DE analysis on sctransformed CROP-seq data
Yifan Zhou
5/21/2019
Load data and functions
library(knitr)
library(kableExtra)
library(gridExtra)
library(Matrix)
library(sctransform)
library(ggplot2)
wd <- '~/Downloads/ASoC/singlecell/'
set.seed(1000)
## Necessary data
load(paste0(wd,'data/cropseq_expression.Rd'))
vst_out <- readRDS(paste0(wd,'data/sctransformed_object.rds'))
# Got 'vst_out' from:
# sctransform::vst(gene.exp,n_genes = NULL,return_gene_attr = TRUE, return_cell_attr = TRUE)
## Necessary functions
source(paste0(wd,'github/code/qq-plot.R'))
summ_pvalues <- function(pvalues){
library(gridExtra)
# p values distribution histogram
plot1 <- histogram(pvalues,col='grey',type="count",xlim=c(0,1),breaks=100, main= "p value distribution")
# pvalues qq-plot
plot2 <- qqunif.plot(pvalues, main="p value qq-plot")
grid.arrange(plot1,plot2, ncol=2)
}
ttest <- function(subset,tg.cells,neg.cells){
pval <- rep(NA,nrow(subset))
for (i in 1:nrow(subset)){
pval[i] = t.test(subset[i,neg.cells],
subset[i,tg.cells])$p.value
}
pval <- data.frame(gene=row.names(subset),pval=pval)
pval$adj <- p.adjust(pval$pval,method = 'fdr')
summ_pvalues(pval$pval)
cat(sum(pval$adj<0.1),'genes passed FDR=0.1 cutoff','\n')
tb <- pval[order(pval$pval),]
tb[,2:3] <- signif(tb[,2:3],digits = 2)
print(kable(tb,row.names = F) %>% kable_styling() %>%
scroll_box(width = '100%', height = '400px'))
return(pval)
}
ttest_loop <- function(subset,tg.cells,neg.cells,nshow){
pval <- rep(NA,nrow(subset))
for (i in 1:nrow(subset)){
pval[i] = t.test(subset[i,neg.cells],
subset[i,tg.cells])$p.value
}
pval <- data.frame(gene=row.names(subset),pval=pval)
pval$adj <- p.adjust(pval$pval,method = 'fdr')
# summ_pvalues(pval$pval)
cat(sum(pval$adj<0.1),'genes passed FDR=0.1 cutoff','\n')
tb <- head(pval[order(pval$pval),],nshow)
tb[,2:3] <- signif(tb[,2:3],digits = 2)
print(kable(tb,row.names = F) %>% kable_styling(position = 'center',full_width = T))
# return(pval)
}sctransform on single-cell UMI data
Before sctransform
- Gene expression mean ~ variance relationship
orig_attr <- data.frame(mean = rowMeans(gene.exp),
detection_rate = rowMeans(gene.exp > 0),
var = apply(gene.exp, 1, var))
ggplot(orig_attr, aes(log10(mean), log10(var))) +
geom_point(alpha=0.3, shape=16) + geom_density_2d(size = 0.3) +
geom_abline(intercept = 0, slope = 1, color='red') + theme_bw()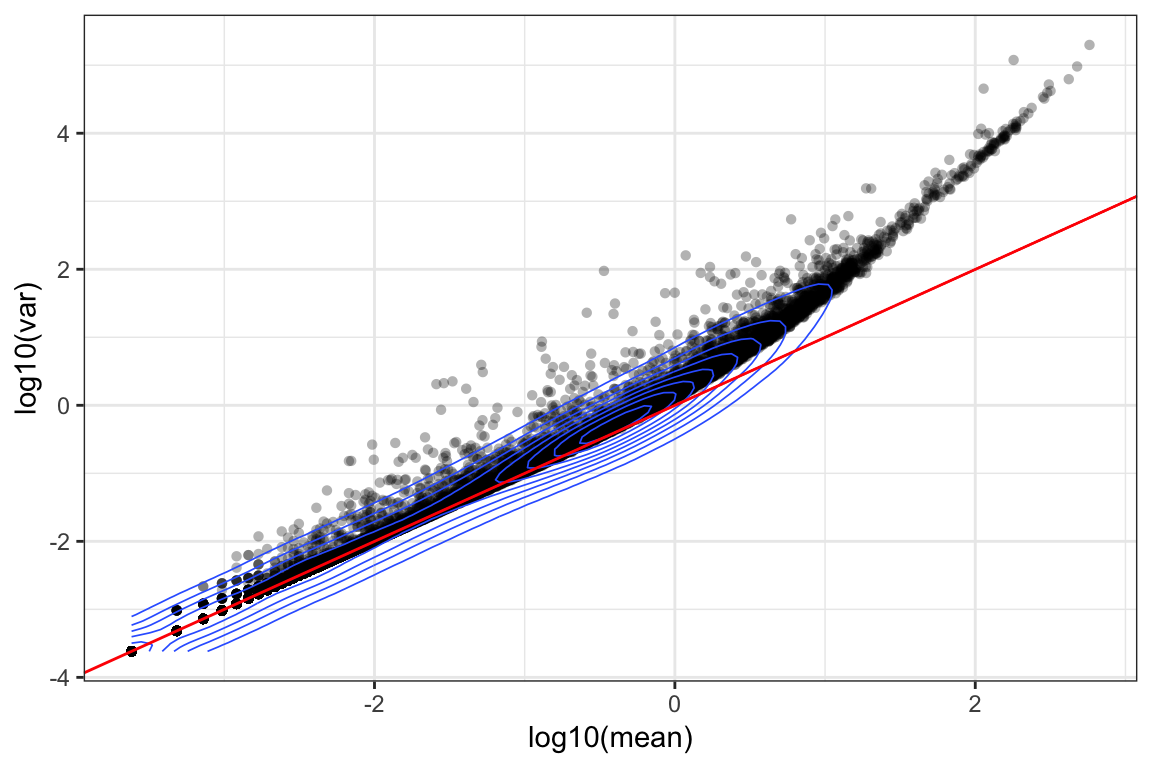
For the genes, we can see that up to a mean UMI count of ~1, the mean-variance follows the 1:1 line, i.e. variance and mean are roughly equal (as expected under a Poisson model). However, genes with a higher average UMI count show overdispersion compared to Poisson.
- Mean ~ detection-rate relationship
# add the expected detection rate under Poisson model
x = seq(from = -3, to = 2, length.out = 1000)
poisson_model <- data.frame(log_mean = x, detection_rate = 1 - dpois(0, lambda = 10^x))
ggplot(orig_attr, aes(log10(mean), detection_rate)) + geom_point(alpha=0.3, shape=16) +
geom_line(data=poisson_model, aes(x=log_mean, y=detection_rate), color='red') +
theme_bw()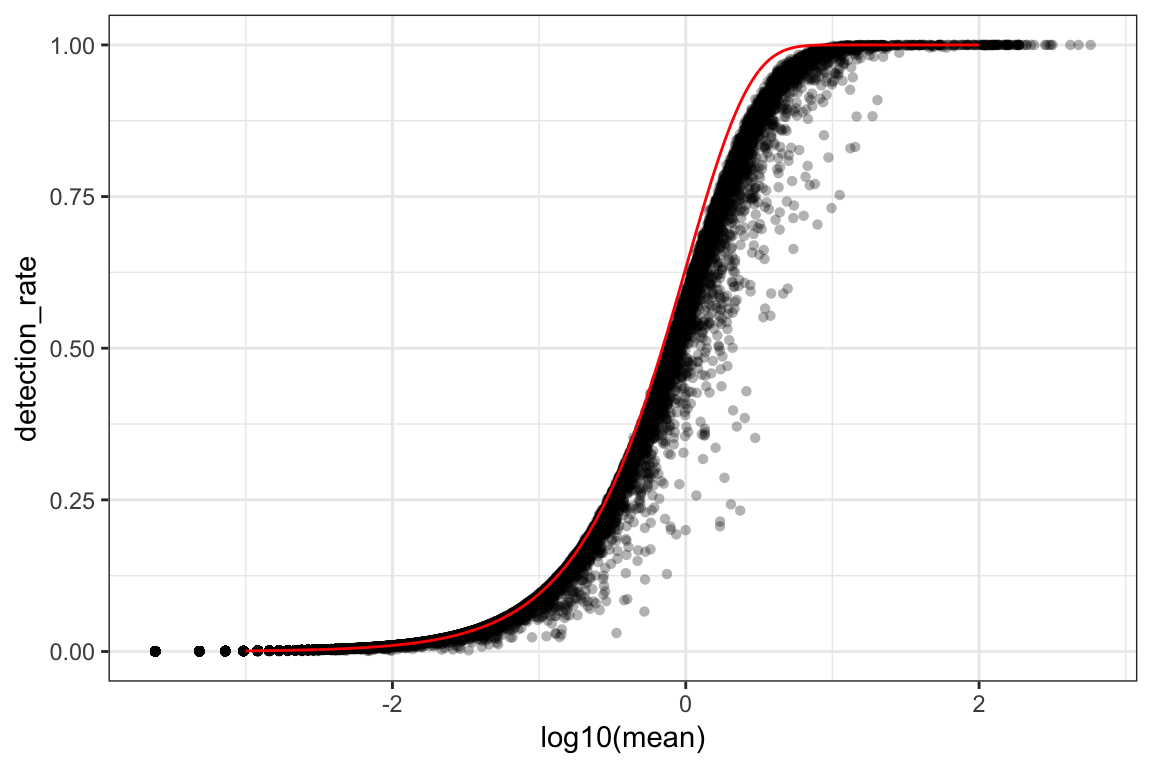
We see a lower than expected detection rate in the medium expression range. However, for the highly expressed genes, the rate is very close to 1.0, suggesting that there is no zero-inflation in the counts for those genes and that zero-inflation is a result of overdispersion, rather than an independent systematic bias.
After sctransform
sctransform models the expression of each gene as a negative binomial random variable with a mean that depends on other variables, such as the sequencing depth for each cell.
Each observed UMI count is transformed into a Pearson residual which can be interpreted as the number of standard deviations an observed count was away from the expected mean. If the model accurately describes the mean-variance relationship and the dependency of mean and latent factors, then the result should have mean zero and a stable variance across the range of expression.
The vst function was used to estimate model parameters and performs the variance stabilizing transformation. Here we use the default – log10 of the total UMI counts of a cell – as the latent variable for sequencing depth for each cell.
vst_out <- sctransform::vst(as.matrix(gene.exp),n_genes = NULL,return_gene_attr = TRUE, return_cell_attr = TRUE)sctransform_data <- vst_out$y
cat('Output dimension (Gene x Cell):',dim(sctransform_data))Output dimension (Gene x Cell): 17281 4144- The model parameters as a function of gene mean (geometric mean)
plot_model_pars(vst_out)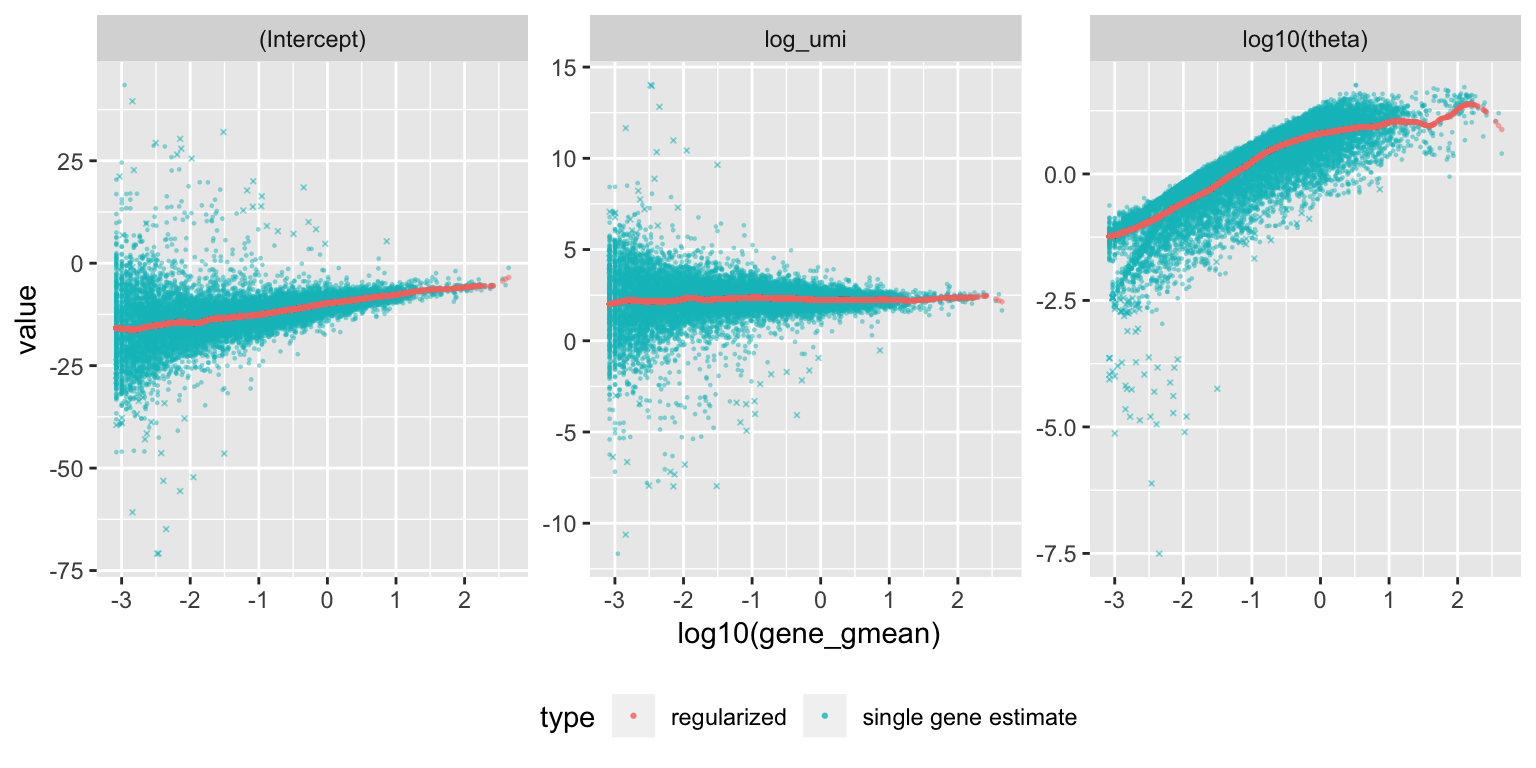
Internally vst performs Poisson regression per gene with \(log(\mu)=\beta_0+\beta_1 x\), where \(x\) is log10(umi), and \(\mu\) is the expected number of UMI counts of the given gene. The above plot shows (Intercept): \(\beta_0\) , log_umi: \(\beta_1\) , and theta: the maximum likelihood estimate of the overdispersion parameter \(\theta\) under the negative binomial model, (where the variance of a gene depends on the expected UMI counts and theta: \(\mu+\frac{\mu^2}{\theta}\).) In the second step, the regularized model parameters are used to turn observed UMI counts into Pearson residuals.
- Overall properties of the transformed data
ggplot(vst_out$gene_attr, aes(residual_mean)) + geom_histogram(binwidth=0.01) +
xlab(label = 'residual mean') + theme_bw() +
theme(axis.text = element_text(size = 14),
axis.title = element_text(size = 14,face = 'bold'))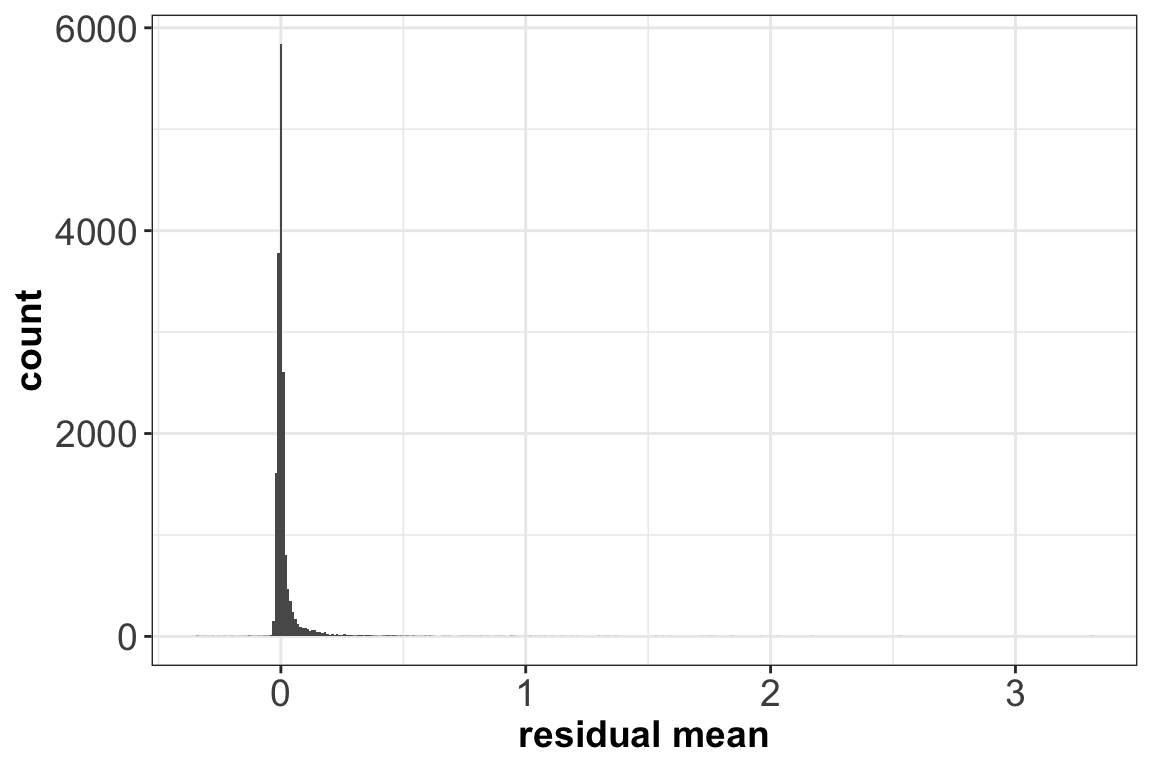
ggplot(vst_out$gene_attr, aes(residual_variance)) + geom_histogram(binwidth=0.1) +
geom_vline(xintercept=1, color='red') + xlim(0, 10) +
xlab(label = 'residual variance') + theme_bw() +
theme(axis.text = element_text(size = 14),
axis.title = element_text(size = 14,face = 'bold'))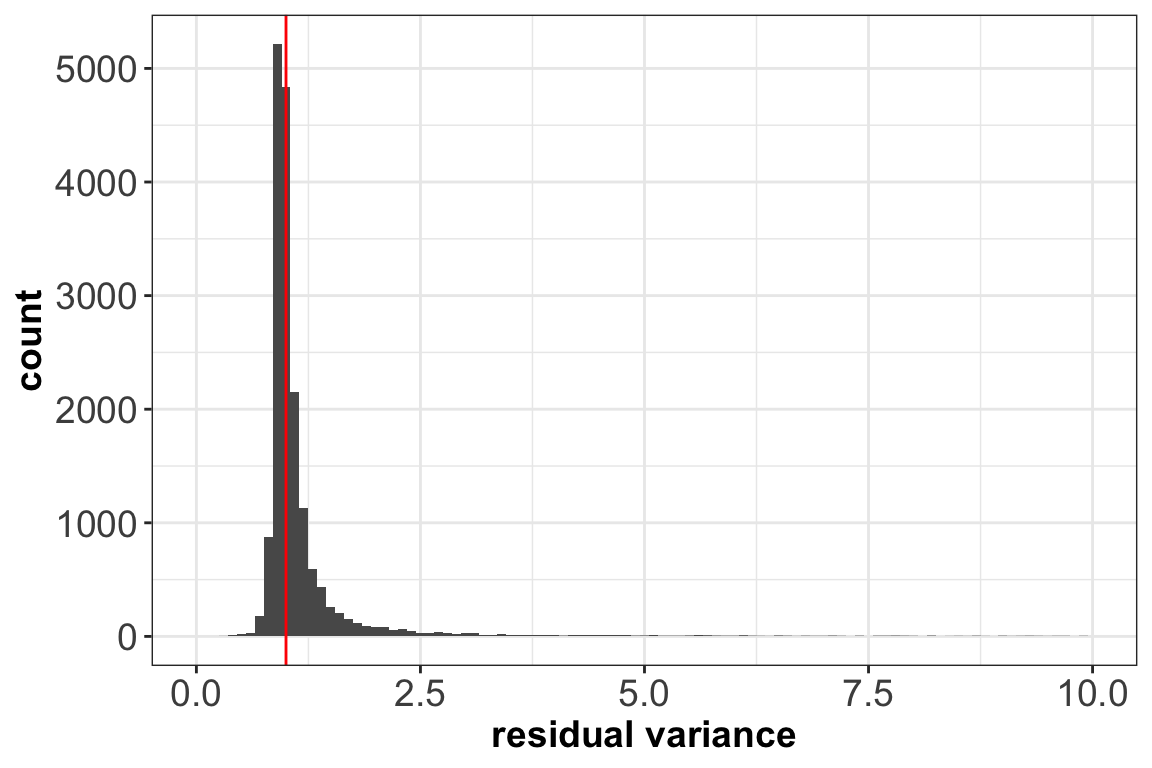
After transformation, the mean of the gene residuals is close to zero, and most genes have a variance around one. This suggests that overall the regularized negative binomial model is a suitable model that describes the effect of sequencing depth on UMI counts. Further, after transformation there is no relationship between gene mean and variance, as the next plot shows:
ggplot(vst_out$gene_attr, aes(log10(gmean), log10(residual_variance))) +
geom_point(alpha=0.3, shape=16) + geom_density_2d(size = 0.3) +
theme_bw() + theme(axis.text = element_text(size = 14),
axis.title = element_text(size = 14,face = 'bold'))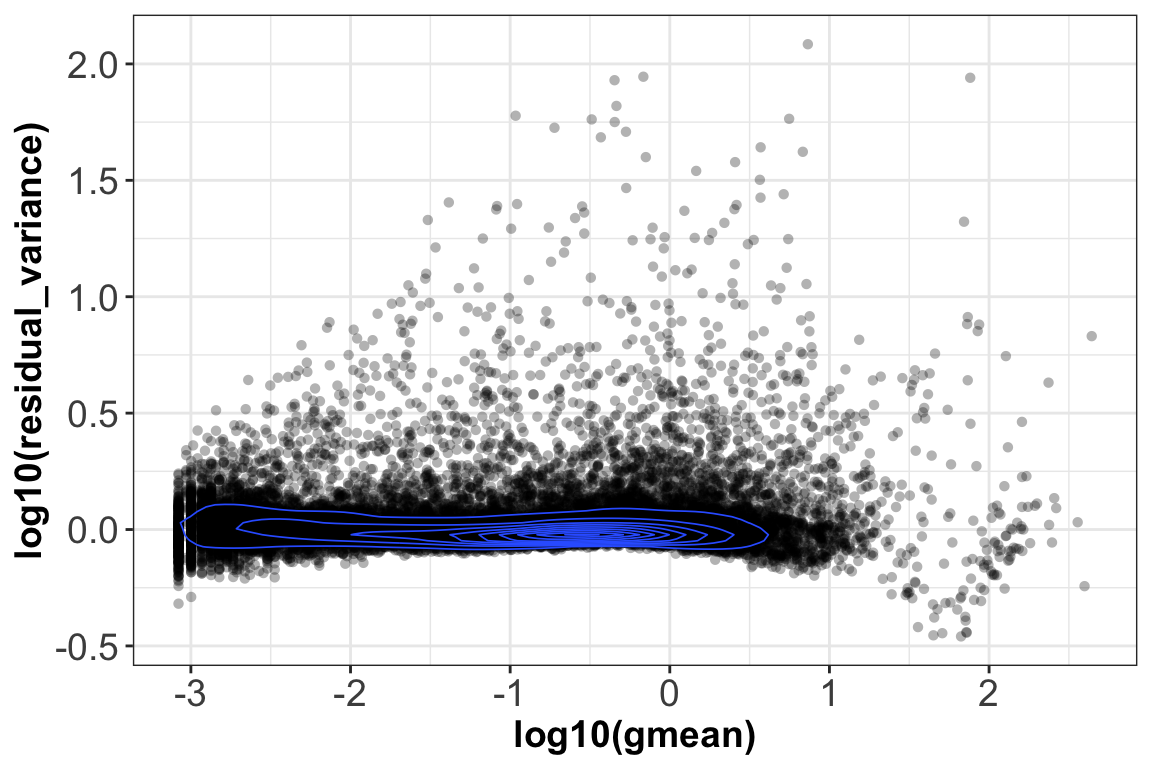
DE analysis using independent t-test
gRNA target: BCL11B locus
We filter the genes to be those present in at least 20% of all cells.
Number of genes that passed the filtering criterion: 8117
For a given target locus, we focus on 2 groups of cells: cells that contain only gRNAs for the target locus, and cells that only contain the negative control gRNAs.
Number of targeted cells with this type of gRNA uniquely: 59 Number of neg control cells: 111
Then, we conduct categorical regression on the Pearson residuals for cells in these 2 conditions to assess how differentially expressed each gene is.
