNB2; CDK5R2; CHRNB2; CPLX1; CPLX2; KCNB1; NAPB; PPFIA3; RAB3A; RIMS2; SCAMP5; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; SYT7; UNC13A |
| GO:0035418 | protein localization to synapse | 4.99 | 1.53e-04 | 7.31e-03 | 8/40 | BSN; CPLX1; GRIN2C; IQSEC2; KIF5A; ARHGAP44; SNAP25; WNT5A |
| GO:0099504 | synaptic vesicle cycle | 4.86 | 6.91e-11 | 5.63e-08 | 24/123 | BRSK1; BRSK2; BSN; CACNB2; CDK5R1; CHRNB2; CPLX1; CPLX2; DNAJC6; NAPB; PPFIA3; RAB3A; RIMS2; SNAP25; SNCB; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; SYT7; UNC13A |
| GO:0099003 | vesicle-mediated transport in synapse | 4.69 | 9.34e-10 | 2.53e-07 | 22/117 | AP3B2; CACNB2; CHRNB2; CPLX1; CPLX2; DNAJC6; KIF5A; NAPB; PPFIA3; RAB3A; RIMS2; SNAP25; SNCB; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; SYT7; UNC13A |
| GO:0022406 | membrane docking | 4.32 | 1.91e-04 | 8.64e-03 | 9/52 | CPLX2; KCNB1; PPFIA3; RAB3A; RAB3C; SNAP25; STX1A; STXBP1; UNC13A |
| GO:0051648 | vesicle localization | 3.93 | 1.38e-08 | 2.25e-06 | 23/146 | AP3B2; BRSK1; BRSK2; CACNB2; CHRNB2; CPLX1; CPLX2; GRIA1; KIF5A; NAPB; PPFIA3; RAB3A; RIMS2; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; SYT7; UNC13A |
| GO:0015844 | monoamine transport | 3.72 | 3.01e-04 | 1.17e-02 | 10/67 | CARTPT; CHGA; CHRNB2; HRH3; KCNB1; STX1A; SYT13; SYT4; SYT5; SYT7 |
| GO:0061025 | membrane fusion | 3.62 | 7.42e-04 | 2.74e-02 | 9/62 | CPLX1; CPLX2; RAB39A; RAB3A; SNAP25; STX1A; STXBP1; SYT4; SYT7 |
| GO:0015837 | amine transport | 3.47 | 2.84e-04 | 1.16e-02 | 11/79 | CARTPT; CHGA; CHRNB2; HRH3; KCNB1; STX1A; STXBP1; SYT13; SYT4; SYT5; SYT7 |
| GO:0099565 | chemical synaptic transmission, postsynaptic | 3.47 | 2.84e-04 | 1.16e-02 | 11/79 | CELF4; CHRNB2; GRIK2; GRIN2C; IGSF9B; MAPK8IP2; MPP2; P2RX6; RIMS2; SEZ6; STX1A |
| GO:0072089 | stem cell proliferation | 3.28 | 8.43e-04 | 2.99e-02 | 10/76 | ACE; ACSL6; ASCL1; EML1; FGFR1; NEUROD4; SMARCD3; WNT5A; YAP1; ZFP36L1 |
| GO:0006836 | neurotransmitter transport | 3.18 | 4.08e-07 | 4.75e-05 | 24/188 | BRSK1; CACNB2; CHRNB2; CPLX1; CPLX2; GRM4; HRH3; NAPB; PPFIA3; PTPRN2; RAB3A; RIMS2; SLC1A2; SLC6A17; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; SYT7; UNC13A |
| GO:0099177 | regulation of trans-synaptic signaling | 3.01 | 3.43e-09 | 6.97e-07 | 35/290 | ACHE; BRSK1; CACNB2; CAMK2B; CELF4; CHRNB2; CPLX1; CPLX2; DGKE; GRIA1; GRIK2; GRIN2C; GRM4; IGSF9B; IQSEC2; JPH3; KCNB1; KIT; MAPK8IP2; MPP2; NAPB; NPTXR; PPFIA3; RAB3A; RIMS2; SCGN; SLC8A2; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT4; SYT7; UNC13A |
| GO:0032386 | regulation of intracellular transport | 2.77 | 1.27e-05 | 9.39e-04 | 22/198 | ABCA2; ANK3; BRSK2; CACNB2; CDK5R1; CHRNB2; CPLX1; CPLX2; KCNB1; NAPB; PIK3R1; RAB3A; REEP2; ARHGAP44; RIMS2; STX1A; STXBP1; SYN1; SYP; SYT4; SYT7; TCF7L2 |
| GO:1990778 | protein localization to cell periphery | 2.77 | 7.91e-05 | 4.29e-03 | 18/162 | ANK1; ANK3; CACNB2; CPLX1; GRIN2C; IQSEC2; KCNB1; KCNIP3; LGALS3; NFASC; PIK3R1; RAB3A; RAB3C; ARHGAP44; SNAP25; SPTBN4; STXBP1; ZDHHC22 |