Distribution of p values from the tests:
BCL11B.pval <- ttest(subset.sctransform_data,tg.cells,neg.cells)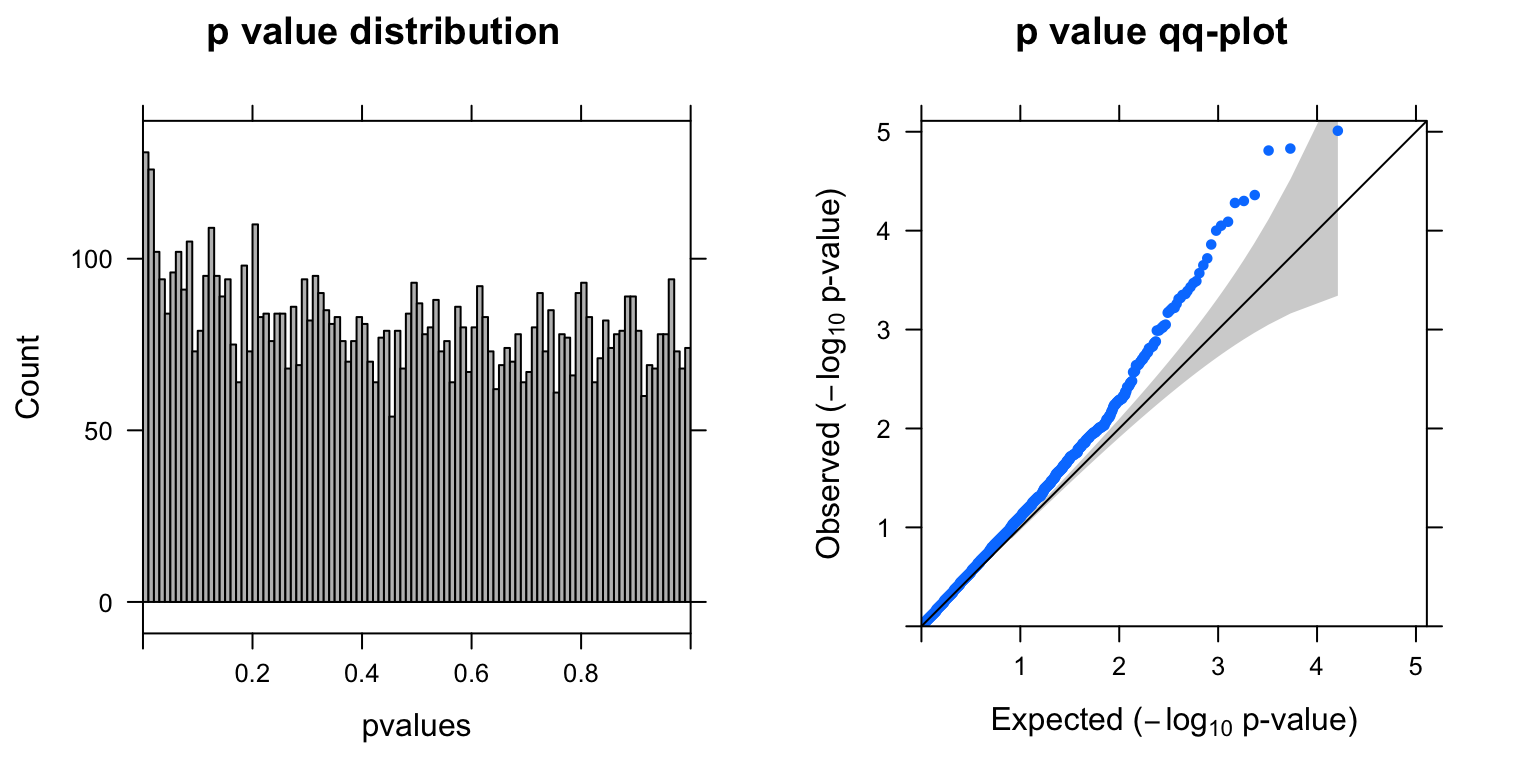 9 genes passed FDR=0.1 cutoff
9 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| NINJ1 | 9.7e-06 | 0.042 |
| DNAJC9 | 1.5e-05 | 0.042 |
| SLC35A1 | 1.6e-05 | 0.042 |
| CCP110 | 4.4e-05 | 0.071 |
| NPHP1 | 5.0e-05 | 0.071 |
| IMPA2 | 5.3e-05 | 0.071 |
| CDC6 | 8.1e-05 | 0.090 |
| CDCA7 | 8.9e-05 | 0.090 |
| XPNPEP3 | 1.0e-04 | 0.091 |
| PCNA | 1.4e-04 | 0.110 |
| MCM5 | 1.9e-04 | 0.140 |
| KEAP1 | 2.2e-04 | 0.150 |
| CACYBP | 2.7e-04 | 0.170 |
| MCM3 | 3.3e-04 | 0.180 |
| RANBP1 | 3.4e-04 | 0.180 |
| RFC5 | 3.7e-04 | 0.190 |
| HAUS1 | 4.0e-04 | 0.190 |
| APEX2 | 4.3e-04 | 0.190 |
| SLBP | 4.4e-04 | 0.190 |
| HPRT1 | 4.8e-04 | 0.190 |
| WDR76 | 4.9e-04 | 0.190 |
| SCML1 | 5.5e-04 | 0.200 |
| CTNNBIP1 | 6.0e-04 | 0.200 |
| TYMS | 6.0e-04 | 0.200 |
| BRI3BP | 6.4e-04 | 0.200 |
| FANCL | 6.6e-04 | 0.200 |
| MRPL12 | 6.8e-04 | 0.200 |
| LIMD2 | 8.8e-04 | 0.240 |
| TFDP1 | 9.1e-04 | 0.240 |
| CTPS1 | 9.5e-04 | 0.240 |
| ASRGL1 | 9.6e-04 | 0.240 |
| B3GALT6 | 1.0e-03 | 0.240 |
| VOPP1 | 1.0e-03 | 0.240 |
| FBXO5 | 1.0e-03 | 0.240 |
| PAQR4 | 1.3e-03 | 0.300 |
| MCM7 | 1.3e-03 | 0.300 |
| HNRNPF | 1.4e-03 | 0.300 |
| HELLS | 1.5e-03 | 0.310 |
| DUT | 1.5e-03 | 0.310 |
| FAM105A | 1.5e-03 | 0.310 |
| MAD2L2 | 1.6e-03 | 0.310 |
| RPS6KB2 | 1.7e-03 | 0.320 |
| PIGBOS1 | 1.7e-03 | 0.320 |
| IER2 | 1.8e-03 | 0.330 |
| TM9SF2 | 1.8e-03 | 0.330 |
| PPIF | 1.9e-03 | 0.330 |
| SNU13 | 2.0e-03 | 0.340 |
| ZNF24 | 2.0e-03 | 0.340 |
| RMI2 | 2.1e-03 | 0.340 |
| ATAD2 | 2.1e-03 | 0.340 |
| ST3GAL4 | 2.2e-03 | 0.340 |
| HN1L | 2.3e-03 | 0.340 |
| GLRX2 | 2.3e-03 | 0.340 |
| AKAP17A | 2.3e-03 | 0.340 |
| SRSF9 | 2.3e-03 | 0.340 |
| PSMC3IP | 2.3e-03 | 0.340 |
| PNPT1 | 2.7e-03 | 0.370 |
| SPATA33 | 2.7e-03 | 0.370 |
| TCF7L2 | 2.7e-03 | 0.370 |
| DSN1 | 3.3e-03 | 0.440 |
| ZMYND19 | 3.3e-03 | 0.440 |
| TPGS2 | 3.4e-03 | 0.440 |
| NUDT22 | 3.5e-03 | 0.450 |
| EXD2 | 3.6e-03 | 0.450 |
| NT5M | 3.7e-03 | 0.460 |
| RRM2 | 3.8e-03 | 0.460 |
| NUP58 | 3.8e-03 | 0.460 |
| SFT2D2 | 3.8e-03 | 0.460 |
| MED24 | 4.2e-03 | 0.490 |
| CWF19L2 | 4.2e-03 | 0.490 |
| COL6A1 | 4.3e-03 | 0.490 |
| SCARA3 | 4.6e-03 | 0.510 |
| MRPL11 | 4.7e-03 | 0.510 |
| NUP155 | 4.7e-03 | 0.510 |
| FAM228B | 4.8e-03 | 0.510 |
| EIF4A3 | 4.9e-03 | 0.510 |
| CHCHD3 | 5.0e-03 | 0.510 |
| IMPDH1 | 5.0e-03 | 0.510 |
| YTHDF3 | 5.1e-03 | 0.510 |
| RAB5C | 5.1e-03 | 0.510 |
| SLC12A5 | 5.2e-03 | 0.510 |
| SDF2 | 5.2e-03 | 0.510 |
| CMC2 | 5.2e-03 | 0.510 |
| POP7 | 5.3e-03 | 0.510 |
| GTF2IRD2 | 5.4e-03 | 0.510 |
| LAMP5 | 5.4e-03 | 0.510 |
| NSMAF | 5.4e-03 | 0.510 |
| SLC38A6 | 5.7e-03 | 0.520 |
| SKP2 | 5.7e-03 | 0.520 |
| MTERF3 | 5.8e-03 | 0.520 |
| POLR3K | 5.8e-03 | 0.520 |
| PSMA6 | 5.9e-03 | 0.520 |
| SGTA | 6.1e-03 | 0.530 |
| RP11-205M20.3 | 6.1e-03 | 0.530 |
| GINS2 | 6.5e-03 | 0.550 |
| GMPR2 | 6.6e-03 | 0.550 |
| ST8SIA1 | 6.7e-03 | 0.560 |
| DDX6 | 6.7e-03 | 0.560 |
| TCF12 | 6.8e-03 | 0.560 |
| ALPL | 7.2e-03 | 0.590 |
| GGA3 | 7.4e-03 | 0.590 |
| MPZL1 | 7.4e-03 | 0.590 |
| RPA1 | 7.7e-03 | 0.590 |
| GCSH | 7.7e-03 | 0.590 |
| POLM | 7.8e-03 | 0.590 |
| CHAF1B | 7.8e-03 | 0.590 |
| KAT6B | 8.0e-03 | 0.590 |
| POLR2E | 8.0e-03 | 0.590 |
| SNAPC3 | 8.0e-03 | 0.590 |
| LDHB | 8.4e-03 | 0.590 |
| YWHAQ | 8.5e-03 | 0.590 |
| RBM41 | 8.6e-03 | 0.590 |
| ARMCX6 | 8.6e-03 | 0.590 |
| DHFR | 9.0e-03 | 0.590 |
| CCDC174 | 9.2e-03 | 0.590 |
| GBF1 | 9.2e-03 | 0.590 |
| DERA | 9.3e-03 | 0.590 |
| TXNDC9 | 9.4e-03 | 0.590 |
| RPA2 | 9.4e-03 | 0.590 |
| MCM6 | 9.5e-03 | 0.590 |
| NASP | 9.5e-03 | 0.590 |
| AMOTL1 | 9.6e-03 | 0.590 |
| GPBP1 | 9.6e-03 | 0.590 |
| STRA13 | 9.7e-03 | 0.590 |
| TIMELESS | 9.7e-03 | 0.590 |
| TPGS1 | 9.7e-03 | 0.590 |
| SRSF2 | 9.8e-03 | 0.590 |
| STK39 | 9.9e-03 | 0.590 |
| GLUD1 | 9.9e-03 | 0.590 |
| BID | 1.0e-02 | 0.590 |
| NTMT1 | 1.0e-02 | 0.590 |
| RNH1 | 1.0e-02 | 0.590 |
| ZNF227 | 1.0e-02 | 0.590 |
| TTF2 | 1.0e-02 | 0.590 |
| FAAP20 | 1.0e-02 | 0.590 |
| IRGQ | 1.0e-02 | 0.590 |
| MPC1 | 1.1e-02 | 0.590 |
| TMEM98 | 1.1e-02 | 0.590 |
| CMTM7 | 1.1e-02 | 0.590 |
| RP3-395M20.12 | 1.1e-02 | 0.590 |
| TOMM5 | 1.1e-02 | 0.590 |
| MCMBP | 1.1e-02 | 0.590 |
| SNRNP25 | 1.1e-02 | 0.590 |
| OLFM1 | 1.1e-02 | 0.590 |
| METTL3 | 1.1e-02 | 0.590 |
| DNAJB9 | 1.1e-02 | 0.590 |
| FAM58A | 1.1e-02 | 0.590 |
| TOMM40L | 1.1e-02 | 0.590 |
| IWS1 | 1.1e-02 | 0.590 |
| ASF1B | 1.1e-02 | 0.590 |
| NAP1L4 | 1.1e-02 | 0.590 |
| ANKRD9 | 1.2e-02 | 0.590 |
| RP11-304L19.5 | 1.2e-02 | 0.590 |
| SKA1 | 1.2e-02 | 0.590 |
| PKMYT1 | 1.2e-02 | 0.590 |
| DPF1 | 1.2e-02 | 0.590 |
| ZNF691 | 1.2e-02 | 0.590 |
| LTBP1 | 1.2e-02 | 0.590 |
| CRKL | 1.2e-02 | 0.590 |
| ZNF562 | 1.2e-02 | 0.590 |
| PPIL4 | 1.2e-02 | 0.590 |
| HIBADH | 1.2e-02 | 0.590 |
| HES5 | 1.2e-02 | 0.590 |
| CCDC124 | 1.2e-02 | 0.590 |
| FABP5 | 1.2e-02 | 0.590 |
| SLC35A5 | 1.2e-02 | 0.590 |
| FOXO3 | 1.3e-02 | 0.590 |
| PLEKHM2 | 1.3e-02 | 0.590 |
| VAMP4 | 1.3e-02 | 0.590 |
| STT3B | 1.3e-02 | 0.590 |
| CPNE3 | 1.3e-02 | 0.590 |
| NPRL2 | 1.3e-02 | 0.590 |
| PVT1 | 1.3e-02 | 0.590 |
| ZNF7 | 1.3e-02 | 0.590 |
| DPH5 | 1.3e-02 | 0.590 |
| CCNE1 | 1.3e-02 | 0.590 |
| CLPTM1L | 1.3e-02 | 0.610 |
| ZNF677 | 1.3e-02 | 0.610 |
| FEN1 | 1.3e-02 | 0.610 |
| PSMG1 | 1.4e-02 | 0.610 |
| TUBG1 | 1.4e-02 | 0.610 |
| PRPF19 | 1.4e-02 | 0.610 |
| UBE2T | 1.4e-02 | 0.610 |
| DCTPP1 | 1.4e-02 | 0.610 |
| UNG | 1.4e-02 | 0.610 |
| TEAD1 | 1.4e-02 | 0.610 |
| THOC6 | 1.4e-02 | 0.610 |
| ACAT2 | 1.4e-02 | 0.610 |
| PDCD5 | 1.4e-02 | 0.610 |
| ENDOG | 1.4e-02 | 0.610 |
| STAMBP | 1.4e-02 | 0.610 |
| PNPLA2 | 1.4e-02 | 0.610 |
| ERVK3-1 | 1.4e-02 | 0.610 |
| CFAP36 | 1.5e-02 | 0.610 |
| RTN3 | 1.5e-02 | 0.620 |
| NUDT3 | 1.5e-02 | 0.620 |
| OBFC1 | 1.5e-02 | 0.620 |
| PPP1R37 | 1.5e-02 | 0.620 |
| COX5A | 1.5e-02 | 0.620 |
| TRIT1 | 1.5e-02 | 0.620 |
| ENTHD2 | 1.5e-02 | 0.620 |
| NSMCE4A | 1.6e-02 | 0.620 |
| GNG4 | 1.6e-02 | 0.620 |
| MYBL2 | 1.6e-02 | 0.620 |
| PLCE1 | 1.6e-02 | 0.620 |
| ZNHIT3 | 1.6e-02 | 0.620 |
| RTCA | 1.6e-02 | 0.620 |
| KDM4B | 1.6e-02 | 0.620 |
| SUCO | 1.6e-02 | 0.620 |
| SLC2A3 | 1.6e-02 | 0.620 |
| CAPN15 | 1.6e-02 | 0.620 |
| MARC1 | 1.6e-02 | 0.620 |
| MED11 | 1.7e-02 | 0.620 |
| CD46 | 1.7e-02 | 0.620 |
| TWF1 | 1.7e-02 | 0.620 |
| ARPC1A | 1.7e-02 | 0.620 |
| RGS17 | 1.7e-02 | 0.620 |
| THYN1 | 1.7e-02 | 0.620 |
| RNPS1 | 1.7e-02 | 0.620 |
| C14orf166 | 1.7e-02 | 0.620 |
| SRD5A1 | 1.8e-02 | 0.620 |
| ARFGEF1 | 1.8e-02 | 0.620 |
| CCDC106 | 1.8e-02 | 0.620 |
| CNRIP1 | 1.8e-02 | 0.620 |
| TRIM27 | 1.8e-02 | 0.620 |
| PTEN | 1.8e-02 | 0.620 |
| SDHAF3 | 1.8e-02 | 0.620 |
| MRPL23 | 1.8e-02 | 0.620 |
| ADGRL2 | 1.8e-02 | 0.620 |
| UBE2H | 1.8e-02 | 0.620 |
| REEP3 | 1.8e-02 | 0.620 |
| BLMH | 1.8e-02 | 0.620 |
| SIVA1 | 1.8e-02 | 0.620 |
| EIF2A | 1.8e-02 | 0.620 |
| RAD23A | 1.9e-02 | 0.620 |
| SCD | 1.9e-02 | 0.620 |
| PTPRN2 | 1.9e-02 | 0.620 |
| TRIP4 | 1.9e-02 | 0.620 |
| CCDC85B | 1.9e-02 | 0.620 |
| SNX4 | 1.9e-02 | 0.620 |
| ALKBH5 | 1.9e-02 | 0.620 |
| TNIP1 | 1.9e-02 | 0.620 |
| SNRPD2 | 1.9e-02 | 0.620 |
| MRPL24 | 1.9e-02 | 0.620 |
| ERLEC1 | 1.9e-02 | 0.620 |
| RPRML | 1.9e-02 | 0.620 |
| PRKD3 | 1.9e-02 | 0.620 |
| LRRC45 | 1.9e-02 | 0.620 |
| ZNF738 | 1.9e-02 | 0.620 |
| IFNGR2 | 1.9e-02 | 0.620 |
| COMMD2 | 2.0e-02 | 0.620 |
| AP1S1 | 2.0e-02 | 0.620 |
| NELFB | 2.0e-02 | 0.620 |
| NUDT2 | 2.0e-02 | 0.620 |
| PPP3CB | 2.0e-02 | 0.620 |
| ARHGEF26 | 2.0e-02 | 0.620 |
| FRZB | 2.0e-02 | 0.620 |
| MAF1 | 2.0e-02 | 0.630 |
| MIS12 | 2.0e-02 | 0.630 |
| CBLB | 2.0e-02 | 0.630 |
| SRSF7 | 2.0e-02 | 0.630 |
| FBXW11 | 2.0e-02 | 0.630 |
| H2AFY2 | 2.0e-02 | 0.630 |
| MID1IP1 | 2.1e-02 | 0.630 |
| PLRG1 | 2.1e-02 | 0.630 |
| SEC11A | 2.1e-02 | 0.630 |
| UQCRH | 2.1e-02 | 0.630 |
| UBAP1 | 2.1e-02 | 0.640 |
| TM2D1 | 2.1e-02 | 0.640 |
| TNRC6A | 2.2e-02 | 0.640 |
| NRDC | 2.2e-02 | 0.640 |
| TSN | 2.2e-02 | 0.640 |
| SLC29A4 | 2.2e-02 | 0.640 |
| WNT7B | 2.2e-02 | 0.640 |
| INO80B | 2.2e-02 | 0.640 |
| PHLDA1 | 2.2e-02 | 0.650 |
| DBI | 2.2e-02 | 0.650 |
| ZRANB1 | 2.2e-02 | 0.650 |
| SPRYD4 | 2.2e-02 | 0.650 |
| RB1CC1 | 2.2e-02 | 0.650 |
| UIMC1 | 2.2e-02 | 0.650 |
| HLA-B | 2.2e-02 | 0.650 |
| FNDC3A | 2.3e-02 | 0.650 |
| SNRNP200 | 2.3e-02 | 0.650 |
| BIVM | 2.3e-02 | 0.650 |
| FLAD1 | 2.3e-02 | 0.650 |
| C19orf68 | 2.3e-02 | 0.650 |
| EPS15 | 2.3e-02 | 0.650 |
| RFC2 | 2.3e-02 | 0.650 |
| ARPIN | 2.3e-02 | 0.650 |
| PDLIM3 | 2.3e-02 | 0.650 |
| POLE3 | 2.3e-02 | 0.650 |
| CARHSP1 | 2.3e-02 | 0.650 |
| BDH2 | 2.4e-02 | 0.650 |
| YTHDF1 | 2.4e-02 | 0.650 |
| WDR13 | 2.4e-02 | 0.650 |
| C9orf40 | 2.4e-02 | 0.650 |
| TSPAN15 | 2.4e-02 | 0.650 |
| DDX46 | 2.4e-02 | 0.650 |
| PRKAB2 | 2.4e-02 | 0.650 |
| GMNN | 2.4e-02 | 0.660 |
| S100PBP | 2.5e-02 | 0.660 |
| KLF3 | 2.5e-02 | 0.660 |
| TRMT11 | 2.5e-02 | 0.660 |
| NOLC1 | 2.5e-02 | 0.660 |
| KLHL42 | 2.5e-02 | 0.660 |
| RP4-758J18.2 | 2.5e-02 | 0.660 |
| PXMP2 | 2.5e-02 | 0.660 |
| FDPS | 2.5e-02 | 0.660 |
| PPP1R7 | 2.6e-02 | 0.660 |
| STXBP3 | 2.6e-02 | 0.660 |
| PROSER1 | 2.6e-02 | 0.660 |
| PRMT7 | 2.6e-02 | 0.660 |
| TP53I13 | 2.6e-02 | 0.660 |
| TOMM40 | 2.6e-02 | 0.660 |
| SFXN1 | 2.6e-02 | 0.660 |
| EIF3L | 2.6e-02 | 0.660 |
| TMEM183A | 2.7e-02 | 0.660 |
| GPATCH2L | 2.7e-02 | 0.660 |
| PKIB | 2.7e-02 | 0.660 |
| CD320 | 2.7e-02 | 0.660 |
| EME1 | 2.7e-02 | 0.660 |
| SNRPD1 | 2.7e-02 | 0.660 |
| RFC1 | 2.7e-02 | 0.660 |
| EPS15L1 | 2.7e-02 | 0.660 |
| BANF1 | 2.7e-02 | 0.660 |
| MCEE | 2.7e-02 | 0.660 |
| AAK1 | 2.7e-02 | 0.660 |
| SEMA3A | 2.7e-02 | 0.660 |
| SAT2 | 2.7e-02 | 0.660 |
| NUDT10 | 2.7e-02 | 0.660 |
| CETN2 | 2.7e-02 | 0.660 |
| C15orf61 | 2.8e-02 | 0.660 |
| TP53RK | 2.8e-02 | 0.660 |
| RFC4 | 2.8e-02 | 0.660 |
| HNRNPD | 2.8e-02 | 0.660 |
| SNRPB | 2.8e-02 | 0.660 |
| RGS5 | 2.8e-02 | 0.660 |
| MUTYH | 2.8e-02 | 0.660 |
| HSD11B1L | 2.8e-02 | 0.660 |
| DHX57 | 2.8e-02 | 0.660 |
| CITED4 | 2.8e-02 | 0.660 |
| JTB | 2.8e-02 | 0.660 |
| ZNF660 | 2.8e-02 | 0.660 |
| MSH6 | 2.8e-02 | 0.660 |
| UGDH | 2.8e-02 | 0.660 |
| GABPB1 | 2.8e-02 | 0.660 |
| RAD54L2 | 2.9e-02 | 0.670 |
| TMEM88 | 2.9e-02 | 0.670 |
| RNF165 | 2.9e-02 | 0.670 |
| PLSCR1 | 2.9e-02 | 0.670 |
| DCLK2 | 2.9e-02 | 0.670 |
| YEATS4 | 2.9e-02 | 0.670 |
| TUBA1B | 3.0e-02 | 0.670 |
| SAP130 | 3.0e-02 | 0.670 |
| TARBP2 | 3.0e-02 | 0.670 |
| MIOS | 3.0e-02 | 0.680 |
| GOLGB1 | 3.0e-02 | 0.680 |
| MRPS26 | 3.0e-02 | 0.680 |
| CC2D1A | 3.0e-02 | 0.680 |
| MEA1 | 3.0e-02 | 0.680 |
| EZR | 3.1e-02 | 0.680 |
| TMEM199 | 3.1e-02 | 0.680 |
| SF3A3 | 3.1e-02 | 0.690 |
| MAPKAPK5 | 3.1e-02 | 0.690 |
| HEXIM1 | 3.1e-02 | 0.690 |
| CD151 | 3.1e-02 | 0.690 |
| ODF2L | 3.1e-02 | 0.690 |
| RAD23B | 3.2e-02 | 0.690 |
| C9orf116 | 3.2e-02 | 0.690 |
| SSBP1 | 3.2e-02 | 0.690 |
| FAM13B | 3.2e-02 | 0.690 |
| RBPMS2 | 3.2e-02 | 0.690 |
| AGPAT1 | 3.2e-02 | 0.690 |
| UBAP2 | 3.2e-02 | 0.690 |
| KLHDC3 | 3.2e-02 | 0.690 |
| PNMA1 | 3.2e-02 | 0.690 |
| SRSF1 | 3.2e-02 | 0.690 |
| QARS | 3.2e-02 | 0.700 |
| EXOSC9 | 3.3e-02 | 0.700 |
| TGS1 | 3.3e-02 | 0.700 |
| TIMM23 | 3.3e-02 | 0.700 |
| RBBP7 | 3.3e-02 | 0.700 |
| MRPL57 | 3.3e-02 | 0.700 |
| GPATCH11 | 3.4e-02 | 0.700 |
| WDR34 | 3.4e-02 | 0.700 |
| MGAT4B | 3.4e-02 | 0.700 |
| PAXBP1 | 3.4e-02 | 0.700 |
| CMTM8 | 3.4e-02 | 0.700 |
| FAM118A | 3.4e-02 | 0.700 |
| HMG20B | 3.4e-02 | 0.700 |
| RPN2 | 3.4e-02 | 0.700 |
| MARCH7 | 3.4e-02 | 0.700 |
| ETNK2 | 3.4e-02 | 0.700 |
| INPP5F | 3.4e-02 | 0.700 |
| C21orf2 | 3.4e-02 | 0.700 |
| CDT1 | 3.4e-02 | 0.700 |
| GRPEL1 | 3.4e-02 | 0.700 |
| ACTR3 | 3.5e-02 | 0.710 |
| FAM108C1 | 3.5e-02 | 0.710 |
| ARPC5L | 3.5e-02 | 0.710 |
| LAMTOR4 | 3.5e-02 | 0.710 |
| MSH2 | 3.5e-02 | 0.710 |
| AP2S1 | 3.5e-02 | 0.710 |
| HDAC8 | 3.6e-02 | 0.710 |
| DYNC1I2 | 3.6e-02 | 0.710 |
| ATL1 | 3.6e-02 | 0.710 |
| ACP2 | 3.6e-02 | 0.710 |
| TMEM251 | 3.6e-02 | 0.710 |
| COA5 | 3.6e-02 | 0.710 |
| CCDC90B | 3.6e-02 | 0.710 |
| SPTBN1 | 3.6e-02 | 0.710 |
| CLSPN | 3.6e-02 | 0.710 |
| NTRK3 | 3.6e-02 | 0.710 |
| BAK1 | 3.6e-02 | 0.710 |
| SF1 | 3.6e-02 | 0.710 |
| EEF1AKMT1 | 3.6e-02 | 0.710 |
| LUC7L | 3.7e-02 | 0.710 |
| INSIG2 | 3.7e-02 | 0.710 |
| WDR33 | 3.7e-02 | 0.710 |
| CCNG1 | 3.7e-02 | 0.710 |
| CDCA4 | 3.7e-02 | 0.710 |
| WASHC4 | 3.7e-02 | 0.710 |
| IER5 | 3.7e-02 | 0.710 |
| ATP6V1C1 | 3.7e-02 | 0.710 |
| SLC44A2 | 3.7e-02 | 0.710 |
| PLEKHA5 | 3.7e-02 | 0.710 |
| PPP2R5E | 3.7e-02 | 0.710 |
| NUDCD1 | 3.8e-02 | 0.710 |
| KAT8 | 3.8e-02 | 0.710 |
| GLE1 | 3.8e-02 | 0.710 |
| UFL1 | 3.8e-02 | 0.710 |
| B3GAT3 | 3.8e-02 | 0.710 |
| PRPS2 | 3.8e-02 | 0.710 |
| AP2A1 | 3.8e-02 | 0.710 |
| SNRNP27 | 3.8e-02 | 0.710 |
| FAM111B | 3.8e-02 | 0.710 |
| BAG2 | 3.8e-02 | 0.710 |
| RAD9A | 3.9e-02 | 0.710 |
| CTSA | 3.9e-02 | 0.710 |
| ZKSCAN1 | 3.9e-02 | 0.710 |
| CCBL2 | 3.9e-02 | 0.710 |
| TCF19 | 3.9e-02 | 0.710 |
| CDC42EP3 | 3.9e-02 | 0.710 |
| BRPF1 | 3.9e-02 | 0.710 |
| CCDC109B | 3.9e-02 | 0.710 |
| MPV17L2 | 3.9e-02 | 0.710 |
| TECR | 3.9e-02 | 0.710 |
| ITGA6 | 4.0e-02 | 0.710 |
| PCLAF | 4.0e-02 | 0.710 |
| USP1 | 4.0e-02 | 0.710 |
| EPHB4 | 4.0e-02 | 0.710 |
| TRIM56 | 4.0e-02 | 0.720 |
| POP1 | 4.0e-02 | 0.720 |
| CPPED1 | 4.0e-02 | 0.720 |
| SAAL1 | 4.0e-02 | 0.720 |
| FKBP1B | 4.0e-02 | 0.720 |
| RP11-21N3.1 | 4.1e-02 | 0.720 |
| TMEM106C | 4.1e-02 | 0.720 |
| C17orf80 | 4.1e-02 | 0.720 |
| HACD2 | 4.1e-02 | 0.720 |
| UXT | 4.1e-02 | 0.720 |
| LARP7 | 4.1e-02 | 0.720 |
| CGRRF1 | 4.1e-02 | 0.720 |
| PQLC1 | 4.1e-02 | 0.720 |
| MBD6 | 4.2e-02 | 0.730 |
| NEK3 | 4.2e-02 | 0.730 |
| ARAF | 4.2e-02 | 0.730 |
| OGT | 4.2e-02 | 0.730 |
| RAB4A | 4.3e-02 | 0.730 |
| CCDC50 | 4.3e-02 | 0.730 |
| ZNF672 | 4.3e-02 | 0.730 |
| VRK3 | 4.3e-02 | 0.730 |
| CERS6 | 4.3e-02 | 0.730 |
| PIGX | 4.3e-02 | 0.730 |
| NUFIP1 | 4.3e-02 | 0.730 |
| CHPF2 | 4.3e-02 | 0.740 |
| PSMA5 | 4.4e-02 | 0.740 |
| MLXIP | 4.4e-02 | 0.740 |
| FAM127C | 4.4e-02 | 0.740 |
| FLII | 4.4e-02 | 0.750 |
| RFX3 | 4.4e-02 | 0.750 |
| CCDC167 | 4.5e-02 | 0.750 |
| LIMK2 | 4.5e-02 | 0.750 |
| NDUFV1 | 4.5e-02 | 0.750 |
| OTULIN | 4.5e-02 | 0.750 |
| ERP29 | 4.5e-02 | 0.750 |
| BNIP3 | 4.5e-02 | 0.750 |
| SMIM13 | 4.5e-02 | 0.750 |
| MIS18A | 4.6e-02 | 0.750 |
| HOMER3 | 4.6e-02 | 0.750 |
| QRICH1 | 4.6e-02 | 0.750 |
| PUS7 | 4.6e-02 | 0.750 |
| ATAD1 | 4.6e-02 | 0.750 |
| DSCC1 | 4.6e-02 | 0.750 |
| BCAT2 | 4.6e-02 | 0.750 |
| ICMT | 4.6e-02 | 0.750 |
| RNF144A | 4.7e-02 | 0.750 |
| PGGT1B | 4.7e-02 | 0.750 |
| LEO1 | 4.7e-02 | 0.750 |