| GO:0001505 | regulation of neurotransmitter levels | 2.70 | 3.24e-06 | 2.93e-04 | 26/240 | ACHE; BRSK1; CACNB2; CHRNB2; CPLX1; CPLX2; GRM4; HRH3; NAPB; PAH; PPFIA3; PTPRN2; RAB3A; RIMS2; SLC1A2; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; SYT7; TPH2; UNC13A |
| GO:0006813 | potassium ion transport | 2.65 | 9.37e-05 | 4.76e-03 | 19/179 | ANK3; ATP1A3; HCN2; HCN3; HCN4; KCNB1; KCNC1; KCNH2; KCNH4; KCNH6; KCNIP3; KCNJ6; KCNK3; KCNQ2; KCNU1; SLC12A5; SLC24A3; SLC24A5; SNAP25 |
| GO:0051656 | establishment of organelle localization | 2.56 | 2.93e-05 | 1.79e-03 | 23/224 | AP3B2; CACNB2; CHGA; CHRNB2; CPLX1; CPLX2; GRIA1; KIF5A; KIT; NAPB; PPFIA3; RAB3A; RIMS2; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; SYT7; UNC13A |
| GO:0023061 | signal release | 2.51 | 1.18e-06 | 1.20e-04 | 32/318 | BRSK1; BRSK2; CACNB2; CARTPT; CHGA; CHRNB2; CPLX1; CPLX2; FGFR1; GRM4; HFE; HRH3; IRS2; KCNB1; NAPB; PPFIA3; PTPRN2; RAB3A; RIMS2; SMPD3; SNAP25; STX1A; STXBP1; SYN1; SYP; SYT13; SYT4; SYT5; SYT7; TCF7L2; UNC13A; VGF |
| GO:0042391 | regulation of membrane potential | 2.38 | 1.53e-05 | 1.03e-03 | 28/293 | ANK3; ATP1A3; CACNB2; CELF4; CHRNB2; FGF12; GABRG2; GRIA1; GRIK2; GRIN2C; HCN2; HCN3; HCN4; IGSF9B; KCNB1; KCNC1; KCNH2; KCNH4; KCNH6; KCNU1; MAPK8IP2; MPP2; NTSR2; P2RX6; RIMS2; SEZ6; SLC26A10; STX1A |
| GO:0060627 | regulation of vesicle-mediated transport | 2.34 | 3.07e-05 | 1.79e-03 | 27/288 | CACNB2; CDK5R2; CHRNB2; CPLX1; CPLX2; DNAJC6; HFE; KCNB1; LGALS3; NAPB; PACSIN3; RAB25; RAB3A; RAB3C; ARHGAP44; RIMS2; SCAMP5; SMPD3; STX1A; STXBP1; SYN1; SYP; SYT4; SYT5; SYT7; VTN; WNT5A |
| GO:0050808 | synapse organization | 2.03 | 9.46e-04 | 3.21e-02 | 23/283 | ACHE; ANK3; BSN; CACNB2; CAMK2B; CDK5R1; CHRNB2; CTNNA2; DLGAP3; IGSF9B; L1CAM; LRTM2; NFASC; RAB39B; RAB3A; ARHGAP44; SEZ6L; SEZ6; SLITRK1; SNCB; SYN1; UNC13A; WNT5A |
PIK3CA_mis : 61 significant GO terms
| GeneSet | description | enrichRatio | pValue | FDR | GeneRatio | geneSymbols |
|---|---|---|---|---|---|---|
| GO:0051383 | kinetochore organization | 7.95 | 3.60e-09 | 9.44e-08 | 10/11 | CDT1; CENPA; CENPE; CENPF; CENPK; CENPN; CENPW; NDC80; NUF2; SMC4 |
| GO:0034502 | protein localization to chromosome | 6.76 | 1.27e-12 | 4.70e-11 | 17/22 | AURKB; BRCA2; BUB1B; KNL1; CDCA5; CDK1; CDT1; CENPA; ESCO2; EZH2; HASPIN; H2AFY2; MTBP; NDC80; PLK1; RAD21; TTK |
| GO:0061641 | CENP-A containing chromatin organization | 6.73 | 7.53e-08 | 1.91e-06 | 10/13 | KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; HJURP; CENPU; OIP5 |
| GO:0050000 | chromosome localization | 6.15 | 9.61e-13 | 3.73e-11 | 19/27 | CCNB1; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; CHMP4C; DLGAP5; FAM83D; KIF14; KIF18A; KIF2C; KIFC1; NDC80; NUF2; PSRC1; RRS1; SPAG5 |
| GO:0036297 | interstrand cross-link repair | 5.83 | 9.19e-06 | 1.87e-04 | 8/12 | EME1; FANCA; FANCB; FANCE; FANCI; RAD51AP1; RAD51; UBE2T |
| GO:0006260 | DNA replication | 5.56 | 0.00e+00 | 0.00e+00 | 61/96 | ACHE; ATAD5; BARD1; BLM; BRCA1; BRCA2; BRIP1; TICRR; RMI2; MCMDC2; CCNA2; CCNE1; CCNE2; CDC45; CDC6; CDC7; CDK1; CDT1; CHAF1B; CHEK1; CLSPN; DBF4; DDX11; DNA2; DSCC1; DTL; E2F7; E2F8; EME1; ESCO2; EXO1; FBXO5; FEN1; FGFR1; GINS1; GINS2; GINS4; GMNN; PCLAF; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; NFIA; TONSL; ORC1; PIF1; POLD1; POLE2; POLQ; RAD51; RECQL4; RFC4; RNASEH2A; RRM2B; RRM2; ALYREF; TOP1MT; WDHD1 |
| GO:0007059 | chromosome segregation | 5.32 | 0.00e+00 | 0.00e+00 | 76/125 | AURKB; BIRC5; BLM; BRCA1; BRIP1; BUB1B; BUB1; KNSTRN; RMI2; MCMDC2; KNL1; CCNB1; CCNE1; CCNE2; CDC20; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CENPN; CENPW; CEP55; CHMP4C; DDX11; DLGAP5; DSCC1; ECT2; EME1; ESCO2; ESPL1; FAM83D; FBXO5; FEN1; HASPIN; HJURP; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; MAD2L1; MKI67; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NUF2; NUSAP1; OIP5; PLK1; PRC1; PSRC1; PTTG1; RACGAP1; RAD21; RAD51C; RRS1; SGO1; SGO2; SKA1; SKA2; SKA3; SMC4; SPAG5; SYCE2; TACC3; NDC1; TOP2A; TRIP13; TTK; ZWINT |
| GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 5.12 | 2.22e-16 | 1.39e-14 | 31/53 | AURKA; AURKB; SAPCD2; CCNB1; CDC20; CENPA; CENPE; CHMP4C; ESPL1; GPSM2; CEP126; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; NUSAP1; PLK1; POC1A; PRC1; PSRC1; RACGAP1; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0032200 | telomere organization | 4.92 | 9.30e-10 | 2.91e-08 | 18/32 | AURKB; BLM; BRCA2; CCNE1; CCNE2; DNA2; EXO1; FEN1; HMBOX1; MYC; NEK2; PIF1; POLD1; PRKDC; RAD51C; RAD51; RECQL4; RFC4 |
| GO:0000075 | cell cycle checkpoint | 4.91 | 0.00e+00 | 0.00e+00 | 41/73 | AURKA; AURKB; BLM; BRCA1; BRIP1; BRSK1; BUB1B; BUB1; TICRR; CCNB1; CDC20; CDC25C; CDC45; CDC6; CDK1; CDKN1A; CDT1; CENPF; CHEK1; CHMP4C; CLSPN; DNA2; DTL; E2F1; E2F7; E2F8; EME1; GTSE1; H2AFX; KNTC1; MAD2L1; NDC80; NEK11; ORC1; PLK1; PRKDC; TOP2A; TRIP13; TTK; WDR76; ZWINT |
| GO:0045023 | G0 to G1 transition | 4.71 | 2.30e-04 | 3.74e-03 | 7/13 | BRCA1; CDC7; CHEK1; E2F1; EZH2; RAD51; RRM2 |
| GO:0071166 | ribonucleoprotein complex localization | 4.71 | 2.30e-04 | 3.74e-03 | 7/13 | DDX25; DDX39A; HHEX; NUP210; RRS1; ALYREF; NDC1 |
| GO:0007051 | spindle organization | 4.64 | 1.11e-16 | 8.22e-15 | 35/66 | ASPM; AURKA; AURKB; KNSTRN; AUNIP; CCNB1; CDC20; CENPE; CEP72; CHMP4C; ESPL1; FBXO5; GPSM2; WASHC5; CEP126; KIF11; KIF23; KIF4A; KIFC1; MYBL2; NDC80; NEK2; NUF2; PLK1; POC1A; PRC1; PSRC1; RACGAP1; SPAG5; STIL; STMN1; TACC3; TPX2; TTK; WDR62 |
| GO:0044839 | cell cycle G2/M phase transition | 4.64 | 0.00e+00 | 0.00e+00 | 44/83 | ATAD5; AURKA; AURKB; BLM; BRCA1; BRSK1; BRSK2; TICRR; CCNA2; CCNB1; CCNB2; CDC25A; CDC25B; CDC25C; CDC6; CDC7; CDK1; CDKN1A; CENPF; CEP72; CHEK1; CHMP4C; CIT; CLSPN; DTL; DYRK3; FBXO5; FOXM1; GTSE1; HMMR; AJUBA; KIF14; MELK; NEK10; NEK2; ORC1; PKMYT1; PLK1; PLK4; RAD21; RAD51C; RECQL4; SKP2; TPX2 |