| AC092296.1 | 4.7e-02 | 0.750 |
| TSEN54 | 4.7e-02 | 0.750 |
| TPRA1 | 4.7e-02 | 0.750 |
| MOK | 4.7e-02 | 0.750 |
| ZNF277 | 4.8e-02 | 0.750 |
| GNAQ | 4.8e-02 | 0.750 |
| ERCC3 | 4.8e-02 | 0.750 |
| SCAF8 | 4.8e-02 | 0.750 |
| PLXNA1 | 4.8e-02 | 0.750 |
| ARHGEF40 | 4.8e-02 | 0.750 |
| NUP214 | 4.8e-02 | 0.750 |
| GBA2 | 4.8e-02 | 0.750 |
| TRIP12 | 4.8e-02 | 0.750 |
| NETO2 | 4.8e-02 | 0.750 |
| GOLGA2 | 4.8e-02 | 0.750 |
| UBQLN4 | 4.8e-02 | 0.750 |
| PLCD3 | 4.9e-02 | 0.750 |
| CLEC11A | 4.9e-02 | 0.750 |
| TRIM24 | 4.9e-02 | 0.750 |
| SMCO4 | 4.9e-02 | 0.750 |
| DXO | 4.9e-02 | 0.750 |
| HSD17B4 | 4.9e-02 | 0.750 |
| RCOR3 | 4.9e-02 | 0.750 |
| FBXO28 | 4.9e-02 | 0.750 |
| TTLL1 | 4.9e-02 | 0.750 |
| TNFRSF10B | 4.9e-02 | 0.750 |
| BAG4 | 4.9e-02 | 0.750 |
| DDX23 | 4.9e-02 | 0.750 |
| HIRIP3 | 4.9e-02 | 0.750 |
| TSPAN31 | 4.9e-02 | 0.750 |
| FPGS | 4.9e-02 | 0.750 |
| RRAS2 | 4.9e-02 | 0.750 |
| PRKAR2B | 4.9e-02 | 0.750 |
| LPIN2 | 4.9e-02 | 0.750 |
| SLC37A4 | 5.0e-02 | 0.750 |
| PDLIM2 | 5.0e-02 | 0.750 |
| FOS | 5.0e-02 | 0.750 |
| ARVCF | 5.0e-02 | 0.750 |
| NDUFAF2 | 5.0e-02 | 0.750 |
| TPX2 | 5.0e-02 | 0.750 |
| CENPJ | 5.0e-02 | 0.750 |
| EFNA5 | 5.0e-02 | 0.750 |
| MSANTD2 | 5.0e-02 | 0.750 |
| TMEM8B | 5.0e-02 | 0.750 |
| PPAN | 5.0e-02 | 0.750 |
| TRNT1 | 5.0e-02 | 0.750 |
| MZT2B | 5.0e-02 | 0.750 |
| CUL2 | 5.1e-02 | 0.750 |
| MPHOSPH6 | 5.1e-02 | 0.750 |
| LNP1 | 5.1e-02 | 0.750 |
| FXYD6 | 5.1e-02 | 0.750 |
| PLCXD1 | 5.1e-02 | 0.750 |
| SRSF6 | 5.1e-02 | 0.750 |
| ANKRD10 | 5.1e-02 | 0.750 |
| TMEM159 | 5.2e-02 | 0.750 |
| TAF5 | 5.2e-02 | 0.750 |
| TMEM70 | 5.2e-02 | 0.750 |
| FAM219B | 5.2e-02 | 0.750 |
| FAHD2A | 5.2e-02 | 0.750 |
| RRP9 | 5.2e-02 | 0.750 |
| ATP5E | 5.2e-02 | 0.750 |
| SCARB2 | 5.2e-02 | 0.750 |
| EFCAB11 | 5.2e-02 | 0.750 |
| PIGU | 5.2e-02 | 0.750 |
| OGDH | 5.2e-02 | 0.750 |
| NDUFA6 | 5.2e-02 | 0.750 |
| OSTC | 5.2e-02 | 0.750 |
| GEMIN4 | 5.2e-02 | 0.750 |
| MFSD14C | 5.2e-02 | 0.750 |
| BLOC1S1 | 5.3e-02 | 0.750 |
| DENND5A | 5.3e-02 | 0.750 |
| MBTPS1 | 5.3e-02 | 0.750 |
| TMCO3 | 5.3e-02 | 0.750 |
| NUDT16L1 | 5.3e-02 | 0.750 |
| DNAJC24 | 5.3e-02 | 0.750 |
| C2orf49 | 5.4e-02 | 0.750 |
| LY6E | 5.4e-02 | 0.750 |
| PTMA | 5.4e-02 | 0.750 |
| WWC3 | 5.4e-02 | 0.750 |
| NLE1 | 5.4e-02 | 0.750 |
| LIG1 | 5.4e-02 | 0.750 |
| TMEM60 | 5.4e-02 | 0.750 |
| RPL36AL | 5.4e-02 | 0.750 |
| NDUFC2 | 5.4e-02 | 0.750 |
| UQCRC1 | 5.4e-02 | 0.750 |
| DTX3 | 5.4e-02 | 0.750 |
| CTNND1 | 5.4e-02 | 0.750 |
| CD24 | 5.5e-02 | 0.750 |
| U2SURP | 5.5e-02 | 0.750 |
| PTCD2 | 5.5e-02 | 0.750 |
| BCL2L12 | 5.5e-02 | 0.750 |
| XRCC3 | 5.5e-02 | 0.750 |
| NET1 | 5.5e-02 | 0.750 |
| GPN1 | 5.5e-02 | 0.750 |
| RP11-398C13.8 | 5.5e-02 | 0.750 |
| RPS6KA1 | 5.6e-02 | 0.750 |
| FBXO21 | 5.6e-02 | 0.750 |
| SPG20 | 5.6e-02 | 0.750 |
| ZNF274 | 5.6e-02 | 0.750 |
| CENPM | 5.6e-02 | 0.750 |
| DHRS1 | 5.6e-02 | 0.750 |
| LPP | 5.6e-02 | 0.750 |
| MRPS34 | 5.6e-02 | 0.750 |
| RNF126 | 5.6e-02 | 0.750 |
| GOLPH3L | 5.6e-02 | 0.750 |
| PHF21A | 5.6e-02 | 0.760 |
| NCAPG | 5.7e-02 | 0.760 |
| VDAC1 | 5.7e-02 | 0.760 |
| ASAP2 | 5.7e-02 | 0.760 |
| NUDT5 | 5.7e-02 | 0.760 |
| PDCD10 | 5.7e-02 | 0.760 |
| ZNF48 | 5.7e-02 | 0.760 |
| THAP10 | 5.7e-02 | 0.760 |
| DLG5 | 5.7e-02 | 0.760 |
| NDUFAF7 | 5.8e-02 | 0.760 |
| PLEKHA1 | 5.8e-02 | 0.760 |
| DCTN3 | 5.8e-02 | 0.760 |
| CSAD | 5.8e-02 | 0.760 |
| CKLF | 5.8e-02 | 0.760 |
| ZNF385A | 5.8e-02 | 0.760 |
| TIMM9 | 5.8e-02 | 0.760 |
| TTI1 | 5.8e-02 | 0.760 |
| THAP1 | 5.8e-02 | 0.760 |
| EIF3B | 5.8e-02 | 0.760 |
| TMEM222 | 5.9e-02 | 0.760 |
| RPAP3 | 5.9e-02 | 0.760 |
| ARMT1 | 5.9e-02 | 0.760 |
| GATAD2A | 5.9e-02 | 0.760 |
| NRBF2 | 5.9e-02 | 0.760 |
| REPS1 | 6.0e-02 | 0.760 |
| ARL13B | 6.0e-02 | 0.760 |
| MRPS27 | 6.0e-02 | 0.760 |
| ZFAND2A | 6.0e-02 | 0.760 |
| GMPPA | 6.0e-02 | 0.760 |
| SNRPC | 6.0e-02 | 0.760 |
| SPC25 | 6.0e-02 | 0.760 |
| C20orf27 | 6.0e-02 | 0.760 |
| SOX21 | 6.0e-02 | 0.760 |
| QTRT1 | 6.0e-02 | 0.760 |
| CENPV | 6.0e-02 | 0.760 |
| SPA17 | 6.0e-02 | 0.760 |
| FMNL2 | 6.0e-02 | 0.760 |
| RBBP5 | 6.1e-02 | 0.770 |
| CEP89 | 6.1e-02 | 0.770 |
| FIBP | 6.1e-02 | 0.770 |
| RHOT1 | 6.1e-02 | 0.770 |
| LRRC8D | 6.1e-02 | 0.770 |
| BCL7A | 6.2e-02 | 0.770 |
| PES1 | 6.2e-02 | 0.770 |
| AKAP1 | 6.2e-02 | 0.770 |
| RAB35 | 6.2e-02 | 0.770 |
| RAF1 | 6.2e-02 | 0.770 |
| NXT1 | 6.2e-02 | 0.770 |
| RBBP6 | 6.2e-02 | 0.770 |
| PHB | 6.2e-02 | 0.770 |
| SHKBP1 | 6.2e-02 | 0.770 |
| HIPK1 | 6.2e-02 | 0.770 |
| ESF1 | 6.2e-02 | 0.770 |
| CHAF1A | 6.3e-02 | 0.770 |
| STARD3 | 6.3e-02 | 0.770 |
| HIST1H4C | 6.3e-02 | 0.770 |
| FAM92A1 | 6.3e-02 | 0.770 |
| ZNF644 | 6.3e-02 | 0.770 |
| USP33 | 6.3e-02 | 0.770 |
| PRKACB | 6.3e-02 | 0.770 |
| SMYD5 | 6.3e-02 | 0.770 |
| NARS2 | 6.3e-02 | 0.770 |
| BAG1 | 6.3e-02 | 0.770 |
| NCOR1 | 6.4e-02 | 0.770 |
| CCNL1 | 6.4e-02 | 0.770 |
| PPIH | 6.4e-02 | 0.770 |
| HACD3 | 6.4e-02 | 0.770 |
| IMPDH2 | 6.4e-02 | 0.770 |
| WDR26 | 6.4e-02 | 0.770 |
| RP11-410L14.2 | 6.4e-02 | 0.770 |
| RBAK-RBAKDN | 6.5e-02 | 0.770 |
| ZNF581 | 6.5e-02 | 0.770 |
| SMPD1 | 6.5e-02 | 0.770 |
| CHAC2 | 6.5e-02 | 0.770 |
| UTP18 | 6.5e-02 | 0.770 |
| PNISR | 6.5e-02 | 0.770 |
| TELO2 | 6.5e-02 | 0.770 |
| TAF8 | 6.5e-02 | 0.770 |
| H2AFY | 6.5e-02 | 0.770 |
| HINFP | 6.5e-02 | 0.770 |
| RP4-614O4.5 | 6.5e-02 | 0.770 |
| ETV1 | 6.6e-02 | 0.770 |
| ZBTB17 | 6.6e-02 | 0.770 |
| PPP2CA | 6.6e-02 | 0.770 |
| SELT | 6.6e-02 | 0.770 |
| ANXA11 | 6.6e-02 | 0.770 |
| STX7 | 6.6e-02 | 0.770 |
| USP10 | 6.6e-02 | 0.770 |
| CBX1 | 6.6e-02 | 0.770 |
| MPND | 6.6e-02 | 0.770 |
| ARMC1 | 6.6e-02 | 0.770 |
| CISD1 | 6.6e-02 | 0.770 |
| TXNDC12 | 6.6e-02 | 0.770 |
| DST | 6.6e-02 | 0.770 |
| RAB6A | 6.7e-02 | 0.770 |
| OXCT1 | 6.7e-02 | 0.770 |
| BAZ1A | 6.7e-02 | 0.770 |
| CENPT | 6.7e-02 | 0.770 |
| DAXX | 6.7e-02 | 0.770 |
| ITSN1 | 6.7e-02 | 0.770 |
| BAP1 | 6.7e-02 | 0.770 |
| ALDH18A1 | 6.7e-02 | 0.770 |
| SIGMAR1 | 6.7e-02 | 0.770 |
| CENPS | 6.7e-02 | 0.770 |
| ZNF746 | 6.8e-02 | 0.770 |
| ZNF524 | 6.8e-02 | 0.770 |
| ZBTB8OS | 6.8e-02 | 0.770 |
| LAPTM4A | 6.8e-02 | 0.770 |
| BORCS8 | 6.8e-02 | 0.770 |
| B4GAT1 | 6.8e-02 | 0.770 |
| HSPG2 | 6.8e-02 | 0.770 |
| EID2B | 6.8e-02 | 0.770 |
| PROSER3 | 6.8e-02 | 0.770 |
| SIAH1 | 6.9e-02 | 0.770 |
| UBE2G2 | 6.9e-02 | 0.770 |
| PRDX2 | 6.9e-02 | 0.770 |
| PPM1A | 6.9e-02 | 0.770 |
| KIAA0195 | 6.9e-02 | 0.770 |
| GTF2F1 | 6.9e-02 | 0.770 |
| TMEM177 | 6.9e-02 | 0.770 |
| FAM188A | 6.9e-02 | 0.770 |
| NAP1L3 | 6.9e-02 | 0.770 |
| PHF5A | 6.9e-02 | 0.770 |
| FZD7 | 6.9e-02 | 0.770 |
| EIF2S3 | 6.9e-02 | 0.770 |
| AP2A2 | 7.0e-02 | 0.770 |
| MSRA | 7.0e-02 | 0.770 |
| CHMP3 | 7.0e-02 | 0.770 |
| PEBP1 | 7.0e-02 | 0.770 |
| MARCKS | 7.0e-02 | 0.770 |
| PTPN18 | 7.0e-02 | 0.770 |
| TIMM50 | 7.1e-02 | 0.770 |
| ANKRD52 | 7.1e-02 | 0.770 |
| GCC2 | 7.1e-02 | 0.770 |
| YES1 | 7.1e-02 | 0.770 |
| NCOA6 | 7.1e-02 | 0.770 |
| POM121 | 7.1e-02 | 0.770 |
| RIOK1 | 7.1e-02 | 0.770 |
| HNRNPA2B1 | 7.1e-02 | 0.770 |
| PLA2G16 | 7.1e-02 | 0.770 |
| CCDC91 | 7.1e-02 | 0.770 |
| IPO11 | 7.1e-02 | 0.770 |
| NEK1 | 7.1e-02 | 0.770 |
| NHLRC3 | 7.1e-02 | 0.770 |
| TMEM186 | 7.1e-02 | 0.770 |
| ZNF605 | 7.1e-02 | 0.770 |
| TRMT10B | 7.2e-02 | 0.770 |
| H2AFZ | 7.2e-02 | 0.770 |
| LMCD1 | 7.2e-02 | 0.770 |
| METRN | 7.2e-02 | 0.770 |
| AGPAT6 | 7.2e-02 | 0.770 |
| ANLN | 7.2e-02 | 0.770 |
| SMAD1 | 7.2e-02 | 0.770 |
| C16orf59 | 7.2e-02 | 0.770 |
| EHD1 | 7.2e-02 | 0.770 |
| ANKHD1 | 7.2e-02 | 0.770 |
| RAD51D | 7.2e-02 | 0.770 |
| SUPV3L1 | 7.2e-02 | 0.770 |
| UBXN4 | 7.3e-02 | 0.770 |
| TFAM | 7.3e-02 | 0.770 |
| SNX17 | 7.3e-02 | 0.770 |
| SVIP | 7.3e-02 | 0.770 |
| DIP2A | 7.3e-02 | 0.770 |
| BTBD3 | 7.3e-02 | 0.770 |
| SWAP70 | 7.3e-02 | 0.770 |
| COPB2 | 7.3e-02 | 0.770 |
| TMEM170A | 7.3e-02 | 0.770 |
| AJUBA | 7.3e-02 | 0.770 |
| C20orf196 | 7.3e-02 | 0.770 |
| HERC1 | 7.4e-02 | 0.770 |
| LZTFL1 | 7.4e-02 | 0.770 |
| HNRNPA3 | 7.4e-02 | 0.770 |
| NEDD9 | 7.4e-02 | 0.770 |
| ITGB3BP | 7.5e-02 | 0.770 |
| ARHGAP33 | 7.5e-02 | 0.770 |
| ZBTB8A | 7.5e-02 | 0.770 |
| TTLL5 | 7.5e-02 | 0.770 |
| JAM2 | 7.5e-02 | 0.770 |
| VPS8 | 7.5e-02 | 0.770 |
| MAGOHB | 7.5e-02 | 0.780 |
| RBM45 | 7.5e-02 | 0.780 |
| CCDC66 | 7.5e-02 | 0.780 |
| FAM126A | 7.5e-02 | 0.780 |
| GNPTG | 7.5e-02 | 0.780 |
| TMEM50B | 7.6e-02 | 0.780 |
| PRKRA | 7.6e-02 | 0.780 |
| TMCC1 | 7.6e-02 | 0.780 |
| FAM219A | 7.6e-02 | 0.780 |
| SNHG16 | 7.6e-02 | 0.780 |
| TMEM41B | 7.6e-02 | 0.780 |
| ZNF268 | 7.6e-02 | 0.780 |
| BECN1 | 7.6e-02 | 0.780 |
| XPO5 | 7.6e-02 | 0.780 |
| MTOR | 7.7e-02 | 0.780 |
| IBA57 | 7.7e-02 | 0.780 |
| TMEM43 | 7.7e-02 | 0.780 |
| CACHD1 | 7.7e-02 | 0.780 |
| B9D1 | 7.7e-02 | 0.780 |
| ADAM10 | 7.7e-02 | 0.780 |
| NUP188 | 7.7e-02 | 0.780 |
| TP53 | 7.7e-02 | 0.780 |
| ATP6AP2 | 7.7e-02 | 0.780 |
| HRAS | 7.8e-02 | 0.780 |
| PREPL | 7.8e-02 | 0.780 |
| MDH1 | 7.8e-02 | 0.780 |
| TUBB | 7.8e-02 | 0.780 |
| C20orf24 | 7.8e-02 | 0.780 |
| NDUFB1 | 7.8e-02 | 0.780 |
| FTX | 7.8e-02 | 0.780 |
| UPF3A | 7.9e-02 | 0.780 |
| HSPE1 | 7.9e-02 | 0.780 |
| SNRPG | 7.9e-02 | 0.780 |
| ZNF205 | 7.9e-02 | 0.780 |
| YBX1 | 7.9e-02 | 0.780 |
| ARMCX1 | 7.9e-02 | 0.780 |
| ITM2C | 8.0e-02 | 0.780 |
| NOP58 | 8.0e-02 | 0.780 |
| TMEM69 | 8.0e-02 | 0.780 |
| SDCCAG3 | 8.0e-02 | 0.780 |
| PLEKHF2 | 8.0e-02 | 0.780 |
| KIAA1715 | 8.0e-02 | 0.780 |
| ASH1L | 8.0e-02 | 0.780 |
| VDAC2 | 8.0e-02 | 0.780 |
| PLEKHG1 | 8.0e-02 | 0.780 |
| GPR107 | 8.0e-02 | 0.780 |
| OSBPL6 | 8.0e-02 | 0.780 |
| DNAJC17 | 8.0e-02 | 0.780 |
| PSMC1 | 8.0e-02 | 0.780 |
| ZNF567 | 8.0e-02 | 0.780 |
| ERH | 8.0e-02 | 0.780 |
| MKNK2 | 8.0e-02 | 0.780 |
| APOO | 8.1e-02 | 0.780 |
| ACP1 | 8.1e-02 | 0.780 |
| SMARCA1 | 8.1e-02 | 0.780 |
| ZNF576 | 8.1e-02 | 0.780 |
| EIF1AD | 8.1e-02 | 0.780 |
| TSPAN18 | 8.1e-02 | 0.780 |
| CEP68 | 8.1e-02 | 0.780 |
| MZF1 | 8.2e-02 | 0.780 |
| TSPAN6 | 8.2e-02 | 0.780 |
| INTU | 8.2e-02 | 0.780 |
| RAD1 | 8.2e-02 | 0.780 |
| ANP32A | 8.2e-02 | 0.780 |
| NAPA | 8.2e-02 | 0.780 |
| MGA | 8.2e-02 | 0.780 |
| SYT11 | 8.2e-02 | 0.780 |
| HINT2 | 8.2e-02 | 0.780 |
| RYK | 8.2e-02 | 0.780 |
| ULK4 | 8.2e-02 | 0.780 |
| NT5DC2 | 8.3e-02 | 0.780 |
| COL2A1 | 8.3e-02 | 0.780 |
| MAP9 | 8.3e-02 | 0.780 |
| NELFE | 8.3e-02 | 0.780 |
| SDHA | 8.3e-02 | 0.780 |
| CDC45 | 8.3e-02 | 0.780 |
| NUB1 | 8.3e-02 | 0.780 |
| PDIA6 | 8.3e-02 | 0.780 |
| TRPT1 | 8.3e-02 | 0.780 |
| GGA2 | 8.3e-02 | 0.780 |
| RNASEH2A | 8.3e-02 | 0.780 |
| FUS | 8.3e-02 | 0.780 |
| METAP2 | 8.3e-02 | 0.780 |
| ELAVL1 | 8.3e-02 | 0.780 |
| QDPR | 8.4e-02 | 0.780 |
| COA1 | 8.4e-02 | 0.780 |
| MEX3A | 8.4e-02 | 0.780 |
| ZDHHC17 | 8.4e-02 | 0.780 |
| EMC4 | 8.4e-02 | 0.780 |
| HEXIM2 | 8.4e-02 | 0.780 |
| OMA1 | 8.4e-02 | 0.780 |
| FNDC3B | 8.4e-02 | 0.780 |
| NCAPD2 | 8.4e-02 | 0.780 |
| NAA50 | 8.4e-02 | 0.780 |
| ABHD3 | 8.5e-02 | 0.780 |
| RPA3 | 8.5e-02 | 0.780 |
| TSFM | 8.5e-02 | 0.780 |
| GPSM2 | 8.5e-02 | 0.780 |
| STYX | 8.5e-02 | 0.780 |
| KPNB1 | 8.5e-02 | 0.780 |
| GNAS | 8.5e-02 | 0.780 |
| PA2G4 | 8.5e-02 | 0.780 |
| HAGH | 8.5e-02 | 0.780 |
| SURF6 | 8.5e-02 | 0.780 |
| FKBPL | 8.5e-02 | 0.780 |
| CEBPZ | 8.6e-02 | 0.780 |
| NCOA4 | 8.6e-02 | 0.780 |
| KRR1 | 8.6e-02 | 0.780 |
| FSD1L | 8.6e-02 | 0.780 |
| FAM149B1 | 8.6e-02 | 0.780 |
| BCL2L1 | 8.6e-02 | 0.780 |
| UTP11 | 8.6e-02 | 0.780 |
| RCE1 | 8.6e-02 | 0.780 |
| SNX27 | 8.6e-02 | 0.780 |
| DDIT4 | 8.7e-02 | 0.780 |
| MEG8 | 8.7e-02 | 0.780 |
| USP39 | 8.7e-02 | 0.780 |
| RP11-758H9.2 | 8.7e-02 | 0.780 |
| RING1 | 8.7e-02 | 0.780 |
| FNDC4 | 8.7e-02 | 0.780 |
| PABPC4 | 8.7e-02 | 0.780 |
| PRDM4 | 8.8e-02 | 0.780 |
| ICE2 | 8.8e-02 | 0.780 |
| KIF5B | 8.8e-02 | 0.780 |
| RRS1 | 8.8e-02 | 0.780 |
| MT2A | 8.8e-02 | 0.780 |
| SCAPER | 8.8e-02 | 0.780 |
| ETFA | 8.8e-02 | 0.780 |
| NDUFA4 | 8.8e-02 | 0.780 |
| TATDN3 | 8.8e-02 | 0.780 |
| MCM3AP | 8.8e-02 | 0.780 |
| ENO1 | 8.9e-02 | 0.780 |
| FAM204A | 8.9e-02 | 0.780 |
| HNRNPAB | 8.9e-02 | 0.780 |
| ZSCAN29 | 8.9e-02 | 0.780 |
| NBR1 | 8.9e-02 | 0.780 |
| COPS3 | 8.9e-02 | 0.780 |
| GPATCH8 | 8.9e-02 | 0.780 |
| FAM96B | 8.9e-02 | 0.780 |
| FIP1L1 | 8.9e-02 | 0.780 |
| ATP1B1 | 8.9e-02 | 0.780 |
| RPL3 | 8.9e-02 | 0.780 |
| E2F1 | 9.0e-02 | 0.780 |
| ZDHHC16 | 9.0e-02 | 0.780 |
| ELP5 | 9.0e-02 | 0.780 |
| SNHG9 | 9.0e-02 | 0.780 |
| THAP2 | 9.0e-02 | 0.780 |
| ILK | 9.0e-02 | 0.780 |
| ZCCHC17 | 9.0e-02 | 0.780 |
| PCDH17 | 9.0e-02 | 0.780 |
| TCTEX1D2 | 9.0e-02 | 0.780 |
| FAM217B | 9.1e-02 | 0.780 |
| CLTC | 9.1e-02 | 0.780 |
| LTN1 | 9.1e-02 | 0.780 |
| CBX8 | 9.1e-02 | 0.780 |
| REXO1 | 9.1e-02 | 0.780 |
| DDB2 | 9.1e-02 | 0.780 |
| ZNF703 | 9.1e-02 | 0.780 |
| POC5 | 9.1e-02 | 0.780 |
| PCED1A | 9.1e-02 | 0.780 |
| MPI | 9.1e-02 | 0.780 |
| CRADD | 9.1e-02 | 0.780 |
| PDXDC1 | 9.2e-02 | 0.790 |
| WDR5 | 9.2e-02 | 0.790 |
| TNRC6B | 9.2e-02 | 0.790 |
| MEIS1 | 9.2e-02 | 0.790 |
| CDC34 | 9.2e-02 | 0.790 |
| FOPNL | 9.2e-02 | 0.790 |
| ZNF85 | 9.2e-02 | 0.790 |
| RRAGC | 9.3e-02 | 0.790 |
| ZNF526 | 9.3e-02 | 0.790 |
| UNC119B | 9.3e-02 | 0.790 |
| PRMT1 | 9.3e-02 | 0.790 |
| KIAA0907 | 9.3e-02 | 0.790 |
| SRGAP2B | 9.3e-02 | 0.790 |
| UNC45A | 9.3e-02 | 0.790 |
| ELMOD3 | 9.3e-02 | 0.790 |
| IFT81 | 9.3e-02 | 0.790 |
| ZNF626 | 9.3e-02 | 0.790 |
| MTIF2 | 9.3e-02 | 0.790 |
| C1orf56 | 9.4e-02 | 0.790 |
| KMT2E | 9.4e-02 | 0.790 |
| SLC35A2 | 9.4e-02 | 0.790 |
| TTC9C | 9.4e-02 | 0.790 |
| MORN2 | 9.5e-02 | 0.790 |
| TCAIM | 9.5e-02 | 0.790 |
| SEPT8 | 9.5e-02 | 0.790 |
| AFF4 | 9.5e-02 | 0.790 |
| VPS26A | 9.5e-02 | 0.790 |
| MALAT1 | 9.5e-02 | 0.790 |
| ERMARD | 9.5e-02 | 0.790 |
| CENPP | 9.5e-02 | 0.790 |
| RP11-143E21.7 | 9.5e-02 | 0.790 |
| TTC31 | 9.6e-02 | 0.790 |
| MRPL45 | 9.6e-02 | 0.790 |
| TRAPPC1 | 9.6e-02 | 0.790 |
| CRK | 9.6e-02 | 0.790 |
| MAPK8IP3 | 9.6e-02 | 0.790 |
| TRIM26 | 9.6e-02 | 0.790 |
| NSUN5 | 9.6e-02 | 0.790 |
| MED31 | 9.6e-02 | 0.790 |
| DOLK | 9.7e-02 | 0.790 |
| MEX3C | 9.7e-02 | 0.800 |
| P4HA1 | 9.7e-02 | 0.800 |
| SEL1L | 9.7e-02 | 0.800 |
| RGS20 | 9.7e-02 | 0.800 |
| ACAA1 | 9.7e-02 | 0.800 |
| HADHA | 9.8e-02 | 0.800 |
| C19orf25 | 9.8e-02 | 0.800 |
| FAT3 | 9.8e-02 | 0.800 |
| SMCHD1 | 9.8e-02 | 0.800 |
| AHCYL1 | 9.9e-02 | 0.800 |
| MANF | 9.9e-02 | 0.800 |
| IFI16 | 9.9e-02 | 0.800 |
| CALM2 | 9.9e-02 | 0.800 |
| CAMTA1 | 9.9e-02 | 0.800 |
| HSCB | 9.9e-02 | 0.800 |
| RP11-182L21.6 | 1.0e-01 | 0.810 |
| POLR1D | 1.0e-01 | 0.810 |
| ST13 | 1.0e-01 | 0.810 |
| PRCC | 1.0e-01 | 0.810 |
| SLC39A3 | 1.0e-01 | 0.810 |
| C11orf24 | 1.0e-01 | 0.810 |