| GO:0000910 | cytokinesis | 4.45 | 7.75e-14 | 3.94e-12 | 30/59 | ANK3; ANLN; AURKA; AURKB; BRCA2; CDC25B; CDC6; CDT1; CENPA; CEP55; CHMP4C; CIT; CKAP2; CSPP1; DRD2; E2F7; E2F8; ECT2; ESPL1; WASHC5; KIF14; KIF20A; KIF23; KIF4A; NUSAP1; PLK1; PRC1; RACGAP1; SEPT3; STMN1 |
| GO:0006302 | double-strand break repair | 4.43 | 2.22e-16 | 1.39e-14 | 36/71 | BARD1; BLM; BRCA1; BRCA2; BRIP1; PARPBP; RMI2; AUNIP; MCMDC2; CDC45; CDC7; CDCA5; CHEK1; DDX11; DEK; DNA2; EME1; ESCO2; EXO1; FANCB; FEN1; FOXM1; GINS2; GINS4; H2AFX; TONSL; POLQ; PRKDC; RAD21; RAD51AP1; RAD51C; RAD51; RAD54B; RAD54L; RECQL4; TRIP13 |
| GO:0001539 | cilium or flagellum-dependent cell motility | 4.37 | 8.77e-06 | 1.83e-04 | 11/22 | DRC1; CCDC65; DNAH2; DNAH3; DNAH6; DNAH7; SPEF1; TEKT1; TEKT2; TEKT3; CFAP57 |
| GO:0051653 | spindle localization | 4.37 | 1.57e-04 | 2.71e-03 | 8/16 | ASPM; SAPCD2; CENPA; ESPL1; GPSM2; LLGL2; NDC80; NUSAP1 |
| GO:0006301 | postreplication repair | 4.37 | 1.10e-03 | 1.50e-02 | 6/12 | BRCA1; DTL; PCLAF; POLD1; POLE2; RFC4 |
| GO:0071103 | DNA conformation change | 4.28 | 0.00e+00 | 0.00e+00 | 48/98 | ASF1B; BLM; BRIP1; KNL1; CCNB1; CDC45; CDCA5; CDK1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; CHD5; DDX11; DNA2; GINS1; GINS2; GINS4; H2AFX; H2AFY2; HELLS; HHEX; HIST2H3C; HJURP; HMGB2; MCM2; MCM4; MCM6; MCM7; CENPU; NCAPD2; NCAPG2; NCAPG; NCAPH; NUSAP1; OIP5; PARP10; PIF1; RAD51; RAD54B; RAD54L; RECQL4; SMC4; TOP1MT; TOP2A |
| GO:0031023 | microtubule organizing center organization | 4.27 | 1.18e-09 | 3.55e-08 | 21/43 | AURKA; BRCA1; BRCA2; BRSK1; CCNF; CDK1; CEP72; CHEK1; CHMP4C; CNTLN; PCLAF; KIF11; NEK2; PARD6A; PLK1; PLK4; POC1A; SGO1; STIL; TRIM37; WDR62 |
| GO:1904029 | regulation of cyclin-dependent protein kinase activity | 4.13 | 3.08e-11 | 1.05e-09 | 26/55 | BLM; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CCNI2; CCNO; CDC25A; CDC25C; CDC6; CDK5R1; CDK5R2; CDKN1A; CDKN2C; CDKN2D; CDKN3; CKS1B; CKS2; HHEX; NR2F2; PDGFB; PKMYT1; PLK1; PSRC1 |
| GO:0000726 | non-recombinational repair | 4.12 | 2.67e-04 | 4.17e-03 | 8/17 | BARD1; BLM; BRCA1; AUNIP; DEK; H2AFX; POLQ; PRKDC |
| GO:0001578 | microtubule bundle formation | 4.07 | 2.01e-11 | 7.13e-10 | 27/58 | ARMC4; CFAP43; DRC1; CFAP206; CFAP157; CCDC103; CCDC114; CCDC65; DNAH7; DNAI2; DNAJB13; HYDIN; KIF20A; LRGUK; LRRC49; DNAAF1; LRRC6; NAV1; NCKAP5; PLK1; PRC1; PSRC1; RSPH1; RSPH4A; TEKT2; TRIM46; ZMYND10 |
| GO:0044772 | mitotic cell cycle phase transition | 4.05 | 0.00e+00 | 0.00e+00 | 89/192 | ANLN; ATAD5; AURKA; AURKB; BLM; BRCA1; BRSK1; BRSK2; BUB1B; BUB1; TICRR; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN1A; CDKN2C; CDKN2D; CDKN3; CDT1; CENPE; CENPF; CEP72; CHMP4C; CIT; CKS1B; CKS2; CLSPN; DBF4; DLGAP5; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FBXO5; FOXM1; GMNN; GTSE1; HMMR; IQGAP3; AJUBA; KIF14; KNTC1; MAD2L1; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MELK; MTBP; MYC; NDC80; NEK11; NEK2; ORC1; PKMYT1; PLK1; PLK4; POLE2; PRKDC; RAD21; RAD51C; RBL1; RECQL4; RRM2; SKP2; TACC3; TPX2; TRIP13; TTK; TYMS; UBE2C; UBE2S; ZFP36L1; ZFP36L2 |