| TNKS | 1.0e-01 | 0.810 |
| PICALM | 1.0e-01 | 0.810 |
| THG1L | 1.0e-01 | 0.810 |
| GPN3 | 1.0e-01 | 0.810 |
| RPUSD3 | 1.0e-01 | 0.810 |
| CNIH2 | 1.0e-01 | 0.810 |
| MRPL14 | 1.0e-01 | 0.810 |
| AP2M1 | 1.0e-01 | 0.810 |
| DNAJB5 | 1.0e-01 | 0.810 |
| EXT1 | 1.0e-01 | 0.810 |
| SDC2 | 1.0e-01 | 0.810 |
| OAZ1 | 1.0e-01 | 0.810 |
| C11orf58 | 1.0e-01 | 0.810 |
| RP11-995C19.3 | 1.0e-01 | 0.810 |
| GYG1 | 1.0e-01 | 0.810 |
| MED12 | 1.0e-01 | 0.810 |
| APRT | 1.0e-01 | 0.810 |
| MAD1L1 | 1.0e-01 | 0.810 |
| TAOK3 | 1.0e-01 | 0.810 |
| ATP6V0D1 | 1.0e-01 | 0.810 |
| CCDC126 | 1.0e-01 | 0.810 |
| NNT | 1.0e-01 | 0.810 |
| ARF5 | 1.0e-01 | 0.810 |
| SRSF5 | 1.0e-01 | 0.810 |
| TK1 | 1.0e-01 | 0.810 |
| TIPRL | 1.0e-01 | 0.810 |
| LGR4 | 1.0e-01 | 0.810 |
| ABCF1 | 1.0e-01 | 0.810 |
| CAPRIN1 | 1.0e-01 | 0.810 |
| ABHD12 | 1.0e-01 | 0.810 |
| BTBD1 | 1.0e-01 | 0.810 |
| GOLGA5 | 1.0e-01 | 0.810 |
| TMED3 | 1.0e-01 | 0.820 |
| CDC73 | 1.0e-01 | 0.820 |
| NACA | 1.0e-01 | 0.820 |
| SFI1 | 1.0e-01 | 0.820 |
| FBXO3 | 1.0e-01 | 0.820 |
| CDCA7L | 1.1e-01 | 0.820 |
| TEAD4 | 1.1e-01 | 0.820 |
| PSMA7 | 1.1e-01 | 0.820 |
| PBX1 | 1.1e-01 | 0.820 |
| FAM3A | 1.1e-01 | 0.820 |
| KMT5A | 1.1e-01 | 0.820 |
| TMEM109 | 1.1e-01 | 0.820 |
| ZNF580 | 1.1e-01 | 0.820 |
| TMEM258 | 1.1e-01 | 0.820 |
| HMBS | 1.1e-01 | 0.820 |
| RP11-12G12.7 | 1.1e-01 | 0.820 |
| WDR25 | 1.1e-01 | 0.820 |
| NPRL3 | 1.1e-01 | 0.820 |
| SURF1 | 1.1e-01 | 0.820 |
| EXOSC2 | 1.1e-01 | 0.820 |
| PRR5 | 1.1e-01 | 0.820 |
| MCM2 | 1.1e-01 | 0.820 |
| PJA1 | 1.1e-01 | 0.820 |
| RAPGEF1 | 1.1e-01 | 0.820 |
| AC069513.3 | 1.1e-01 | 0.820 |
| AP5S1 | 1.1e-01 | 0.820 |
| BCAS2 | 1.1e-01 | 0.820 |
| RRP12 | 1.1e-01 | 0.820 |
| P4HTM | 1.1e-01 | 0.820 |
| PDIA5 | 1.1e-01 | 0.820 |
| ZNF518A | 1.1e-01 | 0.820 |
| TUNAR | 1.1e-01 | 0.820 |
| TMEM134 | 1.1e-01 | 0.820 |
| ISOC1 | 1.1e-01 | 0.820 |
| KCTD3 | 1.1e-01 | 0.820 |
| AC011753.3 | 1.1e-01 | 0.820 |
| MAML1 | 1.1e-01 | 0.820 |
| PUS7L | 1.1e-01 | 0.820 |
| TOP3A | 1.1e-01 | 0.820 |
| ARL3 | 1.1e-01 | 0.820 |
| HMOX2 | 1.1e-01 | 0.820 |
| GYG2 | 1.1e-01 | 0.820 |
| RBM4B | 1.1e-01 | 0.820 |
| NTHL1 | 1.1e-01 | 0.820 |
| METTL14 | 1.1e-01 | 0.820 |
| PRSS23 | 1.1e-01 | 0.820 |
| TFG | 1.1e-01 | 0.820 |
| AHDC1 | 1.1e-01 | 0.820 |
| PKN2 | 1.1e-01 | 0.820 |
| RBM8A | 1.1e-01 | 0.820 |
| ZMIZ1 | 1.1e-01 | 0.820 |
| POR | 1.1e-01 | 0.820 |
| KIAA1586 | 1.1e-01 | 0.820 |
| CPSF3 | 1.1e-01 | 0.820 |
| GCAT | 1.1e-01 | 0.820 |
| NOP2 | 1.1e-01 | 0.820 |
| ADI1 | 1.1e-01 | 0.820 |
| PRR11 | 1.1e-01 | 0.820 |
| SH3KBP1 | 1.1e-01 | 0.820 |
| DUS1L | 1.1e-01 | 0.820 |
| RFX2 | 1.1e-01 | 0.820 |
| COMMD4 | 1.1e-01 | 0.820 |
| TMA7 | 1.1e-01 | 0.820 |
| KCTD10 | 1.1e-01 | 0.820 |
| C14orf119 | 1.1e-01 | 0.820 |
| UXS1 | 1.1e-01 | 0.820 |
| CD3EAP | 1.1e-01 | 0.820 |
| TSEN15 | 1.1e-01 | 0.820 |
| EFCAB2 | 1.1e-01 | 0.820 |
| PANX1 | 1.1e-01 | 0.820 |
| TNKS2 | 1.1e-01 | 0.820 |
| ENO2 | 1.1e-01 | 0.820 |
| SLC7A1 | 1.1e-01 | 0.820 |
| TMEM97 | 1.1e-01 | 0.820 |
| EIF3F | 1.1e-01 | 0.820 |
| PSMA4 | 1.1e-01 | 0.820 |
| LYPLA2 | 1.1e-01 | 0.820 |
| ZBED5 | 1.1e-01 | 0.820 |
| RAE1 | 1.1e-01 | 0.820 |
| LINS1 | 1.1e-01 | 0.820 |
| UQCRC2 | 1.1e-01 | 0.820 |
| DAAM1 | 1.1e-01 | 0.820 |
| KHSRP | 1.1e-01 | 0.820 |
| MAP4K5 | 1.1e-01 | 0.820 |
| PRR13 | 1.1e-01 | 0.820 |
| GART | 1.1e-01 | 0.820 |
| MED17 | 1.1e-01 | 0.820 |
| SAFB2 | 1.1e-01 | 0.820 |
| ZNF331 | 1.1e-01 | 0.820 |
| ALKBH3 | 1.1e-01 | 0.820 |
| CNPY4 | 1.1e-01 | 0.820 |
| TYW5 | 1.1e-01 | 0.820 |
| ZNF720 | 1.1e-01 | 0.820 |
| KTN1 | 1.2e-01 | 0.820 |
| R3HDM4 | 1.2e-01 | 0.820 |
| KNSTRN | 1.2e-01 | 0.820 |
| NRAS | 1.2e-01 | 0.820 |
| LIN7C | 1.2e-01 | 0.820 |
| MYO19 | 1.2e-01 | 0.820 |
| EDEM2 | 1.2e-01 | 0.820 |
| AC093323.3 | 1.2e-01 | 0.820 |
| VPS37B | 1.2e-01 | 0.820 |
| NR2F2 | 1.2e-01 | 0.820 |
| SLC25A22 | 1.2e-01 | 0.820 |
| GLT25D1 | 1.2e-01 | 0.820 |
| IRS1 | 1.2e-01 | 0.820 |
| AC013394.2 | 1.2e-01 | 0.820 |
| UBE2G1 | 1.2e-01 | 0.820 |
| RGL2 | 1.2e-01 | 0.820 |
| PIH1D1 | 1.2e-01 | 0.820 |
| GRWD1 | 1.2e-01 | 0.820 |
| RBM19 | 1.2e-01 | 0.820 |
| THUMPD1 | 1.2e-01 | 0.820 |
| DCUN1D1 | 1.2e-01 | 0.820 |
| SNX9 | 1.2e-01 | 0.820 |
| SEPT11 | 1.2e-01 | 0.820 |
| GAP43 | 1.2e-01 | 0.820 |
| CTNNAL1 | 1.2e-01 | 0.820 |
| LRP1 | 1.2e-01 | 0.820 |
| RNF214 | 1.2e-01 | 0.820 |
| UTP4 | 1.2e-01 | 0.820 |
| VAMP2 | 1.2e-01 | 0.820 |
| TAF6 | 1.2e-01 | 0.820 |
| PSIP1 | 1.2e-01 | 0.820 |
| SPIN3 | 1.2e-01 | 0.820 |
| TMUB2 | 1.2e-01 | 0.820 |
| APMAP | 1.2e-01 | 0.820 |
| TMEM101 | 1.2e-01 | 0.820 |
| TAF10 | 1.2e-01 | 0.820 |
| C2orf68 | 1.2e-01 | 0.820 |
| RP3-412A9.11 | 1.2e-01 | 0.820 |
| DPP3 | 1.2e-01 | 0.820 |
| EXTL3 | 1.2e-01 | 0.820 |
| CSPG5 | 1.2e-01 | 0.820 |
| DUS3L | 1.2e-01 | 0.820 |
| METTL2A | 1.2e-01 | 0.820 |
| FAM110B | 1.2e-01 | 0.820 |
| UBE2J2 | 1.2e-01 | 0.820 |
| SCRIB | 1.2e-01 | 0.820 |
| AP3M2 | 1.2e-01 | 0.820 |
| DRAP1 | 1.2e-01 | 0.820 |
| UBE3B | 1.2e-01 | 0.820 |
| LRRC4B | 1.2e-01 | 0.820 |
| ELF2 | 1.2e-01 | 0.820 |
| SNCA | 1.2e-01 | 0.820 |
| CD99 | 1.2e-01 | 0.820 |
| NFE2L2 | 1.2e-01 | 0.820 |
| METTL22 | 1.2e-01 | 0.820 |
| TMX1 | 1.2e-01 | 0.820 |
| CENPU | 1.2e-01 | 0.820 |
| RCC1 | 1.2e-01 | 0.820 |
| VRK1 | 1.2e-01 | 0.820 |
| PIGM | 1.2e-01 | 0.820 |
| HNRNPK | 1.2e-01 | 0.820 |
| PKP4 | 1.2e-01 | 0.820 |
| RBM42 | 1.2e-01 | 0.820 |
| IREB2 | 1.2e-01 | 0.820 |
| ORC5 | 1.2e-01 | 0.820 |
| PIGH | 1.2e-01 | 0.820 |
| CAP1 | 1.2e-01 | 0.820 |
| ATAD5 | 1.2e-01 | 0.820 |
| DESI1 | 1.2e-01 | 0.820 |
| SLC25A3 | 1.2e-01 | 0.820 |
| SLC39A14 | 1.2e-01 | 0.820 |
| GTF2H2 | 1.2e-01 | 0.820 |
| CDK1 | 1.2e-01 | 0.820 |
| MID1 | 1.2e-01 | 0.820 |
| APTX | 1.2e-01 | 0.820 |
| EPT1 | 1.2e-01 | 0.820 |
| APC | 1.2e-01 | 0.820 |
| ZSWIM7 | 1.2e-01 | 0.820 |
| ATN1 | 1.2e-01 | 0.820 |
| AVL9 | 1.2e-01 | 0.820 |
| HCFC1R1 | 1.2e-01 | 0.820 |
| RPL13A | 1.2e-01 | 0.820 |
| ANKRD26 | 1.2e-01 | 0.820 |
| MEAF6 | 1.2e-01 | 0.820 |
| SMARCA4 | 1.2e-01 | 0.820 |
| RELA | 1.2e-01 | 0.820 |
| HSPB1 | 1.2e-01 | 0.820 |
| METTL9 | 1.2e-01 | 0.820 |
| PCBP2 | 1.2e-01 | 0.820 |
| ANTXR2 | 1.2e-01 | 0.820 |
| FAM181B | 1.2e-01 | 0.820 |
| NDUFB5 | 1.2e-01 | 0.820 |
| COPS7A | 1.2e-01 | 0.820 |
| ENSA | 1.2e-01 | 0.820 |
| TALDO1 | 1.2e-01 | 0.820 |
| BOC | 1.2e-01 | 0.820 |
| SORBS2 | 1.2e-01 | 0.820 |
| NBPF1 | 1.2e-01 | 0.820 |
| CCDC84 | 1.2e-01 | 0.820 |
| MDH2 | 1.2e-01 | 0.820 |
| WASHC2C | 1.2e-01 | 0.820 |
| ELP3 | 1.2e-01 | 0.820 |
| TBC1D16 | 1.2e-01 | 0.820 |
| PSMD10 | 1.2e-01 | 0.820 |
| TMPO | 1.2e-01 | 0.820 |
| ACBD5 | 1.2e-01 | 0.820 |
| BCL10 | 1.3e-01 | 0.820 |
| WASHC2A | 1.3e-01 | 0.820 |
| SP8 | 1.3e-01 | 0.820 |
| CDK4 | 1.3e-01 | 0.820 |
| FKBP3 | 1.3e-01 | 0.820 |
| LAMC1 | 1.3e-01 | 0.820 |
| CFL1 | 1.3e-01 | 0.820 |
| SMARCB1 | 1.3e-01 | 0.820 |
| NAA15 | 1.3e-01 | 0.820 |
| PDLIM7 | 1.3e-01 | 0.820 |
| SPP1 | 1.3e-01 | 0.820 |
| CACUL1 | 1.3e-01 | 0.820 |
| NSMCE2 | 1.3e-01 | 0.820 |
| TCF7L1 | 1.3e-01 | 0.820 |
| BACH1 | 1.3e-01 | 0.820 |
| PRDM2 | 1.3e-01 | 0.820 |
| MTSS1 | 1.3e-01 | 0.820 |
| KDM3A | 1.3e-01 | 0.820 |
| MGST2 | 1.3e-01 | 0.820 |
| GJA1 | 1.3e-01 | 0.820 |
| IDS | 1.3e-01 | 0.820 |
| ERO1A | 1.3e-01 | 0.820 |
| ZC3H8 | 1.3e-01 | 0.820 |
| PANK2 | 1.3e-01 | 0.820 |
| TMSB15A | 1.3e-01 | 0.820 |
| C1orf123 | 1.3e-01 | 0.820 |
| GLRX5 | 1.3e-01 | 0.820 |
| BCL9 | 1.3e-01 | 0.820 |
| DBF4 | 1.3e-01 | 0.820 |
| CDK9 | 1.3e-01 | 0.820 |
| ARID5B | 1.3e-01 | 0.820 |
| TMED4 | 1.3e-01 | 0.820 |
| HSPB11 | 1.3e-01 | 0.820 |
| EIF4EBP1 | 1.3e-01 | 0.820 |
| PPP1R1A | 1.3e-01 | 0.820 |
| MBIP | 1.3e-01 | 0.820 |
| DDX42 | 1.3e-01 | 0.820 |
| PPP2R2A | 1.3e-01 | 0.820 |
| ATMIN | 1.3e-01 | 0.820 |
| HDDC2 | 1.3e-01 | 0.820 |
| SS18L1 | 1.3e-01 | 0.820 |
| NCLN | 1.3e-01 | 0.820 |
| SRP9 | 1.3e-01 | 0.820 |
| NUP88 | 1.3e-01 | 0.820 |
| TDRD3 | 1.3e-01 | 0.820 |
| ARL6IP4 | 1.3e-01 | 0.820 |
| ELOVL4 | 1.3e-01 | 0.820 |
| PTRH1 | 1.3e-01 | 0.820 |
| RAB11B | 1.3e-01 | 0.820 |
| IER3 | 1.3e-01 | 0.820 |
| NSF | 1.3e-01 | 0.820 |
| SYNC | 1.3e-01 | 0.820 |
| PIM3 | 1.3e-01 | 0.820 |
| VASP | 1.3e-01 | 0.820 |
| GMPS | 1.3e-01 | 0.820 |
| FGF19 | 1.3e-01 | 0.820 |
| MND1 | 1.3e-01 | 0.820 |
| PRELID1 | 1.3e-01 | 0.820 |
| CDK2AP1 | 1.3e-01 | 0.820 |
| CTC-444N24.11 | 1.3e-01 | 0.820 |
| FZR1 | 1.3e-01 | 0.820 |
| DPP9 | 1.3e-01 | 0.820 |
| TCP1 | 1.3e-01 | 0.820 |
| ABCD3 | 1.3e-01 | 0.820 |
| NORAD | 1.3e-01 | 0.820 |
| UMPS | 1.3e-01 | 0.820 |
| GPCPD1 | 1.3e-01 | 0.820 |
| HEXDC | 1.3e-01 | 0.820 |
| CNPPD1 | 1.3e-01 | 0.820 |
| TMEM87A | 1.3e-01 | 0.820 |
| COMMD6 | 1.3e-01 | 0.820 |
| ACTR8 | 1.3e-01 | 0.820 |
| CYB561 | 1.3e-01 | 0.820 |
| RFWD2 | 1.3e-01 | 0.820 |
| AXL | 1.3e-01 | 0.820 |
| CCT2 | 1.3e-01 | 0.820 |
| TBC1D20 | 1.3e-01 | 0.820 |
| ATIC | 1.3e-01 | 0.820 |
| CENPE | 1.3e-01 | 0.820 |
| CRABP2 | 1.3e-01 | 0.820 |
| CPD | 1.3e-01 | 0.820 |
| NUDT21 | 1.3e-01 | 0.820 |
| C16orf72 | 1.3e-01 | 0.820 |
| CHRAC1 | 1.3e-01 | 0.820 |
| SLC35G1 | 1.3e-01 | 0.820 |
| C12orf43 | 1.3e-01 | 0.820 |
| CDR2L | 1.3e-01 | 0.820 |
| MIF | 1.3e-01 | 0.820 |
| CCZ1B | 1.3e-01 | 0.820 |
| MON1B | 1.3e-01 | 0.820 |
| TMEM126A | 1.3e-01 | 0.820 |
| FBXO16 | 1.3e-01 | 0.820 |
| WASHC1 | 1.3e-01 | 0.820 |
| ARMC9 | 1.3e-01 | 0.820 |
| BRAP | 1.3e-01 | 0.820 |
| G6PC3 | 1.4e-01 | 0.820 |
| TGDS | 1.4e-01 | 0.820 |
| DTNBP1 | 1.4e-01 | 0.820 |
| LRP2 | 1.4e-01 | 0.820 |
| CHRNA5 | 1.4e-01 | 0.820 |
| LIMS1 | 1.4e-01 | 0.820 |
| DHX30 | 1.4e-01 | 0.820 |
| SF3B1 | 1.4e-01 | 0.820 |
| RRM1 | 1.4e-01 | 0.820 |
| COPA | 1.4e-01 | 0.820 |
| LINC00493 | 1.4e-01 | 0.820 |
| DNM1L | 1.4e-01 | 0.820 |
| ACOT13 | 1.4e-01 | 0.820 |
| SEC23A | 1.4e-01 | 0.820 |
| NHSL1 | 1.4e-01 | 0.820 |
| RNGTT | 1.4e-01 | 0.820 |
| ZNF207 | 1.4e-01 | 0.820 |
| ZDHHC4 | 1.4e-01 | 0.820 |
| ZFP90 | 1.4e-01 | 0.820 |
| C17orf58 | 1.4e-01 | 0.820 |
| DCK | 1.4e-01 | 0.820 |
| ZNF787 | 1.4e-01 | 0.820 |
| DHPS | 1.4e-01 | 0.820 |
| TMEM127 | 1.4e-01 | 0.820 |
| RBM17 | 1.4e-01 | 0.820 |
| SPAG7 | 1.4e-01 | 0.820 |
| SNAPC5 | 1.4e-01 | 0.820 |
| MMADHC | 1.4e-01 | 0.820 |
| SSR3 | 1.4e-01 | 0.820 |
| WNK3 | 1.4e-01 | 0.820 |
| SLC25A25 | 1.4e-01 | 0.820 |
| ST6GALNAC4 | 1.4e-01 | 0.820 |
| ACTA2 | 1.4e-01 | 0.820 |
| DKK3 | 1.4e-01 | 0.820 |
| FUNDC1 | 1.4e-01 | 0.820 |
| TAF12 | 1.4e-01 | 0.820 |
| STARD7 | 1.4e-01 | 0.820 |
| NDC1 | 1.4e-01 | 0.820 |
| CEP131 | 1.4e-01 | 0.820 |
| RABGGTA | 1.4e-01 | 0.820 |
| ANKRD11 | 1.4e-01 | 0.820 |
| KCTD6 | 1.4e-01 | 0.820 |
| CBX2 | 1.4e-01 | 0.820 |
| FIZ1 | 1.4e-01 | 0.820 |
| ERCC4 | 1.4e-01 | 0.820 |
| SPAG5 | 1.4e-01 | 0.820 |
| MIEF1 | 1.4e-01 | 0.820 |
| UBIAD1 | 1.4e-01 | 0.820 |
| SURF4 | 1.4e-01 | 0.820 |
| PNMA2 | 1.4e-01 | 0.820 |
| RARRES2 | 1.4e-01 | 0.820 |
| ZC3H3 | 1.4e-01 | 0.820 |
| SERP1 | 1.4e-01 | 0.820 |
| SUMO2 | 1.4e-01 | 0.820 |
| CIZ1 | 1.4e-01 | 0.820 |
| SLC25A5 | 1.4e-01 | 0.820 |
| ARHGAP1 | 1.4e-01 | 0.820 |
| SLC4A2 | 1.4e-01 | 0.820 |
| PGD | 1.4e-01 | 0.820 |
| PPFIA1 | 1.4e-01 | 0.820 |
| TMEM165 | 1.4e-01 | 0.820 |
| NRCAM | 1.4e-01 | 0.820 |
| PTPN23 | 1.4e-01 | 0.820 |
| SLC38A10 | 1.4e-01 | 0.820 |
| POLD3 | 1.4e-01 | 0.820 |
| NPLOC4 | 1.4e-01 | 0.820 |
| GLI4 | 1.4e-01 | 0.820 |
| DPP8 | 1.4e-01 | 0.820 |
| DCTN5 | 1.4e-01 | 0.820 |
| RALGPS2 | 1.4e-01 | 0.820 |
| ZNF195 | 1.4e-01 | 0.820 |
| PPA2 | 1.4e-01 | 0.820 |
| GBX2 | 1.4e-01 | 0.820 |
| RAD54L | 1.4e-01 | 0.820 |
| C11orf84 | 1.4e-01 | 0.820 |
| SOX1 | 1.4e-01 | 0.820 |
| RP11-113K21.5 | 1.4e-01 | 0.820 |
| CDK19 | 1.4e-01 | 0.820 |
| SLC20A1 | 1.4e-01 | 0.820 |
| MAP3K4 | 1.4e-01 | 0.820 |
| AHI1 | 1.4e-01 | 0.820 |
| FTO | 1.4e-01 | 0.820 |
| N4BP2L2 | 1.4e-01 | 0.820 |
| DHRS7 | 1.4e-01 | 0.820 |
| CFAP20 | 1.4e-01 | 0.820 |
| DACH1 | 1.4e-01 | 0.820 |
| ATF3 | 1.4e-01 | 0.820 |
| ZBTB44 | 1.4e-01 | 0.820 |
| UBTD2 | 1.4e-01 | 0.820 |
| NOL4L | 1.4e-01 | 0.820 |
| DOK1 | 1.4e-01 | 0.820 |
| ACTG1 | 1.4e-01 | 0.820 |
| ATP5J | 1.4e-01 | 0.820 |
| CCDC107 | 1.4e-01 | 0.820 |
| FAU | 1.4e-01 | 0.820 |
| IMPACT | 1.4e-01 | 0.820 |
| PDPK1 | 1.4e-01 | 0.820 |
| CTB-31O20.2 | 1.4e-01 | 0.820 |
| CDK6 | 1.4e-01 | 0.820 |
| CHTF18 | 1.4e-01 | 0.820 |
| TNIK | 1.5e-01 | 0.820 |
| NSG1 | 1.5e-01 | 0.820 |
| DOHH | 1.5e-01 | 0.820 |
| NDFIP2 | 1.5e-01 | 0.820 |
| SHCBP1 | 1.5e-01 | 0.820 |
| COQ4 | 1.5e-01 | 0.820 |
| IKBKAP | 1.5e-01 | 0.820 |
| SREK1 | 1.5e-01 | 0.820 |
| PBX2 | 1.5e-01 | 0.820 |
| STT3A | 1.5e-01 | 0.820 |
| DAP3 | 1.5e-01 | 0.820 |
| POGK | 1.5e-01 | 0.820 |
| NKIRAS1 | 1.5e-01 | 0.820 |
| ANAPC16 | 1.5e-01 | 0.820 |
| RBMX2 | 1.5e-01 | 0.820 |
| CHD6 | 1.5e-01 | 0.820 |
| IQCE | 1.5e-01 | 0.820 |
| SMO | 1.5e-01 | 0.820 |
| MRC2 | 1.5e-01 | 0.820 |
| EEF2 | 1.5e-01 | 0.820 |
| ZC3H18 | 1.5e-01 | 0.820 |
| RABL6 | 1.5e-01 | 0.820 |
| BORCS6 | 1.5e-01 | 0.830 |
| FIG4 | 1.5e-01 | 0.830 |
| TCN2 | 1.5e-01 | 0.830 |
| IFT46 | 1.5e-01 | 0.830 |
| RAB5B | 1.5e-01 | 0.830 |
| TMEM41A | 1.5e-01 | 0.830 |
| MANEAL | 1.5e-01 | 0.830 |
| KDELC1 | 1.5e-01 | 0.830 |
| HERC4 | 1.5e-01 | 0.830 |
| SLC2A6 | 1.5e-01 | 0.830 |
| ZDHHC2 | 1.5e-01 | 0.830 |
| ELOC | 1.5e-01 | 0.830 |
| HSP90AB1 | 1.5e-01 | 0.830 |
| NUP98 | 1.5e-01 | 0.830 |
| SLC41A3 | 1.5e-01 | 0.830 |
| PTPRE | 1.5e-01 | 0.830 |
| RPS19BP1 | 1.5e-01 | 0.830 |
| RPS19 | 1.5e-01 | 0.830 |
| FAM122A | 1.5e-01 | 0.830 |
| TACC2 | 1.5e-01 | 0.830 |
| CENPH | 1.5e-01 | 0.830 |
| PIGC | 1.5e-01 | 0.830 |
| MAPK9 | 1.5e-01 | 0.830 |
| MRPS28 | 1.5e-01 | 0.830 |
| CCT6A | 1.5e-01 | 0.830 |
| RASA1 | 1.5e-01 | 0.830 |
| H2AFV | 1.5e-01 | 0.830 |
| CBY1 | 1.5e-01 | 0.830 |
| PDCL | 1.5e-01 | 0.830 |
| CYB5R3 | 1.5e-01 | 0.830 |
| RBPJ | 1.5e-01 | 0.830 |
| MRPS6 | 1.5e-01 | 0.830 |
| UTP14C | 1.5e-01 | 0.830 |
| MRPL34 | 1.5e-01 | 0.830 |
| SIPA1L1 | 1.5e-01 | 0.830 |
| MTO1 | 1.5e-01 | 0.830 |
| TMEM33 | 1.5e-01 | 0.830 |
| LRPPRC | 1.5e-01 | 0.830 |
| SMARCD2 | 1.5e-01 | 0.830 |
| CSE1L | 1.5e-01 | 0.830 |
| HAUS7 | 1.5e-01 | 0.830 |
| ZNF32 | 1.5e-01 | 0.830 |
| DVL2 | 1.5e-01 | 0.830 |
| SMG9 | 1.5e-01 | 0.830 |
| FBXO25 | 1.5e-01 | 0.830 |
| RPL24 | 1.5e-01 | 0.830 |
| RUFY2 | 1.5e-01 | 0.830 |
| ZNF286A | 1.5e-01 | 0.830 |
| SRBD1 | 1.5e-01 | 0.830 |
| AMPH | 1.5e-01 | 0.830 |
| CDC37 | 1.5e-01 | 0.830 |
| FAM127A | 1.5e-01 | 0.830 |
| RANGAP1 | 1.5e-01 | 0.830 |
| DHX15 | 1.5e-01 | 0.830 |
| SF3B4 | 1.5e-01 | 0.830 |
| GNPDA1 | 1.5e-01 | 0.830 |
| USP34 | 1.6e-01 | 0.830 |
| EI24 | 1.6e-01 | 0.830 |
| NUMB | 1.6e-01 | 0.830 |
| CPE | 1.6e-01 | 0.830 |
| TRIM32 | 1.6e-01 | 0.830 |
| HMGN2 | 1.6e-01 | 0.830 |