| GO:1901987 | regulation of cell cycle phase transition | 3.76 | 0.00e+00 | 0.00e+00 | 64/149 | ANLN; ATAD5; AURKA; AURKB; BLM; BRCA1; BRSK1; BUB1B; BUB1; TICRR; CCNB1; CCNE1; CDC20; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDKN1A; CDKN2D; CDT1; CENPE; CENPF; CEP72; CHEK1; CHMP4C; CLSPN; DLGAP5; DTL; DYRK3; E2F1; E2F7; E2F8; ESPL1; EZH2; FAM83D; FBXO5; GTSE1; HMMR; KIF14; KNTC1; MAD2L1; MTBP; NDC80; NEK10; NEK11; NEK2; ORC1; PLK1; PLK4; PRKDC; RAD21; RAD51C; RBL1; RECQL4; TPX2; TRIP13; TTK; UBE2C; ZFP36L1; ZFP36L2 |
| GO:0006284 | base-excision repair | 3.75 | 2.93e-03 | 3.83e-02 | 6/14 | DNA2; FEN1; NEIL3; POLD1; POLQ; RECQL4 |
| GO:0048285 | organelle fission | 3.72 | 0.00e+00 | 0.00e+00 | 92/216 | ANLN; ASPM; AURKA; AURKB; BLM; BRCA2; BRIP1; BUB1B; BUB1; KNSTRN; MCMDC2; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CCNI2; CCNO; CDC20; CDC25B; CDC25C; CDC6; CDCA5; CDCA8; CDT1; CENPE; CENPF; CEP55; CHEK1; CHMP4C; CKS2; DLGAP5; DSCC1; EME1; ESPL1; MTFR2; FANCA; FBXO5; HASPIN; WASHC5; KIF11; KIF14; KIF18A; KIF18B; KIF23; KIF2C; KIF4A; KIFC1; KNTC1; MAD2L1; MKI67; MND1; MTBP; MTFR1; MYBL1; MYBL2; NCAPD2; NCAPG; NCAPH; NDC80; NEK2; NUF2; NUSAP1; PDGFB; PKMYT1; PLK1; PRC1; PSRC1; PTTG1; RACGAP1; RAD21; RAD51C; RAD51; RAD54B; RRS1; SEC16B; SGO1; SGO2; SMC4; SPAG5; SYCE2; TACC3; NDC1; TOP2A; TPX2; TRIP13; TTK; UBE2C; UBE2S; WNT5A; ZWINT |
| GO:0010948 | negative regulation of cell cycle process | 3.70 | 0.00e+00 | 0.00e+00 | 47/111 | AURKA; AURKB; BLM; BRCA1; BRSK1; BUB1B; BUB1; TICRR; CCNB1; CCNF; CDC20; CDC25C; CDC6; CDC7; CDK1; CDKN1A; CDKN2D; CDT1; CENPF; CHEK1; CHMP4C; CLSPN; DTL; E2F1; E2F7; E2F8; ESPL1; EZH2; FBXO5; GTSE1; MAD2L1; MTBP; NDC80; NEK2; ORC1; PLK1; PRKDC; PTTG1; RAD21; RAD51; RBL1; RRM2; TRIM37; TRIP13; TTK; ZFP36L1; ZFP36L2 |
| GO:0044380 | protein localization to cytoskeleton | 3.58 | 3.99e-04 | 6.02e-03 | 9/22 | ANLN; AURKA; CEP72; FAM83D; FAM83H; PARD6A; SPAG5; STIL; TTK |
| GO:0045930 | negative regulation of mitotic cell cycle | 3.56 | 8.88e-16 | 5.16e-14 | 46/113 | AURKA; AURKB; BLM; BRCA1; BRSK1; BUB1B; BUB1; TICRR; CCNB1; CDC20; CDC25C; CDC6; CDK1; CDKN1A; CDT1; CENPF; CHEK1; CHMP4C; CLSPN; E2F1; E2F7; E2F8; EME1; EZH2; FBXO5; FZD3; GTSE1; KNTC1; MAD2L1; MTBP; NDC80; NEK11; NKX3-1; ORC1; PLK1; PRKDC; PTTG1; RAD21; RBL1; SCRIB; TOP2A; TRIP13; TTK; ZFP36L1; ZFP36L2; ZWINT |
| GO:0006310 | DNA recombination | 3.41 | 7.61e-13 | 3.10e-11 | 39/100 | ATAD5; BLM; BRCA1; BRCA2; BRIP1; PARPBP; RMI2; AUNIP; MCMDC2; CDC45; CDC7; CHEK1; CLCF1; EME1; EXO1; FANCB; FEN1; GINS2; GINS4; H2AFX; HMGB2; KPNA2; MND1; TONSL; PGBD5; PIF1; POLQ; PRKDC; RAD21; RAD51AP1; RAD51C; RAD51; RAD54B; RAD54L; RECQL4; TFRC; ALYREF; TOP2A; TRIP13 |
| GO:0044843 | cell cycle G1/S phase transition | 3.37 | 1.60e-13 | 7.22e-12 | 42/109 | AURKA; CCNA2; CCNB1; CCNE1; CCNE2; CDC25A; CDC25C; CDC45; CDC6; CDC7; CDK1; CDKN1A; CDKN2C; CDKN2D; CDKN3; CDT1; DBF4; E2F1; E2F7; E2F8; EZH2; FAM83D; FBXO5; GMNN; GTSE1; IQGAP3; KIF14; MCM10; MCM2; MCM4; MCM5; MCM6; MCM7; MTBP; MYC; ORC1; POLE2; PRKDC; RBL1; RRM2; SKP2; TYMS |
| GO:0033044 | regulation of chromosome organization | 3.36 | 6.57e-13 | 2.81e-11 | 40/104 | ATAD2; AURKB; BRCA1; BUB1B; BUB1; RMI2; CCNB1; CDC20; CDC45; CDC6; CDCA5; CDT1; CENPE; CENPF; CHEK1; DDX11; DLGAP5; DNMT3B; ESPL1; FBXO5; FEN1; HMBOX1; JDP2; MAD2L1; MCM2; MKI67; MYC; NDC80; NEK2; PARP10; PHF19; PIF1; PLK1; PRDM12; PTTG1; RAD21; TACC3; TOP2A; TRIP13; TTK |
| GO:0042770 | signal transduction in response to DNA damage | 3.36 | 4.12e-07 | 9.58e-06 | 20/52 | ATAD5; AURKA; BRCA1; BRCA2; CCNB1; CDC25C; CDK1; CDKN1A; CHEK1; DTL; DYRK3; E2F1; E2F7; E2F8; FOXM1; GTSE1; NDRG1; PLK1; PMAIP1; PRKDC |
| GO:0007018 | microtubule-based movement | 3.35 | 2.00e-15 | 1.08e-13 | 49/128 | AP3B2; ARMC4; DRC1; MAATS1; CFAP206; CCDC103; CCDC114; CFAP53; CCDC65; CENPE; CNIH2; DLGAP5; DNAH12; DNAH2; DNAH3; DNAH6; DNAH7; DNAI2; HYDIN; KIF11; KIF14; KIF15; KIF18A; KIF18B; KIF19; KIF20A; KIF21A; KIF23; KIF2C; KIF4A; KIF5A; KIFC1; KIFC2; DNAAF1; LRRC6; MAK; NME5; CFAP221; RACGAP1; ROPN1L; RSPH4A; SPA17; TEKT1; TEKT2; TEKT3; TRIM46; WDR34; WDR63; WDR78 |
| GO:0071824 | protein-DNA complex subunit organization | 3.20 | 2.15e-09 | 6.26e-08 | 30/82 | ANP32E; ASF1B; BRF2; MCMDC2; KNL1; CDC45; CDT1; CENPA; CENPE; CENPF; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; CHD5; GMNN; H2AFX; H2AFY2; HELLS; HIST2H3C; HJURP; HMGB2; HNF1B; MCM2; CENPU; MYC; OIP5; RAD51 |
| GO:0022406 | membrane docking | 3.20 | 2.01e-06 | 4.42e-05 | 19/52 | CDK1; CEP72; CPLX2; GRAMD2A; KCNB1; NEK2; NPHP1; PLK1; PLK4; PPFIA3; RAB3C; SNAP25; SNPH; STX1A; STX1B; STXBP1; SYTL2; TSNARE1; UNC13A |
| GO:0006997 | nucleus organization | 3.18 | 1.66e-04 | 2.82e-03 | 12/33 | CCNB1; CCNB2; CDK1; CELF3; CEP55; CHD5; CHMP4C; DYRK3; GOLM1; HMGB2; PLK1; NDC1 |
| GO:0051321 | meiotic cell cycle | 3.16 | 9.24e-14 | 4.42e-12 | 47/130 | ASPM; AURKA; BLM; BRCA2; BRIP1; BUB1B; BUB1; MCMDC2; CCNE1; CCNE2; CDC20; CDC25A; CDC25B; CDC25C; CKS2; EME1; ESPL1; EXO1; FANCA; FBXO5; H2AFX; WASHC5; KIF18A; MND1; MYBL1; NCAPD2; NCAPH; NEK2; NUF2; PKMYT1; PLK1; PTTG1; RAD21; RAD51C; RAD51; RAD54B; RAD54L; RSPH1; SGO1; SGO2; SMC4; SYCE2; NDC1; TOP2A; TRIP13; TTK; WNT5A |
| GO:0045787 | positive regulation of cell cycle | 3.07 | 0.00e+00 | 0.00e+00 | 67/191 | ASCL1; ATAD5; AURKA; AURKB; BRCA1; BRCA2; CCNA2; CCNB1; CCNB2; CCNE1; CCNE2; CCNF; CCNI2; CCNO; CDC25A; CDC25B; CDC25C; CDC45; CDC6; CDC7; CDCA5; CDK1; CDK5R1; CDKN1A; CDT1; CHEK1; CIT; CKS1B; CKS2; CSPP1; DDX11; DLGAP5; DRD2; DTL; DYRK3; E2F1; E2F7; E2F8; ECT2; ESPL1; EZH2; FAM83D; FBXO5; FEN1; FGFR1; GPSM2; GTSE1; INSM1; KIF14; KIF23; MAD2L1; MTBP; NDC80; NKX3-1; NUSAP1; PDGFB; PLK4; POC1A; PSRC1; RACGAP1; RAD21; RAD51C; RECQL4; SLC6A4; SPAG5; UBE2C; WNT5A |