| UBXN8 | 1.6e-01 | 0.830 |
| OAZ2 | 1.6e-01 | 0.830 |
| PLAA | 1.6e-01 | 0.830 |
| FLOT2 | 1.6e-01 | 0.830 |
| RNF111 | 1.6e-01 | 0.830 |
| TRAP1 | 1.6e-01 | 0.830 |
| DNAJC18 | 1.6e-01 | 0.830 |
| LIX1 | 1.6e-01 | 0.830 |
| COPB1 | 1.6e-01 | 0.830 |
| PSPH | 1.6e-01 | 0.830 |
| ZNF335 | 1.6e-01 | 0.830 |
| GNS | 1.6e-01 | 0.830 |
| NDUFB10 | 1.6e-01 | 0.830 |
| SCARB1 | 1.6e-01 | 0.830 |
| TMEM218 | 1.6e-01 | 0.830 |
| C1orf35 | 1.6e-01 | 0.830 |
| LYPD1 | 1.6e-01 | 0.830 |
| SEPHS1 | 1.6e-01 | 0.830 |
| NKAIN1 | 1.6e-01 | 0.830 |
| PRTFDC1 | 1.6e-01 | 0.830 |
| RHOA | 1.6e-01 | 0.830 |
| ZNF263 | 1.6e-01 | 0.830 |
| RNF8 | 1.6e-01 | 0.830 |
| C14orf80 | 1.6e-01 | 0.830 |
| ZFHX3 | 1.6e-01 | 0.830 |
| MRPL47 | 1.6e-01 | 0.830 |
| MAGEF1 | 1.6e-01 | 0.830 |
| MFSD13A | 1.6e-01 | 0.830 |
| GLG1 | 1.6e-01 | 0.830 |
| GIPC1 | 1.6e-01 | 0.830 |
| FNIP2 | 1.6e-01 | 0.830 |
| TCF25 | 1.6e-01 | 0.830 |
| CAV1 | 1.6e-01 | 0.830 |
| IFNAR1 | 1.6e-01 | 0.830 |
| FAM133B | 1.6e-01 | 0.830 |
| SLC5A6 | 1.6e-01 | 0.830 |
| TMEM237 | 1.6e-01 | 0.830 |
| TMEM47 | 1.6e-01 | 0.830 |
| FGFBP3 | 1.6e-01 | 0.830 |
| RECQL4 | 1.6e-01 | 0.830 |
| PFN1 | 1.6e-01 | 0.830 |
| INTS5 | 1.6e-01 | 0.830 |
| NUDT15 | 1.6e-01 | 0.830 |
| BOD1L1 | 1.6e-01 | 0.830 |
| TMX3 | 1.6e-01 | 0.830 |
| DNAH14 | 1.6e-01 | 0.830 |
| ALKBH4 | 1.6e-01 | 0.830 |
| ATP5D | 1.6e-01 | 0.830 |
| MTHFSD | 1.6e-01 | 0.830 |
| HM13 | 1.6e-01 | 0.830 |
| PPT2 | 1.6e-01 | 0.830 |
| HNRNPM | 1.6e-01 | 0.830 |
| SPPL2B | 1.6e-01 | 0.830 |
| PWP1 | 1.6e-01 | 0.830 |
| TBC1D23 | 1.6e-01 | 0.830 |
| CYB5B | 1.6e-01 | 0.830 |
| SH3GLB1 | 1.6e-01 | 0.830 |
| MTHFD1 | 1.6e-01 | 0.830 |
| RIC8B | 1.6e-01 | 0.830 |
| MEX3B | 1.6e-01 | 0.830 |
| PURA | 1.6e-01 | 0.830 |
| ISG15 | 1.6e-01 | 0.830 |
| SAMD4A | 1.6e-01 | 0.830 |
| GMDS | 1.6e-01 | 0.830 |
| MAPK14 | 1.6e-01 | 0.830 |
| DRAM2 | 1.6e-01 | 0.830 |
| TPD52L2 | 1.6e-01 | 0.830 |
| PDCD7 | 1.6e-01 | 0.830 |
| TBP | 1.6e-01 | 0.830 |
| FANCI | 1.6e-01 | 0.830 |
| STRIP1 | 1.6e-01 | 0.830 |
| CRNKL1 | 1.6e-01 | 0.830 |
| ZNF22 | 1.6e-01 | 0.830 |
| TMEM167B | 1.6e-01 | 0.830 |
| KLHDC2 | 1.6e-01 | 0.830 |
| CRLS1 | 1.6e-01 | 0.830 |
| N4BP1 | 1.6e-01 | 0.830 |
| XPA | 1.6e-01 | 0.830 |
| TBKBP1 | 1.6e-01 | 0.830 |
| SHPRH | 1.6e-01 | 0.830 |
| NEBL | 1.6e-01 | 0.830 |
| TMEM160 | 1.6e-01 | 0.830 |
| ZNF143 | 1.6e-01 | 0.830 |
| ERGIC2 | 1.6e-01 | 0.830 |
| AC104655.3 | 1.6e-01 | 0.830 |
| ATOH8 | 1.6e-01 | 0.830 |
| RPL12 | 1.6e-01 | 0.830 |
| FARSA | 1.6e-01 | 0.830 |
| NCOA3 | 1.6e-01 | 0.830 |
| GTF2E1 | 1.6e-01 | 0.830 |
| PPP1CB | 1.6e-01 | 0.830 |
| LDHA | 1.7e-01 | 0.830 |
| CANT1 | 1.7e-01 | 0.830 |
| EPB41L2 | 1.7e-01 | 0.830 |
| XXYLT1 | 1.7e-01 | 0.830 |
| ELP2 | 1.7e-01 | 0.830 |
| RBM7 | 1.7e-01 | 0.830 |
| PGM3 | 1.7e-01 | 0.840 |
| SPARC | 1.7e-01 | 0.840 |
| ATAT1 | 1.7e-01 | 0.840 |
| RP11-887P2.1 | 1.7e-01 | 0.840 |
| AMN1 | 1.7e-01 | 0.840 |
| MCL1 | 1.7e-01 | 0.840 |
| FSIP1 | 1.7e-01 | 0.840 |
| SIRT3 | 1.7e-01 | 0.840 |
| ZMAT5 | 1.7e-01 | 0.840 |
| ZSCAN32 | 1.7e-01 | 0.840 |
| MORF4L2 | 1.7e-01 | 0.840 |
| EPS8L1 | 1.7e-01 | 0.840 |
| HTT | 1.7e-01 | 0.840 |
| SPRED2 | 1.7e-01 | 0.840 |
| GRK6 | 1.7e-01 | 0.840 |
| SNAPC2 | 1.7e-01 | 0.840 |
| GGA1 | 1.7e-01 | 0.840 |
| WIPF2 | 1.7e-01 | 0.840 |
| MLLT11 | 1.7e-01 | 0.840 |
| FOXB1 | 1.7e-01 | 0.840 |
| RPS6KA4 | 1.7e-01 | 0.840 |
| FBXW2 | 1.7e-01 | 0.840 |
| EHMT1 | 1.7e-01 | 0.840 |
| MRPS21 | 1.7e-01 | 0.840 |
| PHYHIPL | 1.7e-01 | 0.840 |
| FARSB | 1.7e-01 | 0.840 |
| FAM134C | 1.7e-01 | 0.840 |
| CHD7 | 1.7e-01 | 0.850 |
| ILKAP | 1.7e-01 | 0.850 |
| ARHGAP17 | 1.7e-01 | 0.850 |
| LSM14B | 1.7e-01 | 0.850 |
| MFSD2B | 1.7e-01 | 0.850 |
| C11orf54 | 1.7e-01 | 0.850 |
| MAP3K20 | 1.7e-01 | 0.850 |
| ZSCAN18 | 1.7e-01 | 0.850 |
| CNBP | 1.7e-01 | 0.850 |
| YAF2 | 1.7e-01 | 0.850 |
| SH3GL1 | 1.7e-01 | 0.850 |
| RPUSD1 | 1.7e-01 | 0.850 |
| TMEM170B | 1.7e-01 | 0.850 |
| CERS4 | 1.7e-01 | 0.850 |
| POP4 | 1.7e-01 | 0.850 |
| QPRT | 1.7e-01 | 0.850 |
| KPNA2 | 1.7e-01 | 0.850 |
| NUDC | 1.7e-01 | 0.850 |
| TCEAL3 | 1.7e-01 | 0.850 |
| WIZ | 1.7e-01 | 0.850 |
| MALSU1 | 1.7e-01 | 0.850 |
| NHLRC2 | 1.7e-01 | 0.850 |
| RDH14 | 1.7e-01 | 0.850 |
| COX16 | 1.7e-01 | 0.850 |
| FKBP11 | 1.7e-01 | 0.850 |
| RP11-329L6.2 | 1.8e-01 | 0.850 |
| BAZ1B | 1.8e-01 | 0.850 |
| ANKRD13D | 1.8e-01 | 0.850 |
| ATG4D | 1.8e-01 | 0.850 |
| ZNF620 | 1.8e-01 | 0.850 |
| ASPM | 1.8e-01 | 0.850 |
| HINT1 | 1.8e-01 | 0.850 |
| ING3 | 1.8e-01 | 0.850 |
| CRCP | 1.8e-01 | 0.850 |
| MARK4 | 1.8e-01 | 0.850 |
| DOT1L | 1.8e-01 | 0.850 |
| SGO2 | 1.8e-01 | 0.850 |
| RNPEP | 1.8e-01 | 0.850 |
| NCL | 1.8e-01 | 0.850 |
| PUSL1 | 1.8e-01 | 0.850 |
| GID8 | 1.8e-01 | 0.850 |
| CEP57 | 1.8e-01 | 0.850 |
| BRSK2 | 1.8e-01 | 0.850 |
| PIN1 | 1.8e-01 | 0.850 |
| C6orf226 | 1.8e-01 | 0.850 |
| NISCH | 1.8e-01 | 0.850 |
| ZNF33A | 1.8e-01 | 0.850 |
| ZNF414 | 1.8e-01 | 0.850 |
| PARP1 | 1.8e-01 | 0.850 |
| TAF1D | 1.8e-01 | 0.850 |
| HBP1 | 1.8e-01 | 0.850 |
| KIF20A | 1.8e-01 | 0.850 |
| TUT1 | 1.8e-01 | 0.850 |
| GOPC | 1.8e-01 | 0.850 |
| MAPK11 | 1.8e-01 | 0.850 |
| TRIB2 | 1.8e-01 | 0.850 |
| BUB1B | 1.8e-01 | 0.850 |
| GNL1 | 1.8e-01 | 0.850 |
| ELK1 | 1.8e-01 | 0.850 |
| PDRG1 | 1.8e-01 | 0.850 |
| SARNP | 1.8e-01 | 0.850 |
| TOR3A | 1.8e-01 | 0.850 |
| UCK1 | 1.8e-01 | 0.850 |
| TENM3 | 1.8e-01 | 0.850 |
| KHDRBS1 | 1.8e-01 | 0.850 |
| TGFBR1 | 1.8e-01 | 0.850 |
| CARS | 1.8e-01 | 0.850 |
| RBM15 | 1.8e-01 | 0.850 |
| GNAI2 | 1.8e-01 | 0.850 |
| CDKN2A | 1.8e-01 | 0.850 |
| PNRC1 | 1.8e-01 | 0.850 |
| IRF3 | 1.8e-01 | 0.850 |
| TRAPPC6A | 1.8e-01 | 0.850 |
| NOA1 | 1.8e-01 | 0.850 |
| G3BP2 | 1.8e-01 | 0.850 |
| AK2 | 1.8e-01 | 0.850 |
| PPP2R2B | 1.8e-01 | 0.850 |
| DLAT | 1.8e-01 | 0.850 |
| MRPS12 | 1.8e-01 | 0.850 |
| SERPINB6 | 1.8e-01 | 0.850 |
| PRDX4 | 1.8e-01 | 0.850 |
| PARP2 | 1.8e-01 | 0.850 |
| RNF2 | 1.8e-01 | 0.850 |
| TBC1D7 | 1.8e-01 | 0.850 |
| GFOD2 | 1.8e-01 | 0.850 |
| POLA2 | 1.8e-01 | 0.850 |
| CD2BP2 | 1.8e-01 | 0.850 |
| RC3H2 | 1.8e-01 | 0.850 |
| MRPL54 | 1.8e-01 | 0.850 |
| C11orf73 | 1.8e-01 | 0.850 |
| MRPS18C | 1.8e-01 | 0.850 |
| STOX2 | 1.8e-01 | 0.850 |
| STK4 | 1.8e-01 | 0.850 |
| CCDC117 | 1.8e-01 | 0.850 |
| KIAA0355 | 1.8e-01 | 0.850 |
| LSM2 | 1.8e-01 | 0.850 |
| EDA2R | 1.8e-01 | 0.850 |
| GFER | 1.8e-01 | 0.850 |
| MSL3 | 1.8e-01 | 0.850 |
| UBR2 | 1.8e-01 | 0.850 |
| KLC2 | 1.8e-01 | 0.850 |
| BCAR1 | 1.8e-01 | 0.850 |
| MZT1 | 1.8e-01 | 0.850 |
| FRG1 | 1.8e-01 | 0.850 |
| AGO2 | 1.8e-01 | 0.850 |
| CARD8 | 1.8e-01 | 0.850 |
| PRSS35 | 1.8e-01 | 0.850 |
| RABL2B | 1.8e-01 | 0.850 |
| SAE1 | 1.8e-01 | 0.850 |
| SHROOM3 | 1.8e-01 | 0.850 |
| TYW3 | 1.8e-01 | 0.850 |
| BANP | 1.8e-01 | 0.850 |
| WDYHV1 | 1.8e-01 | 0.850 |
| CDC37L1 | 1.8e-01 | 0.850 |
| CENPK | 1.8e-01 | 0.850 |
| JOSD1 | 1.8e-01 | 0.850 |
| MAP4K4 | 1.9e-01 | 0.850 |
| PSME3 | 1.9e-01 | 0.850 |
| COX6B1 | 1.9e-01 | 0.850 |
| CCDC86 | 1.9e-01 | 0.850 |
| C6orf62 | 1.9e-01 | 0.850 |
| UBE4B | 1.9e-01 | 0.850 |
| PACS2 | 1.9e-01 | 0.850 |
| DLGAP5 | 1.9e-01 | 0.850 |
| PTK7 | 1.9e-01 | 0.850 |
| GTSE1 | 1.9e-01 | 0.850 |
| RCL1 | 1.9e-01 | 0.850 |
| LARP4B | 1.9e-01 | 0.850 |
| TBL2 | 1.9e-01 | 0.850 |
| ENAH | 1.9e-01 | 0.850 |
| TRMT10C | 1.9e-01 | 0.850 |
| FAS | 1.9e-01 | 0.850 |
| NECAP2 | 1.9e-01 | 0.850 |
| UPF1 | 1.9e-01 | 0.850 |
| NMT1 | 1.9e-01 | 0.850 |
| ZNF174 | 1.9e-01 | 0.850 |
| UQCRFS1 | 1.9e-01 | 0.850 |
| PIGG | 1.9e-01 | 0.850 |
| PAGR1 | 1.9e-01 | 0.850 |
| HTRA1 | 1.9e-01 | 0.850 |
| LASP1 | 1.9e-01 | 0.850 |
| G2E3 | 1.9e-01 | 0.850 |
| ORC6 | 1.9e-01 | 0.850 |
| MMP15 | 1.9e-01 | 0.850 |
| N6AMT1 | 1.9e-01 | 0.850 |
| ALKBH1 | 1.9e-01 | 0.850 |
| GPKOW | 1.9e-01 | 0.850 |
| SRGAP2 | 1.9e-01 | 0.850 |
| PDZD4 | 1.9e-01 | 0.850 |
| HSPA8 | 1.9e-01 | 0.850 |
| CDO1 | 1.9e-01 | 0.850 |
| DNASE1 | 1.9e-01 | 0.850 |
| OXSR1 | 1.9e-01 | 0.850 |
| BRMS1 | 1.9e-01 | 0.850 |
| DPM1 | 1.9e-01 | 0.850 |
| MRPL42 | 1.9e-01 | 0.850 |
| BEX4 | 1.9e-01 | 0.850 |
| CADM4 | 1.9e-01 | 0.850 |
| CHAMP1 | 1.9e-01 | 0.850 |
| IKBKB | 1.9e-01 | 0.850 |
| BRD7 | 1.9e-01 | 0.850 |
| SLC44A3 | 1.9e-01 | 0.850 |
| DHRS11 | 1.9e-01 | 0.850 |
| RNF187 | 1.9e-01 | 0.850 |
| MT1X | 1.9e-01 | 0.850 |
| TOM1 | 1.9e-01 | 0.850 |
| TFB2M | 1.9e-01 | 0.850 |
| RSAD1 | 1.9e-01 | 0.850 |
| LRRC14 | 1.9e-01 | 0.850 |
| GS1-124K5.4 | 1.9e-01 | 0.850 |
| CEP290 | 1.9e-01 | 0.850 |
| USMG5 | 1.9e-01 | 0.850 |
| FEM1B | 1.9e-01 | 0.850 |
| LEPROT | 1.9e-01 | 0.850 |
| RRAGA | 1.9e-01 | 0.850 |
| PNMAL1 | 1.9e-01 | 0.850 |
| APLP1 | 1.9e-01 | 0.850 |
| PHLDB1 | 1.9e-01 | 0.850 |
| SUV39H2 | 1.9e-01 | 0.850 |
| GCDH | 1.9e-01 | 0.850 |
| UBE2K | 1.9e-01 | 0.850 |
| LUC7L2 | 1.9e-01 | 0.850 |
| NT5C3A | 1.9e-01 | 0.850 |
| GEMIN7 | 1.9e-01 | 0.850 |
| CLPX | 1.9e-01 | 0.850 |
| SNX7 | 1.9e-01 | 0.850 |
| TRIM69 | 1.9e-01 | 0.850 |
| FEZ2 | 1.9e-01 | 0.850 |
| ADAT1 | 1.9e-01 | 0.850 |
| GRHPR | 1.9e-01 | 0.850 |
| WDR36 | 1.9e-01 | 0.850 |
| TRIM28 | 1.9e-01 | 0.850 |
| G3BP1 | 1.9e-01 | 0.850 |
| PEX26 | 1.9e-01 | 0.850 |
| PYCRL | 1.9e-01 | 0.850 |
| WSB2 | 1.9e-01 | 0.850 |
| PPM1B | 1.9e-01 | 0.850 |
| ACADVL | 1.9e-01 | 0.850 |
| C11orf31 | 1.9e-01 | 0.850 |
| SSNA1 | 1.9e-01 | 0.850 |
| STMN3 | 1.9e-01 | 0.850 |
| RTN4R | 1.9e-01 | 0.850 |
| DYRK1B | 2.0e-01 | 0.850 |
| RP11-46C20.1 | 2.0e-01 | 0.850 |
| FRMD4A | 2.0e-01 | 0.850 |
| NQO1 | 2.0e-01 | 0.850 |
| STK32C | 2.0e-01 | 0.850 |
| CBWD2 | 2.0e-01 | 0.850 |
| TNIP2 | 2.0e-01 | 0.850 |
| PBDC1 | 2.0e-01 | 0.850 |
| SMUG1 | 2.0e-01 | 0.850 |
| HIST2H2AC | 2.0e-01 | 0.850 |
| CTNNBL1 | 2.0e-01 | 0.850 |
| PDE7A | 2.0e-01 | 0.850 |
| PSMG2 | 2.0e-01 | 0.850 |
| RFWD3 | 2.0e-01 | 0.850 |
| NSL1 | 2.0e-01 | 0.850 |
| TRIM37 | 2.0e-01 | 0.850 |
| IFIT5 | 2.0e-01 | 0.850 |
| RUVBL2 | 2.0e-01 | 0.850 |
| ATP5L | 2.0e-01 | 0.850 |
| FHL3 | 2.0e-01 | 0.850 |
| RBM38 | 2.0e-01 | 0.850 |
| CKB | 2.0e-01 | 0.850 |
| TSPYL1 | 2.0e-01 | 0.850 |
| COMT | 2.0e-01 | 0.850 |
| ARRDC3 | 2.0e-01 | 0.850 |
| CCNL2 | 2.0e-01 | 0.850 |
| NT5DC1 | 2.0e-01 | 0.850 |
| CYB5A | 2.0e-01 | 0.850 |
| AAMDC | 2.0e-01 | 0.850 |
| KHNYN | 2.0e-01 | 0.850 |
| KDELR1 | 2.0e-01 | 0.850 |
| PLS3 | 2.0e-01 | 0.850 |
| PPRC1 | 2.0e-01 | 0.850 |
| RAB11FIP2 | 2.0e-01 | 0.850 |
| LSM7 | 2.0e-01 | 0.850 |
| RP11-5C23.3 | 2.0e-01 | 0.850 |
| LRRC49 | 2.0e-01 | 0.850 |
| CCNG2 | 2.0e-01 | 0.850 |
| GNB4 | 2.0e-01 | 0.850 |
| TRAF2 | 2.0e-01 | 0.850 |
| FAM174A | 2.0e-01 | 0.850 |
| TEX30 | 2.0e-01 | 0.850 |
| GMEB1 | 2.0e-01 | 0.850 |
| TDP1 | 2.0e-01 | 0.850 |
| CDK16 | 2.0e-01 | 0.850 |
| ARL6IP6 | 2.0e-01 | 0.850 |
| MAVS | 2.0e-01 | 0.850 |
| COG4 | 2.0e-01 | 0.850 |
| CWC22 | 2.0e-01 | 0.850 |
| NOTCH3 | 2.0e-01 | 0.850 |
| SKA2 | 2.0e-01 | 0.850 |
| ZBTB20 | 2.0e-01 | 0.850 |
| PTDSS1 | 2.0e-01 | 0.850 |
| GOLT1B | 2.0e-01 | 0.850 |
| ZNF444 | 2.0e-01 | 0.850 |
| ARMC10 | 2.0e-01 | 0.850 |
| EEF1G | 2.0e-01 | 0.850 |
| TADA1 | 2.0e-01 | 0.850 |
| YRDC | 2.0e-01 | 0.850 |
| TYRO3 | 2.0e-01 | 0.850 |
| PODXL | 2.0e-01 | 0.850 |
| TMEM208 | 2.0e-01 | 0.850 |
| YDJC | 2.0e-01 | 0.850 |
| TEAD2 | 2.0e-01 | 0.850 |
| LPIN1 | 2.0e-01 | 0.850 |
| PYGB | 2.0e-01 | 0.850 |
| NSUN6 | 2.0e-01 | 0.850 |
| STUB1 | 2.0e-01 | 0.850 |
| TTC32 | 2.0e-01 | 0.850 |
| BRCA2 | 2.0e-01 | 0.850 |
| DDX39B | 2.0e-01 | 0.850 |
| PLEKHG2 | 2.0e-01 | 0.850 |
| NARF | 2.0e-01 | 0.850 |
| CCDC97 | 2.0e-01 | 0.850 |
| CTC-338M12.3 | 2.0e-01 | 0.850 |
| NGRN | 2.0e-01 | 0.850 |
| ZNF282 | 2.0e-01 | 0.850 |
| ATP11C | 2.0e-01 | 0.850 |
| PNKP | 2.0e-01 | 0.850 |
| CITED1 | 2.0e-01 | 0.850 |
| C18orf25 | 2.0e-01 | 0.850 |
| OGFRL1 | 2.0e-01 | 0.850 |
| SYNE2 | 2.0e-01 | 0.850 |
| GTF2IRD1 | 2.0e-01 | 0.850 |
| MTAP | 2.0e-01 | 0.850 |
| SYNCRIP | 2.0e-01 | 0.850 |
| IRS2 | 2.0e-01 | 0.850 |
| TMEM259 | 2.0e-01 | 0.850 |
| ZNF514 | 2.0e-01 | 0.850 |
| NMB | 2.0e-01 | 0.850 |
| NUP50 | 2.0e-01 | 0.850 |
| PPP2R2D | 2.0e-01 | 0.850 |
| CTC-459F4.2 | 2.1e-01 | 0.850 |
| FBXO9 | 2.1e-01 | 0.850 |
| MRPS36 | 2.1e-01 | 0.850 |
| PALM | 2.1e-01 | 0.850 |
| GALT | 2.1e-01 | 0.850 |
| TRMT1L | 2.1e-01 | 0.850 |
| RNF10 | 2.1e-01 | 0.850 |
| CAMK2G | 2.1e-01 | 0.850 |
| CHMP1B | 2.1e-01 | 0.850 |
| ZFYVE21 | 2.1e-01 | 0.850 |
| DKC1 | 2.1e-01 | 0.850 |
| PKD1 | 2.1e-01 | 0.850 |
| RPL13 | 2.1e-01 | 0.850 |
| RNASEH2C | 2.1e-01 | 0.850 |
| ZNF134 | 2.1e-01 | 0.850 |
| SERINC1 | 2.1e-01 | 0.850 |
| HSPH1 | 2.1e-01 | 0.850 |
| ZNF84 | 2.1e-01 | 0.850 |
| NAV2 | 2.1e-01 | 0.850 |
| CSDE1 | 2.1e-01 | 0.850 |
| TBL3 | 2.1e-01 | 0.850 |
| DEK | 2.1e-01 | 0.850 |
| GRK4 | 2.1e-01 | 0.850 |
| PLEKHH3 | 2.1e-01 | 0.850 |
| MTCH1 | 2.1e-01 | 0.850 |
| ARL6IP5 | 2.1e-01 | 0.850 |
| KDELR2 | 2.1e-01 | 0.850 |
| FLCN | 2.1e-01 | 0.850 |
| NME4 | 2.1e-01 | 0.850 |
| MLH1 | 2.1e-01 | 0.850 |
| HPS5 | 2.1e-01 | 0.850 |
| PRKAR2A | 2.1e-01 | 0.850 |
| DNMT1 | 2.1e-01 | 0.850 |
| ARHGAP11A | 2.1e-01 | 0.850 |
| SRSF12 | 2.1e-01 | 0.850 |
| GSTZ1 | 2.1e-01 | 0.850 |
| IPO4 | 2.1e-01 | 0.850 |
| TOE1 | 2.1e-01 | 0.850 |
| CIRBP | 2.1e-01 | 0.850 |
| CCNA2 | 2.1e-01 | 0.850 |
| TERF2IP | 2.1e-01 | 0.850 |
| CYFIP1 | 2.1e-01 | 0.850 |
| GAPVD1 | 2.1e-01 | 0.850 |
| WDR6 | 2.1e-01 | 0.860 |
| RPL28 | 2.1e-01 | 0.860 |
| OTUB1 | 2.1e-01 | 0.860 |
| RNF169 | 2.1e-01 | 0.860 |
| HEXA | 2.1e-01 | 0.860 |
| USP46 | 2.1e-01 | 0.860 |
| NUP62 | 2.1e-01 | 0.860 |
| GOLGA7 | 2.1e-01 | 0.860 |
| NUDT16 | 2.1e-01 | 0.860 |
| CCDC34 | 2.1e-01 | 0.860 |
| CIR1 | 2.1e-01 | 0.860 |
| DDX3Y | 2.1e-01 | 0.860 |
| SNRPF | 2.1e-01 | 0.860 |
| SLK | 2.1e-01 | 0.860 |
| VEZF1 | 2.1e-01 | 0.860 |
| CYYR1 | 2.1e-01 | 0.860 |
| RP11-706O15.1 | 2.1e-01 | 0.860 |
| CHFR | 2.1e-01 | 0.860 |
| AC004945.2 | 2.1e-01 | 0.860 |
| HCCS | 2.1e-01 | 0.860 |
| MCCC1 | 2.1e-01 | 0.860 |
| CNPY2 | 2.1e-01 | 0.860 |
| CLPP | 2.1e-01 | 0.860 |
| KIAA1191 | 2.1e-01 | 0.860 |
| SMIM12 | 2.1e-01 | 0.860 |
| EXOC4 | 2.1e-01 | 0.860 |
| PCYT1A | 2.1e-01 | 0.860 |
| TRMT1 | 2.1e-01 | 0.860 |
| XPO1 | 2.1e-01 | 0.860 |
| H2AFX | 2.1e-01 | 0.860 |
| CREG1 | 2.1e-01 | 0.860 |
| C12orf75 | 2.1e-01 | 0.860 |
| BIN3 | 2.1e-01 | 0.860 |
| GGT7 | 2.1e-01 | 0.860 |
| DNTTIP2 | 2.1e-01 | 0.860 |
| CAPN7 | 2.1e-01 | 0.860 |
| PPP1R12A | 2.1e-01 | 0.860 |
| POLR2K | 2.1e-01 | 0.860 |
| SAMM50 | 2.1e-01 | 0.860 |
| EIF6 | 2.1e-01 | 0.860 |
| ARL16 | 2.1e-01 | 0.860 |
| RFT1 | 2.1e-01 | 0.860 |
| TRAPPC6B | 2.1e-01 | 0.860 |
| TMED5 | 2.1e-01 | 0.860 |
| IQCG | 2.2e-01 | 0.860 |
| CDH11 | 2.2e-01 | 0.860 |
| HECTD4 | 2.2e-01 | 0.860 |
| TNPO2 | 2.2e-01 | 0.860 |