| GO:0071826 | ribonucleoprotein complex subunit organization | 3.00 | 3.15e-04 | 4.84e-03 | 12/35 | BOP1; TICRR; CELF3; CELF4; DYRK3; AGO2; EIF3H; MTERF3; PIH1D2; PRKAA2; PSIP1; RRS1 |
| GO:0006338 | chromatin remodeling | 2.97 | 2.10e-05 | 4.06e-04 | 17/50 | ACTL6B; ANP32E; KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHD5; CHEK1; HELLS; HJURP; HMGB2; CENPU; MYC; OIP5 |
| GO:0032886 | regulation of microtubule-based process | 2.89 | 2.41e-09 | 6.77e-08 | 35/106 | APC2; ARMC4; AURKA; BRCA1; DRC1; CCDC65; CCNF; CDK5R1; CHEK1; CHMP4C; CKAP2; CNIH2; GPSM2; KIF11; KIF18A; DNAAF1; NEK2; PLK1; PLK4; POC1A; PRKAA2; PSRC1; SKA1; SKA2; SKA3; SPAG5; SPEF1; STIL; STMN1; STMN3; STMN4; TACC3; TPX2; TRIM37; TRIM46 |
| GO:0031109 | microtubule polymerization or depolymerization | 2.81 | 2.88e-05 | 5.33e-04 | 18/56 | APC2; CDK5R1; CKAP2; FBXO5; KIF14; KIF18A; KIF18B; KIF19; KIF2C; NCKAP5; PSRC1; SKA1; SKA2; SKA3; SPEF1; STMN1; STMN3; STMN4 |
| GO:0072331 | signal transduction by p53 class mediator | 2.78 | 3.99e-07 | 9.56e-06 | 27/85 | ATAD5; AURKA; AURKB; BOP1; BRCA1; BRCA2; CCNB1; CDC25C; CDK1; CDKN1A; CHD5; CHEK1; DYRK3; E2F1; E2F2; E2F7; E2F8; ELL3; FOXM1; GTSE1; MAPK11; NDRG1; PMAIP1; PYCARD; RRM2B; RRS1; TPX2 |
| GO:0071897 | DNA biosynthetic process | 2.77 | 2.20e-05 | 4.17e-04 | 19/60 | AURKB; BLM; CDKN2D; CENPF; DSCC1; DTL; HMBOX1; PCLAF; LIN9; MYC; NEK2; PDGFB; PIF1; POLD1; POLE2; POLQ; RFC4; TK1; TYMS |
| GO:2001020 | regulation of response to DNA damage stimulus | 2.69 | 3.32e-06 | 7.11e-05 | 24/78 | ATAD5; BLM; BRCA1; DDIAS; PARPBP; RMI2; AUNIP; CDKN2D; CHEK1; DDX11; DEK; DYRK3; ELL3; FANCB; FBXO5; FOXM1; H2AFX; MYC; NKX3-1; PMAIP1; POLQ; RAD51AP1; RAD51; WDR76 |
| GO:0006333 | chromatin assembly or disassembly | 2.64 | 4.73e-05 | 8.56e-04 | 19/63 | ASF1B; KNL1; CENPA; CENPI; CENPK; CENPM; CENPN; CENPW; CHAF1B; H2AFX; H2AFY2; HELLS; HIST2H3C; HJURP; HMGB2; MCM2; CENPU; OIP5; PARP10 |
| GO:0044782 | cilium organization | 2.62 | 1.96e-10 | 6.37e-09 | 47/157 | ARMC4; CFAP43; CFAP126; DRC1; CATIP; CFAP206; CFAP157; CCDC103; CCDC114; CFAP53; CCDC13; CCDC65; CCNO; CDK1; CELSR3; CEP72; DNAH7; DNAI2; DNAJB13; FOXJ1; HYDIN; IQUB; KCTD17; CEP126; KIF19; LRGUK; LRRC49; DNAAF1; LRRC6; MAK; NEK2; NME5; NPHP1; CFAP221; PLK1; PLK4; POC1A; RSPH1; RSPH4A; SEPT3; TEKT1; TEKT2; TEKT3; VANGL2; WDR34; WDR90; ZMYND10 |
| GO:0140029 | exocytic process | 2.61 | 5.34e-04 | 7.76e-03 | 14/47 | CPLX2; KCNB1; NAPB; PPFIA3; RAB3C; SCRIB; SNAP25; SNPH; STX1A; STX1B; STXBP1; SYT9; SYTL2; UNC13A |
| GO:0051052 | regulation of DNA metabolic process | 2.56 | 2.60e-09 | 7.07e-08 | 43/147 | ATAD5; AURKB; BLM; BRCA1; BRCA2; PARPBP; TICRR; RMI2; AUNIP; CCNA2; CDC6; CDC7; CDK1; CDT1; CHEK1; CLCF1; DBF4; DDX11; DEK; DNA2; DSCC1; E2F7; E2F8; ESCO2; FANCB; FBXO5; FGFR1; FOXM1; GMNN; H2AFX; HMBOX1; KPNA2; MYC; NEK2; PDGFB; PIF1; POLQ; RAD51AP1; RAD51; RECQL4; RFC4; TFRC; ALYREF |
| GO:0051302 | regulation of cell division | 2.37 | 1.74e-05 | 3.46e-04 | 26/96 | ASPM; AURKA; AURKB; BLM; BRCA2; CDC25B; CDC6; CHMP4C; CIT; CSPP1; DRD2; E2F7; E2F8; ECT2; KIF14; KIF18B; KIF20A; KIF23; MACC1; MYC; NKX3-1; PDGFB; PLK1; PRC1; RACGAP1; RBL1 |
| GO:0022613 | ribonucleoprotein complex biogenesis | 2.35 | 1.60e-03 | 2.14e-02 | 14/52 | BOP1; TICRR; CELF3; CELF4; DCAF13; AGO2; EIF3H; EXOSC4; ISG20; MTERF3; PIH1D2; PRKAA2; PSIP1; RRS1 |
| GO:0031503 | protein-containing complex localization | 2.19 | 9.56e-04 | 1.34e-02 | 18/72 | BIRC5; CEP72; CNIH2; DDX25; DDX39A; GPC4; HASPIN; HHEX; KIF5A; MAK; NUP210; RRS1; SCRIB; SNAP25; STX1B; ALYREF; NDC1; WDR34 |
| GO:0090305 | nucleic acid phosphodiester bond hydrolysis | 2.16 | 8.29e-04 | 1.18e-02 | 19/77 | ATAD5; BOP1; DNA2; AGO2; EME1; EXO1; EXOSC4; FEN1; HMGB2; RIDA; ISG20; LACTB2; REXO5; PGBD5; POLD1; POP1; RFC4; RNASEH2A; RRS1 |
| GO:0010639 | negative regulation of organelle organization | 2.15 | 1.66e-06 | 3.76e-05 | 40/163 | APC2; ARFGEF1; ATAD2; AURKB; BRCA1; BUB1B; BUB1; ARPIN; CCNB1; CCNF; CDC20; CDT1; CENPF; CHEK1; CKAP2; DNMT3B; ESPL1; FBXO5; MAD2L1; MAK; MCM2; MTBP; NDC80; NEK2; PIF1; PLEKHH2; PLK1; PTTG1; RAD21; CARMIL2; SPEF1; STMN1; SYT4; TACSTD2; TMOD2; TMSB15A; TMSB15B; TOP2A; TRIM37; TRIP13; TTK |
| GO:0051656 | establishment of organelle localization | 2.07 | 1.23e-07 | 3.03e-06 | 53/224 | AP3B2; SAPCD2; CACNB2; CCNB1; CDCA5; CDCA8; CDT1; CENPA; CENPE; CENPF; CEP55; CHGA; CHMP4C; CHRNB2; CNIH2; CPLX2; DLGAP5; DOC2A; DRD2; ESPL1; FAM83D; GPSM2; KIF14; KIF18A; KIF23; KIF2C; KIF5A; KIFC1; LLGL2; NAPB; NDC80; NUF2; NUSAP1; PPFIA3; PSRC1; RIMS2; RRS1; SCRIB; SEC16B; SLIT1; SNAP25; SNPH; SPAG5; STX1A; STX1B; STXBP1; SYN1; SYP; SYT4; SYT5; SYT9; TRIM46; UNC13A |
| GO:0099504 | synaptic vesicle cycle | 2.06 | 9.66e-05 | 1.71e-03 | 29/123 | BRSK1; BRSK2; BSN; CACNB2; CDK5R1; CHRNB2; CPLX2; DDC; DNAJC6; DOC2A; DRD2; NAPB; NRXN1; PPFIA3; RIMS2; SCRIB; SNAP25; SNCB; SNPH; STON2; STX1A; STX1B; STXBP1; SYN1; SYP; SYT4; SYT5; SYT9; UNC13A |
| GO:0017156 | calcium ion regulated exocytosis | 2.04 | 4.55e-04 | 6.73e-03 | 24/103 | BAIAP3; CACNB2; CDK5R2; CHRNB2; CPLX2; DOC2A; DRD2; KCNB1; NAPB; PPFIA3; RIMS2; SCAMP5; SCRIB; SNAP25; SNPH; STX1A; STX1B; STXBP1; SYN1; SYP; SYT4; SYT5; SYT9; UNC13A |
| GO:0099003 | vesicle-mediated transport in synapse | 2.02 | 2.43e-04 | 3.88e-03 | 27/117 | AP3B2; CACNB2; CHRNB2; CNIH2; CPLX2; DNAJC6; DOC2A; DRD2; KIF5A; NAPB; PPFIA3; RIMS2; SCRIB; SNAP25; SNCB; SNPH; STON2; STX1A; STX1B; STXBP1; SYN1; SYP; SYT4; SYT5; SYT9; TRIM46; UNC13A |
| Marker | TP53_LOF | TP53_mis | CDH1_LOF | GATA3_LOF | PIK3CA_mis |
| Signif_GO_terms | 62 | 44 | 31 | 24 | 61 |