| RNF14 | 2.2e-01 | 0.860 |
| CNOT6 | 2.2e-01 | 0.860 |
| HS3ST3A1 | 2.2e-01 | 0.860 |
| CHST10 | 2.2e-01 | 0.860 |
| EEF1A1 | 2.2e-01 | 0.860 |
| KIAA1958 | 2.2e-01 | 0.860 |
| CDK2 | 2.2e-01 | 0.860 |
| PDS5B | 2.2e-01 | 0.860 |
| NUBP1 | 2.2e-01 | 0.860 |
| C11orf74 | 2.2e-01 | 0.860 |
| TMEM5 | 2.2e-01 | 0.860 |
| YIPF6 | 2.2e-01 | 0.860 |
| SYMPK | 2.2e-01 | 0.860 |
| BRIX1 | 2.2e-01 | 0.860 |
| NDUFS3 | 2.2e-01 | 0.860 |
| IGF2BP3 | 2.2e-01 | 0.860 |
| PIGW | 2.2e-01 | 0.860 |
| GOLPH3 | 2.2e-01 | 0.860 |
| TRIM33 | 2.2e-01 | 0.860 |
| ATP5I | 2.2e-01 | 0.860 |
| PEX14 | 2.2e-01 | 0.860 |
| PIF1 | 2.2e-01 | 0.860 |
| JMJD4 | 2.2e-01 | 0.860 |
| EEF1E1 | 2.2e-01 | 0.860 |
| AGPAT5 | 2.2e-01 | 0.860 |
| PPM1D | 2.2e-01 | 0.860 |
| UBE2L3 | 2.2e-01 | 0.860 |
| NELL2 | 2.2e-01 | 0.860 |
| EXOC2 | 2.2e-01 | 0.860 |
| PKNOX1 | 2.2e-01 | 0.860 |
| PXYLP1 | 2.2e-01 | 0.860 |
| MAP3K3 | 2.2e-01 | 0.860 |
| SEPT10 | 2.2e-01 | 0.860 |
| DDHD1 | 2.2e-01 | 0.860 |
| ACTL6A | 2.2e-01 | 0.860 |
| GTF3A | 2.2e-01 | 0.860 |
| WASHC5 | 2.2e-01 | 0.860 |
| TGFB1I1 | 2.2e-01 | 0.860 |
| CYB5RL | 2.2e-01 | 0.860 |
| MAT2A | 2.2e-01 | 0.860 |
| CEP350 | 2.2e-01 | 0.860 |
| NAF1 | 2.2e-01 | 0.860 |
| LCORL | 2.2e-01 | 0.860 |
| DCUN1D5 | 2.2e-01 | 0.860 |
| AUP1 | 2.2e-01 | 0.860 |
| PRPF18 | 2.2e-01 | 0.860 |
| PHIP | 2.2e-01 | 0.860 |
| DCBLD2 | 2.2e-01 | 0.860 |
| PTDSS2 | 2.2e-01 | 0.860 |
| MAN2C1 | 2.2e-01 | 0.860 |
| EMP3 | 2.2e-01 | 0.860 |
| PRDX6 | 2.2e-01 | 0.860 |
| ANKS3 | 2.2e-01 | 0.860 |
| TAF11 | 2.2e-01 | 0.860 |
| NSRP1 | 2.2e-01 | 0.860 |
| APP | 2.2e-01 | 0.860 |
| ARMCX3 | 2.2e-01 | 0.860 |
| GLI3 | 2.2e-01 | 0.860 |
| CDC123 | 2.2e-01 | 0.860 |
| CSTB | 2.2e-01 | 0.860 |
| ZNF711 | 2.2e-01 | 0.860 |
| CEP19 | 2.2e-01 | 0.860 |
| TMED2 | 2.2e-01 | 0.860 |
| CAMK2N1 | 2.2e-01 | 0.860 |
| NOM1 | 2.2e-01 | 0.860 |
| PNP | 2.2e-01 | 0.860 |
| MYCL | 2.2e-01 | 0.860 |
| HIF1AN | 2.2e-01 | 0.860 |
| NME1 | 2.2e-01 | 0.860 |
| ATP6V0B | 2.3e-01 | 0.860 |
| RNF113A | 2.3e-01 | 0.860 |
| CNOT8 | 2.3e-01 | 0.860 |
| TOMM20 | 2.3e-01 | 0.860 |
| VANGL2 | 2.3e-01 | 0.860 |
| OST4 | 2.3e-01 | 0.860 |
| H3F3B | 2.3e-01 | 0.860 |
| SSH2 | 2.3e-01 | 0.860 |
| FAM200A | 2.3e-01 | 0.860 |
| C6orf52 | 2.3e-01 | 0.860 |
| RPL10 | 2.3e-01 | 0.860 |
| LRRC40 | 2.3e-01 | 0.860 |
| TLK2 | 2.3e-01 | 0.860 |
| IFFO1 | 2.3e-01 | 0.860 |
| CCDC93 | 2.3e-01 | 0.860 |
| OLA1 | 2.3e-01 | 0.860 |
| LYAR | 2.3e-01 | 0.860 |
| ECD | 2.3e-01 | 0.860 |
| PITHD1 | 2.3e-01 | 0.860 |
| MTHFS | 2.3e-01 | 0.860 |
| MTDH | 2.3e-01 | 0.860 |
| FLNA | 2.3e-01 | 0.860 |
| PPP1R15A | 2.3e-01 | 0.860 |
| TNRC6C | 2.3e-01 | 0.860 |
| SALL1 | 2.3e-01 | 0.860 |
| GLB1 | 2.3e-01 | 0.860 |
| MED22 | 2.3e-01 | 0.860 |
| PABPC1 | 2.3e-01 | 0.860 |
| TEX10 | 2.3e-01 | 0.860 |
| CYFIP2 | 2.3e-01 | 0.860 |
| LTV1 | 2.3e-01 | 0.860 |
| ZNF362 | 2.3e-01 | 0.860 |
| HIC2 | 2.3e-01 | 0.860 |
| EEF1D | 2.3e-01 | 0.860 |
| ECT2 | 2.3e-01 | 0.860 |
| ZNF131 | 2.3e-01 | 0.860 |
| CCDC115 | 2.3e-01 | 0.860 |
| NRSN2 | 2.3e-01 | 0.860 |
| GPBP1L1 | 2.3e-01 | 0.860 |
| FAM168B | 2.3e-01 | 0.860 |
| PTCD1 | 2.3e-01 | 0.860 |
| HILPDA | 2.3e-01 | 0.860 |
| TRMT12 | 2.3e-01 | 0.860 |
| PPP6R2 | 2.3e-01 | 0.860 |
| NDUFB7 | 2.3e-01 | 0.860 |
| GDI2 | 2.3e-01 | 0.860 |
| S100A16 | 2.3e-01 | 0.860 |
| CAB39 | 2.3e-01 | 0.860 |
| ATF6B | 2.3e-01 | 0.860 |
| PTGES3 | 2.3e-01 | 0.860 |
| PFN2 | 2.3e-01 | 0.860 |
| DUSP1 | 2.3e-01 | 0.860 |
| NME7 | 2.3e-01 | 0.860 |
| R3HCC1L | 2.3e-01 | 0.860 |
| KLF6 | 2.3e-01 | 0.860 |
| CCS | 2.3e-01 | 0.860 |
| TRABD | 2.3e-01 | 0.860 |
| PRR3 | 2.3e-01 | 0.860 |
| RRN3 | 2.3e-01 | 0.860 |
| RPL6 | 2.3e-01 | 0.860 |
| TNFAIP1 | 2.3e-01 | 0.860 |
| RUNDC1 | 2.3e-01 | 0.860 |
| THAP8 | 2.3e-01 | 0.860 |
| MAGOH | 2.3e-01 | 0.860 |
| TXLNA | 2.3e-01 | 0.860 |
| MED9 | 2.3e-01 | 0.860 |
| MSL1 | 2.3e-01 | 0.860 |
| MFSD3 | 2.3e-01 | 0.860 |
| COPS6 | 2.3e-01 | 0.860 |
| KCNJ6 | 2.3e-01 | 0.860 |
| OXR1 | 2.3e-01 | 0.860 |
| TMEM128 | 2.3e-01 | 0.860 |
| FAM208B | 2.3e-01 | 0.860 |
| CTD-2006C1.2 | 2.3e-01 | 0.860 |
| DRG2 | 2.3e-01 | 0.860 |
| LRRC75A | 2.3e-01 | 0.860 |
| LRP6 | 2.3e-01 | 0.860 |
| DDX3X | 2.3e-01 | 0.870 |
| ATP6V0E1 | 2.3e-01 | 0.870 |
| ZFAND6 | 2.3e-01 | 0.870 |
| DNAJC4 | 2.3e-01 | 0.870 |
| ZSWIM8 | 2.3e-01 | 0.870 |
| MRPS16 | 2.3e-01 | 0.870 |
| AGA | 2.3e-01 | 0.870 |
| NDUFS5 | 2.3e-01 | 0.870 |
| KRBOX4 | 2.3e-01 | 0.870 |
| HES4 | 2.3e-01 | 0.870 |
| WDR45 | 2.3e-01 | 0.870 |
| CD9 | 2.3e-01 | 0.870 |
| NRDE2 | 2.3e-01 | 0.870 |
| DCTD | 2.3e-01 | 0.870 |
| ID3 | 2.3e-01 | 0.870 |
| SEC62 | 2.3e-01 | 0.870 |
| YAP1 | 2.3e-01 | 0.870 |
| RPN1 | 2.3e-01 | 0.870 |
| GOT2 | 2.3e-01 | 0.870 |
| GPT2 | 2.3e-01 | 0.870 |
| CLASP1 | 2.3e-01 | 0.870 |
| CCDC25 | 2.4e-01 | 0.870 |
| KLHL25 | 2.4e-01 | 0.870 |
| SH3D19 | 2.4e-01 | 0.870 |
| CNTNAP2 | 2.4e-01 | 0.870 |
| DUSP3 | 2.4e-01 | 0.870 |
| METTL8 | 2.4e-01 | 0.870 |
| NPEPPS | 2.4e-01 | 0.870 |
| DCAF8 | 2.4e-01 | 0.870 |
| NDUFAF5 | 2.4e-01 | 0.870 |
| TRMT61A | 2.4e-01 | 0.870 |
| PPM1G | 2.4e-01 | 0.870 |
| PPCS | 2.4e-01 | 0.870 |
| PPWD1 | 2.4e-01 | 0.870 |
| MYCBP2 | 2.4e-01 | 0.870 |
| RNMT | 2.4e-01 | 0.870 |
| GAA | 2.4e-01 | 0.870 |
| NFKBIA | 2.4e-01 | 0.870 |
| UQCC2 | 2.4e-01 | 0.870 |
| AARS | 2.4e-01 | 0.870 |
| KIAA0895L | 2.4e-01 | 0.870 |
| TNFRSF12A | 2.4e-01 | 0.870 |
| PRPF4 | 2.4e-01 | 0.870 |
| TMEM57 | 2.4e-01 | 0.870 |
| OXA1L | 2.4e-01 | 0.870 |
| PFAS | 2.4e-01 | 0.870 |
| BDP1 | 2.4e-01 | 0.870 |
| LCMT1 | 2.4e-01 | 0.870 |
| ATP5C1 | 2.4e-01 | 0.870 |
| GBAS | 2.4e-01 | 0.870 |
| ZNF655 | 2.4e-01 | 0.870 |
| HNRNPA0 | 2.4e-01 | 0.870 |
| MKLN1 | 2.4e-01 | 0.870 |
| STK24 | 2.4e-01 | 0.870 |
| INTS4 | 2.4e-01 | 0.870 |
| ARMCX2 | 2.4e-01 | 0.870 |
| RP11-92K2.2 | 2.4e-01 | 0.870 |
| CTBP2 | 2.4e-01 | 0.870 |
| GTF2F2 | 2.4e-01 | 0.870 |
| STX18 | 2.4e-01 | 0.870 |
| ABHD17A | 2.4e-01 | 0.870 |
| IMP3 | 2.4e-01 | 0.870 |
| VIM | 2.4e-01 | 0.870 |
| NCK2 | 2.4e-01 | 0.870 |
| GAN | 2.4e-01 | 0.870 |
| TRAF4 | 2.4e-01 | 0.870 |
| PPP1R2 | 2.4e-01 | 0.870 |
| PBX3 | 2.4e-01 | 0.870 |
| PHF13 | 2.4e-01 | 0.870 |
| LRRC58 | 2.4e-01 | 0.870 |
| FAM69B | 2.4e-01 | 0.870 |
| CD99L2 | 2.4e-01 | 0.870 |
| ZNF248 | 2.4e-01 | 0.870 |
| DHCR24 | 2.4e-01 | 0.870 |
| SLC37A3 | 2.4e-01 | 0.870 |
| BUB1 | 2.4e-01 | 0.870 |
| MCRS1 | 2.4e-01 | 0.870 |
| CCDC71 | 2.4e-01 | 0.870 |
| PRELID3A | 2.4e-01 | 0.870 |
| ARL4C | 2.4e-01 | 0.870 |
| ZBTB21 | 2.4e-01 | 0.870 |
| PUS3 | 2.4e-01 | 0.870 |
| ENHO | 2.4e-01 | 0.870 |
| STAM | 2.4e-01 | 0.870 |
| LRRN1 | 2.4e-01 | 0.870 |
| MTG2 | 2.4e-01 | 0.870 |
| DHX37 | 2.4e-01 | 0.870 |
| DTYMK | 2.4e-01 | 0.870 |
| DZIP3 | 2.4e-01 | 0.870 |
| TULP3 | 2.4e-01 | 0.870 |
| NCAM1 | 2.4e-01 | 0.870 |
| INTS1 | 2.4e-01 | 0.870 |
| ZNRF3 | 2.4e-01 | 0.870 |
| C7orf73 | 2.4e-01 | 0.870 |
| DHRS13 | 2.4e-01 | 0.870 |
| MYL6B | 2.4e-01 | 0.870 |
| MTFR1 | 2.4e-01 | 0.870 |
| REST | 2.4e-01 | 0.870 |
| METAP1 | 2.4e-01 | 0.870 |
| NCKIPSD | 2.4e-01 | 0.870 |
| CCT4 | 2.4e-01 | 0.870 |
| UGCG | 2.5e-01 | 0.870 |
| CDK7 | 2.5e-01 | 0.870 |
| AAED1 | 2.5e-01 | 0.870 |
| MED7 | 2.5e-01 | 0.870 |
| TOMM34 | 2.5e-01 | 0.870 |
| LMBR1 | 2.5e-01 | 0.870 |
| UGP2 | 2.5e-01 | 0.870 |
| PTGR1 | 2.5e-01 | 0.870 |
| RAB8A | 2.5e-01 | 0.870 |
| RQCD1 | 2.5e-01 | 0.870 |
| SUMO1 | 2.5e-01 | 0.870 |
| SLC6A8 | 2.5e-01 | 0.870 |
| NDUFV2 | 2.5e-01 | 0.870 |
| SIX5 | 2.5e-01 | 0.870 |
| REV3L | 2.5e-01 | 0.870 |
| CLK1 | 2.5e-01 | 0.870 |
| RBBP8 | 2.5e-01 | 0.870 |
| CLDN12 | 2.5e-01 | 0.870 |
| RBM3 | 2.5e-01 | 0.870 |
| TMEM51 | 2.5e-01 | 0.870 |
| MIAT | 2.5e-01 | 0.870 |
| ISG20L2 | 2.5e-01 | 0.870 |
| NMD3 | 2.5e-01 | 0.870 |
| DNALI1 | 2.5e-01 | 0.870 |
| ZNF814 | 2.5e-01 | 0.870 |
| UTP20 | 2.5e-01 | 0.870 |
| CTDSPL2 | 2.5e-01 | 0.870 |
| FANCF | 2.5e-01 | 0.870 |
| RABGAP1 | 2.5e-01 | 0.870 |
| SKAP2 | 2.5e-01 | 0.870 |
| AC090421.1 | 2.5e-01 | 0.870 |
| RIT1 | 2.5e-01 | 0.870 |
| SS18L2 | 2.5e-01 | 0.870 |
| CTB-50L17.10 | 2.5e-01 | 0.870 |
| SHMT1 | 2.5e-01 | 0.870 |
| PCBD2 | 2.5e-01 | 0.870 |
| PRPF8 | 2.5e-01 | 0.870 |
| PLPP5 | 2.5e-01 | 0.880 |
| NUP54 | 2.5e-01 | 0.880 |
| USP24 | 2.5e-01 | 0.880 |
| C7orf13 | 2.5e-01 | 0.880 |
| ZNF264 | 2.5e-01 | 0.880 |
| C17orf62 | 2.5e-01 | 0.880 |
| SLC1A5 | 2.5e-01 | 0.880 |
| SMAP1 | 2.5e-01 | 0.880 |
| SPIDR | 2.5e-01 | 0.880 |
| BLOC1S5 | 2.5e-01 | 0.880 |
| GGCX | 2.5e-01 | 0.880 |
| TIMM21 | 2.5e-01 | 0.880 |
| RP11-16E18.4 | 2.5e-01 | 0.880 |
| UBE3D | 2.5e-01 | 0.880 |
| FDFT1 | 2.5e-01 | 0.880 |
| SNX6 | 2.5e-01 | 0.880 |
| AUTS2 | 2.5e-01 | 0.880 |
| SNRPA | 2.5e-01 | 0.880 |
| GTF3C6 | 2.5e-01 | 0.880 |
| RAB31 | 2.5e-01 | 0.880 |
| ANAPC15 | 2.5e-01 | 0.880 |
| MALT1 | 2.5e-01 | 0.880 |
| DHRS12 | 2.5e-01 | 0.880 |
| MTERFD3 | 2.5e-01 | 0.880 |
| ZSCAN9 | 2.5e-01 | 0.880 |
| RANBP3 | 2.5e-01 | 0.880 |
| LENG8 | 2.5e-01 | 0.880 |
| INAFM1 | 2.5e-01 | 0.880 |
| RAB10 | 2.5e-01 | 0.880 |
| DPYSL5 | 2.5e-01 | 0.880 |
| MAN1B1 | 2.5e-01 | 0.880 |
| ZYG11B | 2.5e-01 | 0.880 |
| PSMD7 | 2.5e-01 | 0.880 |
| PHKG2 | 2.5e-01 | 0.880 |
| PGRMC2 | 2.5e-01 | 0.880 |
| TSR2 | 2.5e-01 | 0.880 |
| SLC1A4 | 2.5e-01 | 0.880 |
| LETM1 | 2.5e-01 | 0.880 |
| MLLT6 | 2.5e-01 | 0.880 |
| ZNRF1 | 2.5e-01 | 0.880 |
| AGTRAP | 2.5e-01 | 0.880 |
| ADD2 | 2.5e-01 | 0.880 |
| RP11-206L10.11 | 2.6e-01 | 0.880 |
| TP53BP1 | 2.6e-01 | 0.880 |
| PSMD2 | 2.6e-01 | 0.880 |
| SPTLC1 | 2.6e-01 | 0.880 |
| BLOC1S4 | 2.6e-01 | 0.880 |
| RABL3 | 2.6e-01 | 0.880 |
| PROX1 | 2.6e-01 | 0.880 |
| COQ10A | 2.6e-01 | 0.880 |
| WRNIP1 | 2.6e-01 | 0.880 |
| CASK | 2.6e-01 | 0.880 |
| ZCCHC8 | 2.6e-01 | 0.880 |
| ELP4 | 2.6e-01 | 0.880 |
| SMIM10L1 | 2.6e-01 | 0.880 |
| PRKDC | 2.6e-01 | 0.880 |
| FBLN1 | 2.6e-01 | 0.880 |
| R3HDM1 | 2.6e-01 | 0.880 |
| RIOX2 | 2.6e-01 | 0.880 |
| EIF5AL1 | 2.6e-01 | 0.880 |
| UBQLN1 | 2.6e-01 | 0.880 |
| CBX4 | 2.6e-01 | 0.880 |
| CENPL | 2.6e-01 | 0.880 |
| EXOSC10 | 2.6e-01 | 0.880 |
| TMEM158 | 2.6e-01 | 0.880 |
| ZNF639 | 2.6e-01 | 0.880 |
| PON2 | 2.6e-01 | 0.880 |
| SSBP2 | 2.6e-01 | 0.880 |
| WRAP73 | 2.6e-01 | 0.880 |
| RAB5A | 2.6e-01 | 0.880 |
| CBWD1 | 2.6e-01 | 0.880 |
| MPDU1 | 2.6e-01 | 0.880 |
| ELOVL6 | 2.6e-01 | 0.880 |
| GDF15 | 2.6e-01 | 0.880 |
| PCIF1 | 2.6e-01 | 0.880 |
| TM7SF3 | 2.6e-01 | 0.880 |
| NUP35 | 2.6e-01 | 0.880 |
| ALG3 | 2.6e-01 | 0.880 |
| ZNF687 | 2.6e-01 | 0.880 |
| TP53I3 | 2.6e-01 | 0.880 |
| FUBP3 | 2.6e-01 | 0.880 |
| WASF1 | 2.6e-01 | 0.880 |
| ARFIP2 | 2.6e-01 | 0.880 |
| DSTN | 2.6e-01 | 0.880 |
| CBFA2T2 | 2.6e-01 | 0.880 |
| SNHG15 | 2.6e-01 | 0.880 |
| MEGF8 | 2.6e-01 | 0.880 |
| RNF7 | 2.6e-01 | 0.880 |
| MBOAT7 | 2.6e-01 | 0.880 |
| SLC38A2 | 2.6e-01 | 0.880 |
| SGCE | 2.6e-01 | 0.880 |
| SLC20A2 | 2.6e-01 | 0.880 |
| SPEN | 2.6e-01 | 0.880 |
| SRP19 | 2.6e-01 | 0.880 |
| TLN2 | 2.6e-01 | 0.880 |
| SIAH2 | 2.6e-01 | 0.880 |
| AADAT | 2.6e-01 | 0.880 |
| UBL4A | 2.6e-01 | 0.880 |
| SLC25A24 | 2.6e-01 | 0.880 |
| NRIP1 | 2.6e-01 | 0.880 |
| AZIN1 | 2.6e-01 | 0.880 |
| RPRD1A | 2.6e-01 | 0.880 |
| LSM5 | 2.6e-01 | 0.880 |
| TAF9B | 2.6e-01 | 0.880 |
| STRN4 | 2.6e-01 | 0.880 |
| USP9X | 2.6e-01 | 0.880 |
| BRD8 | 2.6e-01 | 0.880 |
| TRAPPC3 | 2.6e-01 | 0.880 |
| RBM12 | 2.6e-01 | 0.880 |
| KCNJ8 | 2.6e-01 | 0.880 |
| CTCF | 2.6e-01 | 0.880 |
| S100A10 | 2.6e-01 | 0.880 |
| TIMM8B | 2.6e-01 | 0.880 |
| KIF21A | 2.6e-01 | 0.880 |
| TRMT112 | 2.6e-01 | 0.880 |
| SLC9A3 | 2.6e-01 | 0.880 |
| NCAPH | 2.6e-01 | 0.880 |
| TENM2 | 2.6e-01 | 0.880 |
| EPHA2 | 2.6e-01 | 0.880 |
| MIB2 | 2.7e-01 | 0.880 |
| GTF3C2 | 2.7e-01 | 0.880 |
| TMEM254 | 2.7e-01 | 0.880 |
| ARGLU1 | 2.7e-01 | 0.880 |
| C6orf136 | 2.7e-01 | 0.880 |
| ISY1 | 2.7e-01 | 0.880 |
| TAF2 | 2.7e-01 | 0.880 |
| PLK1 | 2.7e-01 | 0.880 |
| EXOSC3 | 2.7e-01 | 0.890 |
| ATAD2B | 2.7e-01 | 0.890 |
| CALM1 | 2.7e-01 | 0.890 |
| MDN1 | 2.7e-01 | 0.890 |
| PISD | 2.7e-01 | 0.890 |
| COX19 | 2.7e-01 | 0.890 |
| GSX1 | 2.7e-01 | 0.890 |
| STK11IP | 2.7e-01 | 0.890 |
| C21orf59 | 2.7e-01 | 0.890 |
| PKN3 | 2.7e-01 | 0.890 |
| ZWINT | 2.7e-01 | 0.890 |
| ILF2 | 2.7e-01 | 0.890 |
| MTX1 | 2.7e-01 | 0.890 |
| OPA3 | 2.7e-01 | 0.890 |
| STXBP6 | 2.7e-01 | 0.890 |
| CIB2 | 2.7e-01 | 0.890 |
| ZFAND5 | 2.7e-01 | 0.890 |
| RDH11 | 2.7e-01 | 0.890 |
| AKAP11 | 2.7e-01 | 0.890 |
| LAGE3 | 2.7e-01 | 0.890 |
| POLR2G | 2.7e-01 | 0.890 |
| ACO2 | 2.7e-01 | 0.890 |
| LGALSL | 2.7e-01 | 0.890 |
| DCLRE1B | 2.7e-01 | 0.890 |
| P3H1 | 2.7e-01 | 0.890 |
| EEF1B2 | 2.7e-01 | 0.890 |
| POU3F2 | 2.7e-01 | 0.890 |
| TM2D2 | 2.7e-01 | 0.890 |
| SEPT3 | 2.7e-01 | 0.890 |
| CASP9 | 2.7e-01 | 0.890 |
| IRX2 | 2.7e-01 | 0.890 |
| RAB13 | 2.7e-01 | 0.890 |
| CNOT11 | 2.7e-01 | 0.890 |
| TMSB4X | 2.7e-01 | 0.890 |
| ZIK1 | 2.7e-01 | 0.890 |
| TAF9 | 2.7e-01 | 0.890 |
| SNAP23 | 2.7e-01 | 0.890 |
| TMEM214 | 2.7e-01 | 0.890 |
| MXRA7 | 2.7e-01 | 0.890 |
| MTCH2 | 2.7e-01 | 0.890 |
| SUN1 | 2.7e-01 | 0.890 |
| MPP5 | 2.7e-01 | 0.890 |
| HSD17B7 | 2.7e-01 | 0.890 |
| TNPO1 | 2.7e-01 | 0.890 |
| C9orf9 | 2.7e-01 | 0.890 |
| PLOD1 | 2.7e-01 | 0.890 |
| BEND5 | 2.7e-01 | 0.890 |
| PTPN11 | 2.7e-01 | 0.890 |
| MRTO4 | 2.7e-01 | 0.890 |
| RFNG | 2.7e-01 | 0.890 |
| NBDY | 2.7e-01 | 0.890 |
| RAN | 2.7e-01 | 0.890 |
| INF2 | 2.7e-01 | 0.890 |
| CEBPZOS | 2.7e-01 | 0.890 |
| CCAR1 | 2.7e-01 | 0.890 |
| EFHD2 | 2.7e-01 | 0.890 |
| LINC00338 | 2.7e-01 | 0.890 |
| LMNA | 2.7e-01 | 0.890 |
| RNF219 | 2.7e-01 | 0.890 |
| PSEN1 | 2.7e-01 | 0.890 |
| MTFP1 | 2.7e-01 | 0.890 |
| MRPL40 | 2.7e-01 | 0.890 |
| EVI5 | 2.7e-01 | 0.890 |
| DSTYK | 2.7e-01 | 0.890 |
| SMG1 | 2.7e-01 | 0.890 |
| FRA10AC1 | 2.7e-01 | 0.890 |
| CTXN1 | 2.8e-01 | 0.890 |
| B2M | 2.8e-01 | 0.890 |
| NSMF | 2.8e-01 | 0.890 |
| RP11-835E18.2 | 2.8e-01 | 0.890 |
| CAMK1D | 2.8e-01 | 0.890 |
| ZNF618 | 2.8e-01 | 0.890 |
| BARD1 | 2.8e-01 | 0.890 |
| ANAPC5 | 2.8e-01 | 0.890 |
| ILF3 | 2.8e-01 | 0.890 |
| UQCR11 | 2.8e-01 | 0.890 |
| CTD-3116E22.4 | 2.8e-01 | 0.890 |
| KLF10 | 2.8e-01 | 0.890 |
| TMEM181 | 2.8e-01 | 0.890 |
| CLK4 | 2.8e-01 | 0.890 |
| CLNS1A | 2.8e-01 | 0.890 |
| SMG5 | 2.8e-01 | 0.890 |
| FARP1 | 2.8e-01 | 0.890 |
| BRF2 | 2.8e-01 | 0.890 |
| DTL | 2.8e-01 | 0.890 |
| CEP162 | 2.8e-01 | 0.890 |
| PMP22 | 2.8e-01 | 0.890 |
| SMNDC1 | 2.8e-01 | 0.890 |
| PLP1 | 2.8e-01 | 0.890 |
| STX2 | 2.8e-01 | 0.890 |
| ATP5O | 2.8e-01 | 0.890 |
| SLC7A5 | 2.8e-01 | 0.890 |
| KDM5C | 2.8e-01 | 0.890 |
| METTL10 | 2.8e-01 | 0.890 |
| PUS1 | 2.8e-01 | 0.890 |
| LRRFIP1 | 2.8e-01 | 0.890 |
| EAPP | 2.8e-01 | 0.890 |
| ZPR1 | 2.8e-01 | 0.890 |
| RWDD1 | 2.8e-01 | 0.890 |
| CSTF2T | 2.8e-01 | 0.890 |
| WLS | 2.8e-01 | 0.890 |
| LDB1 | 2.8e-01 | 0.890 |
| UVRAG | 2.8e-01 | 0.890 |
| MARCH2 | 2.8e-01 | 0.890 |
| WDR83 | 2.8e-01 | 0.890 |
| GALK2 | 2.8e-01 | 0.890 |
| PIDD1 | 2.8e-01 | 0.890 |
| SEC61B | 2.8e-01 | 0.890 |
| ZNF507 | 2.8e-01 | 0.890 |
| PRPF38B | 2.8e-01 | 0.890 |
| CDK14 | 2.8e-01 | 0.890 |
| DEAF1 | 2.8e-01 | 0.890 |
| PPP1R12C | 2.8e-01 | 0.890 |
| NR1H2 | 2.8e-01 | 0.890 |
| PEX2 | 2.8e-01 | 0.890 |
| RPL7 | 2.8e-01 | 0.890 |
| MAEA | 2.8e-01 | 0.890 |
| FAM200B | 2.8e-01 | 0.890 |
| CRYZL1 | 2.8e-01 | 0.890 |
| WDFY2 | 2.8e-01 | 0.890 |
| SEC16A | 2.8e-01 | 0.890 |
| PPP2R5B | 2.8e-01 | 0.890 |
| IDH2 | 2.8e-01 | 0.890 |
| DDRGK1 | 2.8e-01 | 0.890 |
| PDZD11 | 2.8e-01 | 0.900 |
| CORO1C | 2.8e-01 | 0.900 |
| CEP295 | 2.8e-01 | 0.900 |
| DNAJC2 | 2.8e-01 | 0.900 |
| MON1A | 2.8e-01 | 0.900 |
| FCF1 | 2.8e-01 | 0.900 |
| NFATC3 | 2.8e-01 | 0.900 |
| IGBP1 | 2.8e-01 | 0.900 |
| EGR1 | 2.8e-01 | 0.900 |
| TBRG1 | 2.8e-01 | 0.900 |
| LANCL2 | 2.8e-01 | 0.900 |
| VPS25 | 2.8e-01 | 0.900 |
| TCEA2 | 2.8e-01 | 0.900 |
| BOP1 | 2.8e-01 | 0.900 |
| MAFG | 2.8e-01 | 0.900 |
| MRPL46 | 2.8e-01 | 0.900 |
| ODC1 | 2.8e-01 | 0.900 |
| IGDCC3 | 2.9e-01 | 0.900 |
| MDFI | 2.9e-01 | 0.900 |
| PCDH8 | 2.9e-01 | 0.900 |
| LRRC8A | 2.9e-01 | 0.900 |
| NCK1 | 2.9e-01 | 0.900 |
| CEBPG | 2.9e-01 | 0.900 |
| KCTD13 | 2.9e-01 | 0.900 |
| HOMEZ | 2.9e-01 | 0.900 |
| ARL6IP1 | 2.9e-01 | 0.900 |
| LAMB2 | 2.9e-01 | 0.900 |
| ALDH1B1 | 2.9e-01 | 0.900 |
| SS18 | 2.9e-01 | 0.900 |
| SPDL1 | 2.9e-01 | 0.900 |
| SESTD1 | 2.9e-01 | 0.900 |
| RAC1 | 2.9e-01 | 0.900 |
| RPS27 | 2.9e-01 | 0.900 |
| WBSCR17 | 2.9e-01 | 0.900 |
| PIM1 | 2.9e-01 | 0.900 |
| ISCA1 | 2.9e-01 | 0.900 |
| EMC7 | 2.9e-01 | 0.900 |
| CAPZA1 | 2.9e-01 | 0.900 |
| SNX8 | 2.9e-01 | 0.900 |
| RNPC3 | 2.9e-01 | 0.900 |
| NIPA2 | 2.9e-01 | 0.900 |
| POFUT2 | 2.9e-01 | 0.900 |
| TLDC1 | 2.9e-01 | 0.900 |
| USP12 | 2.9e-01 | 0.900 |
| DCAF7 | 2.9e-01 | 0.900 |
| KANSL1 | 2.9e-01 | 0.900 |
| RBM4 | 2.9e-01 | 0.900 |
| TCEAL8 | 2.9e-01 | 0.900 |
| NDUFB9 | 2.9e-01 | 0.900 |
| YY1 | 2.9e-01 | 0.900 |
| ABAT | 2.9e-01 | 0.900 |
| RP11-11N9.4 | 2.9e-01 | 0.900 |
| THOC3 | 2.9e-01 | 0.900 |
| PPA1 | 2.9e-01 | 0.900 |
| TTC3 | 2.9e-01 | 0.900 |
| TANC2 | 2.9e-01 | 0.900 |
| SPPL3 | 2.9e-01 | 0.900 |
| CRBN | 2.9e-01 | 0.900 |
| CMAS | 2.9e-01 | 0.900 |
| SERBP1 | 2.9e-01 | 0.900 |
| OTUD7B | 2.9e-01 | 0.900 |
| WSB1 | 2.9e-01 | 0.900 |
| CERK | 2.9e-01 | 0.900 |
| SLC25A33 | 2.9e-01 | 0.900 |
| SNN | 2.9e-01 | 0.900 |
| LRRC59 | 2.9e-01 | 0.900 |
| RNF168 | 2.9e-01 | 0.900 |
| GPI | 2.9e-01 | 0.900 |
| GABARAPL1 | 2.9e-01 | 0.900 |
| RP4-717I23.3 | 2.9e-01 | 0.900 |
| OSBPL2 | 2.9e-01 | 0.900 |
| REEP6 | 2.9e-01 | 0.900 |
| VWCE | 2.9e-01 | 0.900 |
| YWHAH | 2.9e-01 | 0.900 |
| DCTN6 | 2.9e-01 | 0.900 |
| UBB | 2.9e-01 | 0.900 |
| ABL1 | 2.9e-01 | 0.900 |
| SPPL2A | 2.9e-01 | 0.900 |
| SDF2L1 | 2.9e-01 | 0.900 |
| SPAST | 2.9e-01 | 0.900 |
| RNF13 | 2.9e-01 | 0.900 |
| DNAJC14 | 2.9e-01 | 0.900 |
| BRAT1 | 2.9e-01 | 0.900 |
| IPO9 | 2.9e-01 | 0.900 |
| SMAD3 | 2.9e-01 | 0.900 |
| DDX54 | 2.9e-01 | 0.900 |
| ARRB2 | 2.9e-01 | 0.900 |
| SARM1 | 2.9e-01 | 0.900 |
| TMBIM4 | 2.9e-01 | 0.900 |
| RNF115 | 2.9e-01 | 0.900 |
| C1GALT1 | 2.9e-01 | 0.900 |
| ITGB1BP1 | 2.9e-01 | 0.900 |
| LINC00632 | 2.9e-01 | 0.900 |
| KDM6B | 2.9e-01 | 0.900 |
| CLU | 2.9e-01 | 0.900 |
| DNAJB6 | 2.9e-01 | 0.900 |
| SCPEP1 | 2.9e-01 | 0.900 |
| TP53I11 | 2.9e-01 | 0.900 |
| MACF1 | 2.9e-01 | 0.900 |
| FGD4 | 2.9e-01 | 0.900 |
| GLRX3 | 2.9e-01 | 0.900 |
| AC003029.1 | 2.9e-01 | 0.900 |
| LSM12 | 2.9e-01 | 0.900 |
| ELOF1 | 2.9e-01 | 0.900 |
| FKBP9 | 2.9e-01 | 0.900 |
| PDCD6 | 2.9e-01 | 0.900 |
| ZNF76 | 2.9e-01 | 0.900 |
| TRUB1 | 3.0e-01 | 0.900 |
| DGCR14 | 3.0e-01 | 0.900 |
| MAX | 3.0e-01 | 0.900 |
| TTTY15 | 3.0e-01 | 0.900 |
| EIF1AY | 3.0e-01 | 0.900 |
| RPP40 | 3.0e-01 | 0.900 |
| CCNF | 3.0e-01 | 0.900 |
| RRP1B | 3.0e-01 | 0.900 |
| CLK3 | 3.0e-01 | 0.900 |
| TIMM17B | 3.0e-01 | 0.900 |
| RFXANK | 3.0e-01 | 0.900 |
| TAF1A | 3.0e-01 | 0.900 |
| A2M | 3.0e-01 | 0.900 |
| METAP1D | 3.0e-01 | 0.900 |
| HNRNPU | 3.0e-01 | 0.900 |
| EMC9 | 3.0e-01 | 0.900 |
| HMBOX1 | 3.0e-01 | 0.900 |
| YKT6 | 3.0e-01 | 0.900 |
| NEK4 | 3.0e-01 | 0.900 |
| OSGEP | 3.0e-01 | 0.900 |
| WBP1L | 3.0e-01 | 0.900 |
| ANXA6 | 3.0e-01 | 0.900 |
| TMEM194A | 3.0e-01 | 0.900 |
| USP48 | 3.0e-01 | 0.900 |
| NUDT9 | 3.0e-01 | 0.900 |
| VPS37A | 3.0e-01 | 0.900 |
| DPH7 | 3.0e-01 | 0.900 |
| MYL12B | 3.0e-01 | 0.900 |
| OSER1 | 3.0e-01 | 0.900 |
| UBE3C | 3.0e-01 | 0.900 |
| ZSCAN2 | 3.0e-01 | 0.900 |
| TOLLIP | 3.0e-01 | 0.900 |
| ZNF254 | 3.0e-01 | 0.900 |
| PCBP1 | 3.0e-01 | 0.900 |
| CHEK1 | 3.0e-01 | 0.900 |
| SCYL1 | 3.0e-01 | 0.900 |
| BTRC | 3.0e-01 | 0.900 |
| BAZ2B | 3.0e-01 | 0.900 |
| EDNRB | 3.0e-01 | 0.900 |
| PIK3R1 | 3.0e-01 | 0.900 |
| JAG1 | 3.0e-01 | 0.900 |
| CEP164 | 3.0e-01 | 0.900 |
| MTERF4 | 3.0e-01 | 0.900 |
| BAIAP2 | 3.0e-01 | 0.900 |
| GFPT1 | 3.0e-01 | 0.900 |
| WDR54 | 3.0e-01 | 0.900 |
| FBXL19 | 3.0e-01 | 0.900 |
| SF3A2 | 3.0e-01 | 0.900 |
| AHCY | 3.0e-01 | 0.900 |
| SLC30A1 | 3.0e-01 | 0.900 |
| FUBP1 | 3.0e-01 | 0.900 |
| REL | 3.0e-01 | 0.900 |
| TGIF1 | 3.0e-01 | 0.900 |
| RAD18 | 3.0e-01 | 0.900 |
| SLC25A10 | 3.0e-01 | 0.900 |
| CYTH2 | 3.0e-01 | 0.900 |
| IQGAP1 | 3.0e-01 | 0.900 |
| DDX5 | 3.0e-01 | 0.900 |
| HSPA5 | 3.0e-01 | 0.900 |
| TRIM13 | 3.0e-01 | 0.900 |
| GAR1 | 3.0e-01 | 0.900 |
| RPE | 3.0e-01 | 0.900 |
| KIAA1324L | 3.0e-01 | 0.900 |
| GIT1 | 3.0e-01 | 0.900 |
| RP11-151N17.2 | 3.0e-01 | 0.900 |
| TRIM11 | 3.0e-01 | 0.900 |
| CCDC41 | 3.0e-01 | 0.900 |
| PRC1 | 3.0e-01 | 0.900 |
| MED21 | 3.0e-01 | 0.900 |
| MMD | 3.0e-01 | 0.900 |
| GOLGA4 | 3.0e-01 | 0.900 |
| C9orf72 | 3.0e-01 | 0.900 |
| TM9SF4 | 3.0e-01 | 0.900 |
| EIF2S1 | 3.0e-01 | 0.900 |
| TDP2 | 3.0e-01 | 0.900 |
| AAMP | 3.0e-01 | 0.900 |
| MFAP1 | 3.0e-01 | 0.900 |
| COPS4 | 3.0e-01 | 0.900 |
| FAM89A | 3.0e-01 | 0.900 |
| DCAF5 | 3.0e-01 | 0.900 |
| GAS5 | 3.0e-01 | 0.900 |
| AC004951.6 | 3.0e-01 | 0.900 |
| PYGO2 | 3.1e-01 | 0.900 |
| HDDC3 | 3.1e-01 | 0.900 |
| FAM45A | 3.1e-01 | 0.900 |
| HSD17B11 | 3.1e-01 | 0.900 |
| PDLIM1 | 3.1e-01 | 0.900 |
| MRPL10 | 3.1e-01 | 0.900 |
| VPS33A | 3.1e-01 | 0.900 |
| RPS6 | 3.1e-01 | 0.900 |
| GNB2 | 3.1e-01 | 0.900 |
| SOX2 | 3.1e-01 | 0.900 |
| CAT | 3.1e-01 | 0.900 |
| SPIN1 | 3.1e-01 | 0.900 |
| C12orf49 | 3.1e-01 | 0.900 |
| ZBTB45 | 3.1e-01 | 0.900 |
| CUTC | 3.1e-01 | 0.900 |
| PIGP | 3.1e-01 | 0.900 |
| ATM | 3.1e-01 | 0.900 |
| B3GLCT | 3.1e-01 | 0.900 |
| ACAP2 | 3.1e-01 | 0.900 |
| ARL8A | 3.1e-01 | 0.900 |
| EIF1 | 3.1e-01 | 0.900 |
| IFRD1 | 3.1e-01 | 0.900 |
| IGF2R | 3.1e-01 | 0.900 |
| MFSD12 | 3.1e-01 | 0.900 |
| BRK1 | 3.1e-01 | 0.900 |
| PITPNB | 3.1e-01 | 0.900 |
| ZFHX4 | 3.1e-01 | 0.900 |
| GLIPR1 | 3.1e-01 | 0.900 |
| CPSF7 | 3.1e-01 | 0.900 |
| MED1 | 3.1e-01 | 0.900 |
| FANCB | 3.1e-01 | 0.900 |
| MAPKAPK3 | 3.1e-01 | 0.900 |
| ADO | 3.1e-01 | 0.900 |
| ATG3 | 3.1e-01 | 0.900 |
| SACM1L | 3.1e-01 | 0.900 |
| TSR3 | 3.1e-01 | 0.900 |
| KMT2A | 3.1e-01 | 0.900 |
| RP11-51J9.5 | 3.1e-01 | 0.900 |
| ADPRHL2 | 3.1e-01 | 0.900 |
| RUFY1 | 3.1e-01 | 0.900 |
| BTAF1 | 3.1e-01 | 0.900 |
| CHCHD7 | 3.1e-01 | 0.900 |
| MLEC | 3.1e-01 | 0.900 |
| PTPRA | 3.1e-01 | 0.900 |
| DDX27 | 3.1e-01 | 0.900 |
| NAGK | 3.1e-01 | 0.900 |
| TOPORS | 3.1e-01 | 0.900 |
| ADPRM | 3.1e-01 | 0.900 |
| TKTL1 | 3.1e-01 | 0.900 |
| USP8 | 3.1e-01 | 0.900 |
| CHN1 | 3.1e-01 | 0.900 |
| STX5 | 3.1e-01 | 0.900 |
| BTF3L4 | 3.1e-01 | 0.900 |
| CYHR1 | 3.1e-01 | 0.900 |
| ASH2L | 3.1e-01 | 0.900 |
| RTN2 | 3.1e-01 | 0.900 |
| NXN | 3.1e-01 | 0.900 |
| KLF13 | 3.1e-01 | 0.900 |
| TMEM59 | 3.1e-01 | 0.900 |
| MRPL37 | 3.1e-01 | 0.900 |
| PCNX1 | 3.1e-01 | 0.900 |
| WDR60 | 3.1e-01 | 0.900 |
| SNRPN | 3.1e-01 | 0.900 |
| MBOAT2 | 3.1e-01 | 0.900 |
| WDR11 | 3.1e-01 | 0.900 |
| MRPS25 | 3.1e-01 | 0.900 |
| ZXDC | 3.1e-01 | 0.900 |
| DEF8 | 3.1e-01 | 0.900 |
| PRPSAP1 | 3.1e-01 | 0.900 |
| ANKLE2 | 3.1e-01 | 0.900 |
| KLHL23 | 3.1e-01 | 0.900 |
| PPP1R11 | 3.1e-01 | 0.900 |
| UMAD1 | 3.1e-01 | 0.900 |
| EFTUD2 | 3.1e-01 | 0.900 |
| RNF149 | 3.1e-01 | 0.900 |
| SCRN1 | 3.1e-01 | 0.900 |
| CAP2 | 3.1e-01 | 0.900 |
| INTS13 | 3.1e-01 | 0.900 |
| EMD | 3.1e-01 | 0.900 |
| ZNF189 | 3.1e-01 | 0.900 |
| RPL38 | 3.1e-01 | 0.900 |
| FAM103A1 | 3.1e-01 | 0.900 |
| CXXC1 | 3.1e-01 | 0.900 |
| TOB2 | 3.1e-01 | 0.900 |
| SOCS4 | 3.1e-01 | 0.900 |
| CDKN1B | 3.1e-01 | 0.900 |
| RSBN1L | 3.1e-01 | 0.900 |
| CKAP2 | 3.1e-01 | 0.900 |
| ASCL1 | 3.1e-01 | 0.900 |
| CTD-2270L9.4 | 3.1e-01 | 0.900 |
| MED28 | 3.1e-01 | 0.900 |
| TSKU | 3.1e-01 | 0.900 |
| BRCC3 | 3.1e-01 | 0.900 |
| RPL22L1 | 3.1e-01 | 0.900 |
| RBM39 | 3.1e-01 | 0.900 |
| NOMO1 | 3.2e-01 | 0.900 |
| DYM | 3.2e-01 | 0.900 |
| CHPF | 3.2e-01 | 0.900 |
| PTPN13 | 3.2e-01 | 0.900 |
| SLC25A19 | 3.2e-01 | 0.900 |
| NDUFA8 | 3.2e-01 | 0.900 |
| RHBDD3 | 3.2e-01 | 0.900 |
| FNIP1 | 3.2e-01 | 0.900 |
| CALM3 | 3.2e-01 | 0.900 |
| USP14 | 3.2e-01 | 0.900 |
| SKI | 3.2e-01 | 0.900 |
| ANKS1A | 3.2e-01 | 0.900 |
| RP11-75C9.1 | 3.2e-01 | 0.900 |
| HUWE1 | 3.2e-01 | 0.900 |
| HJURP | 3.2e-01 | 0.900 |
| CDC25A | 3.2e-01 | 0.900 |
| ABRACL | 3.2e-01 | 0.900 |
| CD164 | 3.2e-01 | 0.900 |
| DDA1 | 3.2e-01 | 0.900 |
| ENC1 | 3.2e-01 | 0.900 |
| NDUFA5 | 3.2e-01 | 0.900 |
| MRRF | 3.2e-01 | 0.900 |
| KIF2C | 3.2e-01 | 0.900 |
| PPP5C | 3.2e-01 | 0.900 |
| USE1 | 3.2e-01 | 0.900 |
| MEPCE | 3.2e-01 | 0.900 |
| RPP30 | 3.2e-01 | 0.900 |
| CHTF8 | 3.2e-01 | 0.900 |
| BCL7B | 3.2e-01 | 0.900 |
| BZW2 | 3.2e-01 | 0.900 |
| JUN | 3.2e-01 | 0.900 |
| PPP1CC | 3.2e-01 | 0.900 |
| JAGN1 | 3.2e-01 | 0.900 |
| RAB33A | 3.2e-01 | 0.900 |
| ZNF880 | 3.2e-01 | 0.900 |
| DVL3 | 3.2e-01 | 0.900 |
| PGK1 | 3.2e-01 | 0.900 |
| ERCC6L2 | 3.2e-01 | 0.900 |
| CELF1 | 3.2e-01 | 0.900 |
| PPP1R18 | 3.2e-01 | 0.900 |
| TIAM2 | 3.2e-01 | 0.900 |
| ANAPC11 | 3.2e-01 | 0.900 |
| GOLGA3 | 3.2e-01 | 0.900 |
| THAP11 | 3.2e-01 | 0.900 |
| WBP11 | 3.2e-01 | 0.900 |
| PHF6 | 3.2e-01 | 0.900 |
| WDR75 | 3.2e-01 | 0.900 |
| FOXRED1 | 3.2e-01 | 0.900 |
| TXNRD1 | 3.2e-01 | 0.900 |
| SLC7A11 | 3.2e-01 | 0.900 |
| DDAH1 | 3.2e-01 | 0.900 |
| HNRNPH2 | 3.2e-01 | 0.900 |
| CUL4A | 3.2e-01 | 0.900 |
| EIF5A | 3.2e-01 | 0.900 |
| E4F1 | 3.2e-01 | 0.900 |
| CXCR4 | 3.2e-01 | 0.900 |
| CCNI | 3.2e-01 | 0.900 |
| CWC27 | 3.2e-01 | 0.900 |
| BPGM | 3.2e-01 | 0.900 |
| BSG | 3.2e-01 | 0.900 |
| CNTROB | 3.2e-01 | 0.900 |
| TNFRSF1A | 3.2e-01 | 0.900 |
| IFT88 | 3.2e-01 | 0.900 |
| PAFAH1B3 | 3.2e-01 | 0.900 |
| AKAP9 | 3.2e-01 | 0.900 |
| CHKA | 3.2e-01 | 0.900 |
| EBPL | 3.2e-01 | 0.900 |
| CARD19 | 3.2e-01 | 0.900 |
| TRAFD1 | 3.2e-01 | 0.900 |
| IMMT | 3.2e-01 | 0.900 |
| WDR83OS | 3.2e-01 | 0.900 |
| RNF141 | 3.2e-01 | 0.900 |
| ATP2A2 | 3.2e-01 | 0.900 |
| DFFA | 3.2e-01 | 0.900 |
| ZNF789 | 3.2e-01 | 0.900 |
| NDUFA2 | 3.2e-01 | 0.900 |
| TRIM9 | 3.2e-01 | 0.900 |
| ARFGAP1 | 3.2e-01 | 0.900 |
| EFNB3 | 3.2e-01 | 0.900 |
| CMBL | 3.2e-01 | 0.900 |
| EIF1B | 3.2e-01 | 0.900 |
| GRPEL2 | 3.2e-01 | 0.900 |
| RMND1 | 3.2e-01 | 0.900 |
| LETMD1 | 3.2e-01 | 0.900 |
| EHBP1 | 3.2e-01 | 0.900 |
| MAN1A2 | 3.2e-01 | 0.900 |
| PTRH2 | 3.2e-01 | 0.900 |
| AGPAT2 | 3.2e-01 | 0.900 |
| CDC23 | 3.2e-01 | 0.900 |
| LBR | 3.2e-01 | 0.900 |
| TMED7 | 3.2e-01 | 0.900 |
| POLE | 3.3e-01 | 0.900 |
| RMDN3 | 3.3e-01 | 0.900 |
| SERTAD1 | 3.3e-01 | 0.900 |
| EBAG9 | 3.3e-01 | 0.900 |
| C4orf48 | 3.3e-01 | 0.900 |
| DAPL1 | 3.3e-01 | 0.900 |
| DDX19B | 3.3e-01 | 0.900 |
| CMSS1 | 3.3e-01 | 0.900 |
| ZBTB5 | 3.3e-01 | 0.900 |
| CNTRL | 3.3e-01 | 0.900 |
| KIF23 | 3.3e-01 | 0.900 |
| ENKD1 | 3.3e-01 | 0.900 |
| CIC | 3.3e-01 | 0.900 |
| DYNC1LI1 | 3.3e-01 | 0.900 |
| GLTP | 3.3e-01 | 0.900 |
| HMGA2 | 3.3e-01 | 0.900 |
| MAP2K7 | 3.3e-01 | 0.900 |
| GGCT | 3.3e-01 | 0.900 |
| SERINC3 | 3.3e-01 | 0.900 |
| ZNF512 | 3.3e-01 | 0.900 |
| ROGDI | 3.3e-01 | 0.900 |
| KATNB1 | 3.3e-01 | 0.900 |
| COQ9 | 3.3e-01 | 0.900 |
| CCND3 | 3.3e-01 | 0.900 |
| PPIL1 | 3.3e-01 | 0.900 |
| EXOSC1 | 3.3e-01 | 0.900 |
| CPSF4 | 3.3e-01 | 0.900 |
| SKA3 | 3.3e-01 | 0.900 |
| XPNPEP1 | 3.3e-01 | 0.900 |
| SEC31A | 3.3e-01 | 0.900 |
| CEP57L1 | 3.3e-01 | 0.900 |
| YPEL5 | 3.3e-01 | 0.900 |
| TSPAN14 | 3.3e-01 | 0.900 |
| AP1B1 | 3.3e-01 | 0.900 |
| YIF1A | 3.3e-01 | 0.900 |
| ACD | 3.3e-01 | 0.900 |
| MRPL13 | 3.3e-01 | 0.900 |
| PCGF1 | 3.3e-01 | 0.900 |
| ITFG1 | 3.3e-01 | 0.900 |
| PKIG | 3.3e-01 | 0.900 |
| ANXA5 | 3.3e-01 | 0.900 |
| ADD3 | 3.3e-01 | 0.900 |
| TTC17 | 3.3e-01 | 0.900 |
| CCDC9 | 3.3e-01 | 0.900 |
| LLPH | 3.3e-01 | 0.900 |
| RPLP1 | 3.3e-01 | 0.900 |
| UHMK1 | 3.3e-01 | 0.900 |
| CDKN1A | 3.3e-01 | 0.900 |
| RPRD2 | 3.3e-01 | 0.900 |
| TRA2A | 3.3e-01 | 0.900 |
| SPATA7 | 3.3e-01 | 0.900 |
| SUPT16H | 3.3e-01 | 0.900 |
| KCNC1 | 3.3e-01 | 0.900 |
| BTG3 | 3.3e-01 | 0.900 |
| MESDC1 | 3.3e-01 | 0.900 |
| BLZF1 | 3.3e-01 | 0.900 |
| SLC35F6 | 3.3e-01 | 0.900 |
| YEATS2 | 3.3e-01 | 0.900 |
| ZFP62 | 3.3e-01 | 0.900 |
| ZFP36L1 | 3.3e-01 | 0.900 |
| DNAJB14 | 3.3e-01 | 0.900 |
| REEP4 | 3.3e-01 | 0.900 |
| POLD2 | 3.3e-01 | 0.900 |
| AC084018.1 | 3.3e-01 | 0.900 |
| HPS1 | 3.3e-01 | 0.900 |
| TMEM205 | 3.3e-01 | 0.900 |
| CPSF2 | 3.3e-01 | 0.900 |
| DDX19A | 3.3e-01 | 0.900 |
| ZNF397 | 3.3e-01 | 0.900 |
| RP11-231E4.4 | 3.3e-01 | 0.900 |
| GADD45G | 3.3e-01 | 0.900 |
| PSENEN | 3.3e-01 | 0.900 |
| SSFA2 | 3.3e-01 | 0.900 |
| MMAB | 3.3e-01 | 0.900 |
| NDC80 | 3.3e-01 | 0.900 |
| COX20 | 3.3e-01 | 0.900 |
| SLC25A51 | 3.3e-01 | 0.900 |
| HIPK2 | 3.3e-01 | 0.900 |
| FAM46A | 3.3e-01 | 0.900 |
| AGRN | 3.3e-01 | 0.900 |
| C5orf28 | 3.3e-01 | 0.900 |
| H3F3A | 3.3e-01 | 0.900 |
| C1orf27 | 3.3e-01 | 0.900 |
| PPAT | 3.4e-01 | 0.900 |
| CNOT10 | 3.4e-01 | 0.900 |
| FNBP1L | 3.4e-01 | 0.900 |
| RBM15B | 3.4e-01 | 0.900 |
| NAGPA | 3.4e-01 | 0.900 |
| FARS2 | 3.4e-01 | 0.900 |
| SMC6 | 3.4e-01 | 0.900 |
| MRPL30 | 3.4e-01 | 0.900 |
| CUL4B | 3.4e-01 | 0.900 |
| PAICS | 3.4e-01 | 0.900 |
| TPRKB | 3.4e-01 | 0.900 |
| C8orf41 | 3.4e-01 | 0.900 |
| MAPK7 | 3.4e-01 | 0.900 |
| SLC12A2 | 3.4e-01 | 0.900 |
| SFT2D1 | 3.4e-01 | 0.900 |
| ALKBH2 | 3.4e-01 | 0.900 |
| SUCLG1 | 3.4e-01 | 0.900 |
| POLD1 | 3.4e-01 | 0.900 |
| SRRM2 | 3.4e-01 | 0.900 |
| NOC4L | 3.4e-01 | 0.900 |
| CDYL | 3.4e-01 | 0.900 |
| DPH3 | 3.4e-01 | 0.900 |
| PAIP2 | 3.4e-01 | 0.900 |
| CCNB2 | 3.4e-01 | 0.900 |
| CENPB | 3.4e-01 | 0.900 |
| DEDD2 | 3.4e-01 | 0.900 |
| COX15 | 3.4e-01 | 0.900 |
| MCOLN1 | 3.4e-01 | 0.900 |
| ADAR | 3.4e-01 | 0.900 |
| MIDN | 3.4e-01 | 0.900 |
| ZBED1 | 3.4e-01 | 0.900 |
| CLCN7 | 3.4e-01 | 0.900 |
| DNAJB4 | 3.4e-01 | 0.900 |
| TXNL4A | 3.4e-01 | 0.900 |
| MOCS3 | 3.4e-01 | 0.900 |
| RDH13 | 3.4e-01 | 0.900 |
| TSG101 | 3.4e-01 | 0.900 |
| IGFBP4 | 3.4e-01 | 0.900 |
| FTH1 | 3.4e-01 | 0.900 |
| CUL3 | 3.4e-01 | 0.900 |
| COX17 | 3.4e-01 | 0.900 |
| SIRT2 | 3.4e-01 | 0.900 |
| HDAC1 | 3.4e-01 | 0.900 |
| SLC25A38 | 3.4e-01 | 0.900 |
| NME5 | 3.4e-01 | 0.900 |
| ORC2 | 3.4e-01 | 0.900 |
| ISOC2 | 3.4e-01 | 0.900 |
| CHD3 | 3.4e-01 | 0.900 |
| LRRN3 | 3.4e-01 | 0.900 |
| TSPAN3 | 3.4e-01 | 0.900 |
| EMID1 | 3.4e-01 | 0.900 |
| COMMD5 | 3.4e-01 | 0.900 |
| ARHGAP32 | 3.4e-01 | 0.900 |
| RBFA | 3.4e-01 | 0.900 |
| B4GALT3 | 3.4e-01 | 0.900 |
| ANKRD16 | 3.4e-01 | 0.900 |
| RALA | 3.4e-01 | 0.900 |
| PANK3 | 3.4e-01 | 0.900 |
| NUP93 | 3.4e-01 | 0.900 |
| CHCHD4 | 3.4e-01 | 0.900 |
| GOSR1 | 3.4e-01 | 0.900 |
| SLC27A5 | 3.4e-01 | 0.900 |
| WDR46 | 3.4e-01 | 0.900 |
| RTCB | 3.4e-01 | 0.900 |
| ST20 | 3.4e-01 | 0.900 |
| AES | 3.4e-01 | 0.900 |
| CWF19L1 | 3.4e-01 | 0.900 |
| SEC22A | 3.4e-01 | 0.900 |
| NFAT5 | 3.4e-01 | 0.900 |
| PRPF31 | 3.4e-01 | 0.900 |
| NFATC2IP | 3.4e-01 | 0.900 |
| SOCS2 | 3.4e-01 | 0.900 |
| CSNK1G1 | 3.4e-01 | 0.900 |
| RPS4Y1 | 3.4e-01 | 0.900 |
| BFAR | 3.4e-01 | 0.900 |
| SPRYD7 | 3.4e-01 | 0.900 |
| RPS27A | 3.4e-01 | 0.900 |
| TCEA1 | 3.4e-01 | 0.900 |
| TSPAN11 | 3.4e-01 | 0.900 |
| MICU1 | 3.4e-01 | 0.900 |
| SPATA2 | 3.4e-01 | 0.900 |
| MIEN1 | 3.4e-01 | 0.900 |
| CSNK1G2 | 3.4e-01 | 0.900 |
| RALBP1 | 3.4e-01 | 0.900 |
| ADK | 3.4e-01 | 0.900 |
| HGS | 3.5e-01 | 0.900 |
| MRPL18 | 3.5e-01 | 0.900 |
| YBEY | 3.5e-01 | 0.900 |
| ZRANB2 | 3.5e-01 | 0.900 |
| TARSL2 | 3.5e-01 | 0.900 |
| VPS16 | 3.5e-01 | 0.900 |
| TMEM167A | 3.5e-01 | 0.900 |
| LSM14A | 3.5e-01 | 0.900 |
| UBE2C | 3.5e-01 | 0.900 |
| APC2 | 3.5e-01 | 0.900 |
| PLEKHA3 | 3.5e-01 | 0.900 |
| ILDR2 | 3.5e-01 | 0.900 |
| CNOT1 | 3.5e-01 | 0.900 |
| KCTD1 | 3.5e-01 | 0.900 |
| COX7C | 3.5e-01 | 0.900 |
| C12orf10 | 3.5e-01 | 0.900 |
| TOB1 | 3.5e-01 | 0.900 |
| NECTIN2 | 3.5e-01 | 0.900 |
| PRPF38A | 3.5e-01 | 0.900 |
| ZC3H7B | 3.5e-01 | 0.900 |
| PDCL3 | 3.5e-01 | 0.900 |
| TMEM200C | 3.5e-01 | 0.900 |
| UBE2J1 | 3.5e-01 | 0.900 |
| ASXL1 | 3.5e-01 | 0.900 |
| C1orf109 | 3.5e-01 | 0.900 |
| UBTD1 | 3.5e-01 | 0.900 |
| TMEM38B | 3.5e-01 | 0.900 |
| GLI2 | 3.5e-01 | 0.900 |
| ATG10 | 3.5e-01 | 0.900 |
| MAPK3 | 3.5e-01 | 0.900 |
| RP11-620J15.3 | 3.5e-01 | 0.900 |
| RASSF1 | 3.5e-01 | 0.900 |
| KATNA1 | 3.5e-01 | 0.900 |
| PLPP3 | 3.5e-01 | 0.900 |
| CENPC | 3.5e-01 | 0.900 |
| RP11-701H24.2 | 3.5e-01 | 0.900 |
| SCAF1 | 3.5e-01 | 0.900 |
| WDR43 | 3.5e-01 | 0.900 |
| SPATA24 | 3.5e-01 | 0.900 |
| MED25 | 3.5e-01 | 0.900 |
| ERICH1 | 3.5e-01 | 0.900 |
| SGTB | 3.5e-01 | 0.900 |
| LDAH | 3.5e-01 | 0.900 |
| IST1 | 3.5e-01 | 0.900 |
| ZNF516 | 3.5e-01 | 0.900 |
| INTS12 | 3.5e-01 | 0.900 |
| CEP78 | 3.5e-01 | 0.900 |
| HSPA14 | 3.5e-01 | 0.900 |
| GARS | 3.5e-01 | 0.900 |
| KIAA1429 | 3.5e-01 | 0.900 |
| CLPB | 3.5e-01 | 0.900 |
| SRP14 | 3.5e-01 | 0.900 |
| POLG | 3.5e-01 | 0.900 |
| LIMA1 | 3.5e-01 | 0.900 |
| SRSF10 | 3.5e-01 | 0.900 |
| NFS1 | 3.5e-01 | 0.900 |
| RANBP17 | 3.5e-01 | 0.900 |
| LHFP | 3.5e-01 | 0.900 |
| MTHFD2 | 3.5e-01 | 0.900 |
| RECQL | 3.5e-01 | 0.900 |
| FLYWCH1 | 3.5e-01 | 0.900 |
| MED6 | 3.5e-01 | 0.900 |
| RP11-332H14.2 | 3.5e-01 | 0.900 |
| AEBP2 | 3.5e-01 | 0.900 |
| PXN | 3.5e-01 | 0.900 |
| DDX11 | 3.5e-01 | 0.900 |
| KIAA0232 | 3.5e-01 | 0.900 |
| PGRMC1 | 3.5e-01 | 0.900 |
| GEMIN8 | 3.5e-01 | 0.900 |
| CASP8AP2 | 3.5e-01 | 0.900 |
| VCAN | 3.5e-01 | 0.900 |
| SETBP1 | 3.5e-01 | 0.900 |
| TDG | 3.5e-01 | 0.900 |
| SYNGR1 | 3.5e-01 | 0.900 |
| GRIA1 | 3.5e-01 | 0.900 |
| GLUL | 3.5e-01 | 0.900 |
| CBX5 | 3.5e-01 | 0.900 |
| BIRC6 | 3.5e-01 | 0.900 |
| DNAJB1 | 3.5e-01 | 0.900 |
| CPVL | 3.5e-01 | 0.900 |
| CEBPD | 3.5e-01 | 0.900 |
| CCDC127 | 3.5e-01 | 0.900 |
| MAPKAPK2 | 3.6e-01 | 0.900 |
| PPIA | 3.6e-01 | 0.900 |
| PLGRKT | 3.6e-01 | 0.900 |
| WASL | 3.6e-01 | 0.900 |
| CKAP4 | 3.6e-01 | 0.900 |
| LSM4 | 3.6e-01 | 0.900 |
| MRPS10 | 3.6e-01 | 0.900 |
| ZNF566 | 3.6e-01 | 0.900 |
| B3GNT5 | 3.6e-01 | 0.900 |
| MAP6 | 3.6e-01 | 0.900 |
| ZFPL1 | 3.6e-01 | 0.900 |
| HERPUD1 | 3.6e-01 | 0.900 |
| FAT4 | 3.6e-01 | 0.900 |
| TPI1 | 3.6e-01 | 0.900 |
| ACTR10 | 3.6e-01 | 0.900 |
| BORCS7 | 3.6e-01 | 0.900 |
| ARPP19 | 3.6e-01 | 0.900 |
| RHBDL3 | 3.6e-01 | 0.900 |
| SPTLC2 | 3.6e-01 | 0.900 |
| RP11-448A19.1 | 3.6e-01 | 0.900 |
| UFD1L | 3.6e-01 | 0.900 |
| MED13 | 3.6e-01 | 0.900 |
| CRTAP | 3.6e-01 | 0.900 |
| HP1BP3 | 3.6e-01 | 0.900 |
| NAA30 | 3.6e-01 | 0.900 |
| PTCD3 | 3.6e-01 | 0.900 |
| ZNF3 | 3.6e-01 | 0.900 |
| MED29 | 3.6e-01 | 0.900 |
| TAF6L | 3.6e-01 | 0.900 |
| MCAT | 3.6e-01 | 0.900 |
| LMF2 | 3.6e-01 | 0.900 |
| MARCKSL1 | 3.6e-01 | 0.900 |
| ACSS2 | 3.6e-01 | 0.900 |
| UTP3 | 3.6e-01 | 0.900 |
| DMWD | 3.6e-01 | 0.900 |
| ROBO3 | 3.6e-01 | 0.900 |
| GPRIN1 | 3.6e-01 | 0.900 |
| FSCN1 | 3.6e-01 | 0.900 |
| WDR55 | 3.6e-01 | 0.900 |
| ADAM9 | 3.6e-01 | 0.900 |
| RP11-352M15.2 | 3.6e-01 | 0.900 |
| ANK3 | 3.6e-01 | 0.900 |
| GOSR2 | 3.6e-01 | 0.900 |
| USP15 | 3.6e-01 | 0.910 |
| SCG3 | 3.6e-01 | 0.910 |
| RAP2B | 3.6e-01 | 0.910 |
| ZNF800 | 3.6e-01 | 0.910 |
| SMAD2 | 3.6e-01 | 0.910 |
| PMM2 | 3.6e-01 | 0.910 |
| NMU | 3.6e-01 | 0.910 |
| SFXN5 | 3.6e-01 | 0.910 |
| TMEM106B | 3.6e-01 | 0.910 |
| SLC30A5 | 3.6e-01 | 0.910 |
| ENPP2 | 3.6e-01 | 0.910 |
| CCT7 | 3.6e-01 | 0.910 |
| PTS | 3.6e-01 | 0.910 |
| TSEN34 | 3.6e-01 | 0.910 |
| MRPL17 | 3.6e-01 | 0.910 |
| TMUB1 | 3.6e-01 | 0.910 |
| TPD52 | 3.6e-01 | 0.910 |
| HK1 | 3.6e-01 | 0.910 |
| MSANTD3 | 3.6e-01 | 0.910 |
| SNRNP70 | 3.6e-01 | 0.910 |
| SLC25A32 | 3.6e-01 | 0.910 |
| IFT22 | 3.6e-01 | 0.910 |
| KLHL9 | 3.6e-01 | 0.910 |
| CCDC51 | 3.6e-01 | 0.910 |
| TPM4 | 3.6e-01 | 0.910 |
| NXF1 | 3.6e-01 | 0.910 |
| ZSCAN26 | 3.6e-01 | 0.910 |
| EBNA1BP2 | 3.6e-01 | 0.910 |
| HELZ | 3.6e-01 | 0.910 |
| KIN | 3.6e-01 | 0.910 |
| PCDH9 | 3.6e-01 | 0.910 |
| DCTN2 | 3.6e-01 | 0.910 |
| POMT1 | 3.6e-01 | 0.910 |
| KLHL22 | 3.6e-01 | 0.910 |
| ZNF146 | 3.6e-01 | 0.910 |
| PPP1R15B | 3.7e-01 | 0.910 |
| RRNAD1 | 3.7e-01 | 0.910 |
| RNPEPL1 | 3.7e-01 | 0.910 |
| NAE1 | 3.7e-01 | 0.910 |
| NAA35 | 3.7e-01 | 0.910 |
| ZNF394 | 3.7e-01 | 0.910 |
| GAMT | 3.7e-01 | 0.910 |
| STRADB | 3.7e-01 | 0.910 |
| FN1 | 3.7e-01 | 0.910 |
| RIOK2 | 3.7e-01 | 0.910 |
| CAPN2 | 3.7e-01 | 0.910 |
| AC004696.2 | 3.7e-01 | 0.910 |
| RMND5A | 3.7e-01 | 0.910 |
| YY1AP1 | 3.7e-01 | 0.910 |
| ATG4C | 3.7e-01 | 0.910 |
| PSMB6 | 3.7e-01 | 0.910 |
| SNRPA1 | 3.7e-01 | 0.910 |
| TSEN2 | 3.7e-01 | 0.910 |
| IFT57 | 3.7e-01 | 0.910 |
| RPS21 | 3.7e-01 | 0.910 |
| RAB23 | 3.7e-01 | 0.910 |
| KIF11 | 3.7e-01 | 0.910 |
| SPATA13 | 3.7e-01 | 0.910 |
| TOP2A | 3.7e-01 | 0.910 |
| KIAA0368 | 3.7e-01 | 0.910 |
| SNUPN | 3.7e-01 | 0.910 |
| CCDC137 | 3.7e-01 | 0.910 |
| ARHGEF7 | 3.7e-01 | 0.910 |
| INO80D | 3.7e-01 | 0.910 |
| PSTPIP2 | 3.7e-01 | 0.910 |
| RACK1 | 3.7e-01 | 0.910 |
| EIF4ENIF1 | 3.7e-01 | 0.910 |
| C10orf35 | 3.7e-01 | 0.910 |
| TLE4 | 3.7e-01 | 0.910 |
| SMARCC1 | 3.7e-01 | 0.910 |
| KIF15 | 3.7e-01 | 0.910 |
| FAM104A | 3.7e-01 | 0.910 |
| NXPE3 | 3.7e-01 | 0.910 |
| MTSS1L | 3.7e-01 | 0.910 |
| ATAD3A | 3.7e-01 | 0.910 |
| NAXD | 3.7e-01 | 0.910 |
| ZNRD1 | 3.7e-01 | 0.910 |
| ICE1 | 3.7e-01 | 0.910 |
| GNA11 | 3.7e-01 | 0.910 |
| SOAT1 | 3.7e-01 | 0.910 |
| PAK1IP1 | 3.7e-01 | 0.910 |
| CASC10 | 3.7e-01 | 0.910 |
| POLR2F | 3.7e-01 | 0.910 |
| NAT14 | 3.7e-01 | 0.910 |
| SDHC | 3.7e-01 | 0.910 |
| ACADM | 3.7e-01 | 0.910 |
| YIPF4 | 3.7e-01 | 0.910 |
| NFRKB | 3.7e-01 | 0.910 |
| RAD51C | 3.7e-01 | 0.910 |
| DET1 | 3.7e-01 | 0.910 |
| PDCD2L | 3.7e-01 | 0.910 |
| PTPN2 | 3.7e-01 | 0.910 |
| FAM131A | 3.7e-01 | 0.910 |
| KIAA0319L | 3.7e-01 | 0.910 |
| STRBP | 3.7e-01 | 0.910 |
| SLC48A1 | 3.7e-01 | 0.910 |
| RNF139 | 3.7e-01 | 0.910 |
| DMTF1 | 3.7e-01 | 0.910 |
| SAMD1 | 3.7e-01 | 0.910 |
| BTG1 | 3.7e-01 | 0.910 |
| GPR19 | 3.7e-01 | 0.910 |
| MAP7D1 | 3.7e-01 | 0.910 |
| ADGRA3 | 3.7e-01 | 0.910 |
| KDM1A | 3.7e-01 | 0.910 |
| EEFSEC | 3.7e-01 | 0.910 |
| MTA2 | 3.7e-01 | 0.910 |
| NANS | 3.7e-01 | 0.910 |
| LRRC28 | 3.8e-01 | 0.910 |
| NSFL1C | 3.8e-01 | 0.910 |
| FZD3 | 3.8e-01 | 0.910 |
| TOR1AIP2 | 3.8e-01 | 0.910 |
| SURF2 | 3.8e-01 | 0.910 |
| SYNJ2BP | 3.8e-01 | 0.910 |
| RFC3 | 3.8e-01 | 0.910 |
| COX11 | 3.8e-01 | 0.910 |
| ZC3HC1 | 3.8e-01 | 0.910 |
| ALDH3A2 | 3.8e-01 | 0.910 |
| NBAS | 3.8e-01 | 0.910 |
| C16orf87 | 3.8e-01 | 0.910 |
| ADAL | 3.8e-01 | 0.910 |
| EIF3A | 3.8e-01 | 0.910 |
| ATRX | 3.8e-01 | 0.910 |
| FAM35A | 3.8e-01 | 0.910 |
| RP13-104F24.2 | 3.8e-01 | 0.910 |
| PEMT | 3.8e-01 | 0.910 |
| PJA2 | 3.8e-01 | 0.910 |
| GAS2L3 | 3.8e-01 | 0.910 |
| C19orf12 | 3.8e-01 | 0.910 |
| PITRM1 | 3.8e-01 | 0.910 |
| LDLRAD3 | 3.8e-01 | 0.910 |
| AP1M1 | 3.8e-01 | 0.910 |
| SUPT7L | 3.8e-01 | 0.910 |
| ASPH | 3.8e-01 | 0.910 |
| CCDC8 | 3.8e-01 | 0.910 |
| CMTM3 | 3.8e-01 | 0.910 |
| GNL3 | 3.8e-01 | 0.910 |
| RABGGTB | 3.8e-01 | 0.910 |
| PAPD4 | 3.8e-01 | 0.910 |
| CTPS2 | 3.8e-01 | 0.910 |
| RPL31 | 3.8e-01 | 0.910 |
| PCBD1 | 3.8e-01 | 0.910 |
| EEA1 | 3.8e-01 | 0.910 |
| CLN5 | 3.8e-01 | 0.910 |
| C7orf49 | 3.8e-01 | 0.910 |
| RAB2B | 3.8e-01 | 0.910 |
| TPR | 3.8e-01 | 0.910 |
| PSMG4 | 3.8e-01 | 0.910 |
| HAUS6 | 3.8e-01 | 0.910 |
| ZNF273 | 3.8e-01 | 0.910 |
| PDCD11 | 3.8e-01 | 0.910 |
| CKAP5 | 3.8e-01 | 0.910 |
| MPPE1 | 3.8e-01 | 0.910 |
| NPM1 | 3.8e-01 | 0.910 |
| ZNF74 | 3.8e-01 | 0.910 |
| GRSF1 | 3.8e-01 | 0.910 |
| IRX5 | 3.8e-01 | 0.910 |
| STK16 | 3.8e-01 | 0.910 |
| ROMO1 | 3.8e-01 | 0.910 |
| RTF1 | 3.8e-01 | 0.910 |
| ZNF649 | 3.8e-01 | 0.910 |
| SMAP2 | 3.8e-01 | 0.910 |
| NADK2 | 3.8e-01 | 0.910 |
| KANSL2 | 3.8e-01 | 0.910 |
| RPL35 | 3.8e-01 | 0.910 |
| ZZZ3 | 3.8e-01 | 0.910 |
| OSBPL8 | 3.8e-01 | 0.910 |
| ALG1 | 3.8e-01 | 0.920 |
| CRB2 | 3.8e-01 | 0.920 |
| COPS7B | 3.8e-01 | 0.920 |
| PFKP | 3.8e-01 | 0.920 |
| FAR1 | 3.8e-01 | 0.920 |
| SLC50A1 | 3.8e-01 | 0.920 |
| ZMYND11 | 3.8e-01 | 0.920 |
| SHISA5 | 3.8e-01 | 0.920 |
| BACE2 | 3.8e-01 | 0.920 |
| VPS29 | 3.8e-01 | 0.920 |
| FAM126B | 3.9e-01 | 0.920 |
| B9D2 | 3.9e-01 | 0.920 |
| BYSL | 3.9e-01 | 0.920 |
| ZNF536 | 3.9e-01 | 0.920 |
| NFX1 | 3.9e-01 | 0.920 |
| RP11-395G23.3 | 3.9e-01 | 0.920 |
| C12orf65 | 3.9e-01 | 0.920 |
| WDR18 | 3.9e-01 | 0.920 |
| NDUFS7 | 3.9e-01 | 0.920 |
| UBE2E2 | 3.9e-01 | 0.920 |
| RAB6B | 3.9e-01 | 0.920 |
| PROM1 | 3.9e-01 | 0.920 |
| NR2C2AP | 3.9e-01 | 0.920 |
| NOTCH2 | 3.9e-01 | 0.920 |
| C19orf43 | 3.9e-01 | 0.920 |
| DHCR7 | 3.9e-01 | 0.920 |
| TXN2 | 3.9e-01 | 0.920 |
| FAHD2B | 3.9e-01 | 0.920 |
| MRPL27 | 3.9e-01 | 0.920 |
| CAMKV | 3.9e-01 | 0.920 |
| WASHC3 | 3.9e-01 | 0.920 |
| WDR73 | 3.9e-01 | 0.920 |
| TMEM243 | 3.9e-01 | 0.920 |
| SLC31A1 | 3.9e-01 | 0.920 |
| PM20D2 | 3.9e-01 | 0.920 |
| TMEM219 | 3.9e-01 | 0.920 |
| ZNF423 | 3.9e-01 | 0.920 |
| HACL1 | 3.9e-01 | 0.920 |
| SCCPDH | 3.9e-01 | 0.920 |
| IGF2BP1 | 3.9e-01 | 0.920 |
| FAM32A | 3.9e-01 | 0.920 |
| BZW1 | 3.9e-01 | 0.920 |
| GRN | 3.9e-01 | 0.920 |
| RPL7L1 | 3.9e-01 | 0.920 |
| HARS | 3.9e-01 | 0.920 |
| EGLN2 | 3.9e-01 | 0.920 |
| GNB1 | 3.9e-01 | 0.920 |
| JMJD8 | 3.9e-01 | 0.920 |
| MAGT1 | 3.9e-01 | 0.920 |
| EPHB1 | 3.9e-01 | 0.920 |
| FBXO42 | 3.9e-01 | 0.920 |
| AP3B1 | 3.9e-01 | 0.920 |
| INTS6 | 3.9e-01 | 0.920 |
| XRCC1 | 3.9e-01 | 0.920 |
| PLK3 | 3.9e-01 | 0.920 |
| UCP2 | 3.9e-01 | 0.920 |
| ENTPD4 | 3.9e-01 | 0.920 |
| RSU1 | 3.9e-01 | 0.920 |
| CAMK1 | 3.9e-01 | 0.920 |
| TROVE2 | 3.9e-01 | 0.920 |
| AHCTF1 | 3.9e-01 | 0.920 |
| RANGRF | 3.9e-01 | 0.920 |
| CTNS | 3.9e-01 | 0.920 |
| C2orf69 | 3.9e-01 | 0.920 |
| SP1 | 3.9e-01 | 0.920 |
| CDK13 | 3.9e-01 | 0.920 |
| CERCAM | 3.9e-01 | 0.920 |
| CISD2 | 3.9e-01 | 0.920 |
| PIGK | 3.9e-01 | 0.920 |
| DCAF12 | 3.9e-01 | 0.920 |
| CALU | 3.9e-01 | 0.920 |
| NAV1 | 3.9e-01 | 0.920 |
| TTC28-AS1 | 3.9e-01 | 0.920 |
| RHBDD2 | 3.9e-01 | 0.920 |
| PCTP | 3.9e-01 | 0.920 |
| TAGLN | 3.9e-01 | 0.920 |
| TRMT2A | 3.9e-01 | 0.920 |
| UCKL1 | 3.9e-01 | 0.920 |
| NFYB | 3.9e-01 | 0.920 |
| RAB32 | 3.9e-01 | 0.920 |
| PDS5A | 3.9e-01 | 0.920 |
| RSL1D1 | 3.9e-01 | 0.920 |
| UTP6 | 3.9e-01 | 0.920 |
| C19orf52 | 3.9e-01 | 0.920 |
| PDSS2 | 3.9e-01 | 0.920 |
| PHF8 | 3.9e-01 | 0.920 |
| SNX1 | 3.9e-01 | 0.920 |
| NFE2L1 | 3.9e-01 | 0.920 |
| WRN | 3.9e-01 | 0.920 |
| ZNF559 | 3.9e-01 | 0.920 |
| MFN1 | 3.9e-01 | 0.920 |
| CBL | 3.9e-01 | 0.920 |
| ELAVL2 | 3.9e-01 | 0.920 |
| FAM86B1 | 3.9e-01 | 0.920 |
| TLN1 | 3.9e-01 | 0.920 |
| FAM177A1 | 3.9e-01 | 0.920 |
| CSRP2BP | 3.9e-01 | 0.920 |
| C12orf73 | 3.9e-01 | 0.920 |
| HAT1 | 3.9e-01 | 0.920 |
| NOVA1 | 4.0e-01 | 0.920 |
| TRIM71 | 4.0e-01 | 0.920 |
| NARS | 4.0e-01 | 0.920 |
| CCDC22 | 4.0e-01 | 0.920 |
| PTPRD | 4.0e-01 | 0.920 |
| TBCC | 4.0e-01 | 0.920 |
| LRRCC1 | 4.0e-01 | 0.920 |
| SCOC | 4.0e-01 | 0.920 |
| FOXN3 | 4.0e-01 | 0.920 |
| CSTF1 | 4.0e-01 | 0.920 |
| GLRX | 4.0e-01 | 0.920 |
| PHGDH | 4.0e-01 | 0.920 |
| CYC1 | 4.0e-01 | 0.920 |
| INO80 | 4.0e-01 | 0.920 |
| ETAA1 | 4.0e-01 | 0.920 |
| MAP1B | 4.0e-01 | 0.920 |
| CCT3 | 4.0e-01 | 0.920 |
| RPS18 | 4.0e-01 | 0.920 |
| NDUFB6 | 4.0e-01 | 0.920 |
| EMC3 | 4.0e-01 | 0.920 |
| RAD51AP1 | 4.0e-01 | 0.920 |
| ZNF124 | 4.0e-01 | 0.920 |
| SGSM2 | 4.0e-01 | 0.920 |
| MYEF2 | 4.0e-01 | 0.920 |
| BNIP1 | 4.0e-01 | 0.920 |
| IMPAD1 | 4.0e-01 | 0.920 |
| CD59 | 4.0e-01 | 0.920 |
| MAPK10 | 4.0e-01 | 0.920 |
| RP11-566E18.3 | 4.0e-01 | 0.920 |
| CYTOR | 4.0e-01 | 0.920 |
| OSBPL9 | 4.0e-01 | 0.920 |
| NUFIP2 | 4.0e-01 | 0.920 |
| DDIT3 | 4.0e-01 | 0.920 |
| NDUFB8 | 4.0e-01 | 0.920 |
| MTG1 | 4.0e-01 | 0.920 |
| MTF1 | 4.0e-01 | 0.920 |
| WNK1 | 4.0e-01 | 0.920 |
| RP11-374P20.5 | 4.0e-01 | 0.920 |
| ZDHHC12 | 4.0e-01 | 0.920 |
| COX6C | 4.0e-01 | 0.920 |
| LMF1 | 4.0e-01 | 0.920 |
| VIPAS39 | 4.0e-01 | 0.920 |
| TMEM8A | 4.0e-01 | 0.920 |
| DCHS1 | 4.0e-01 | 0.920 |
| ZC4H2 | 4.0e-01 | 0.920 |
| RPL23A | 4.0e-01 | 0.920 |
| EARS2 | 4.0e-01 | 0.920 |
| PRIMPOL | 4.0e-01 | 0.920 |
| LDLR | 4.0e-01 | 0.920 |
| BOLA3 | 4.0e-01 | 0.920 |
| BCLAF1 | 4.0e-01 | 0.920 |
| ACTN1 | 4.0e-01 | 0.920 |
| POMGNT2 | 4.0e-01 | 0.920 |
| POLR3C | 4.0e-01 | 0.920 |
| ERI3 | 4.0e-01 | 0.920 |
| F2R | 4.0e-01 | 0.920 |
| ARHGAP5 | 4.0e-01 | 0.920 |
| ARPC3 | 4.0e-01 | 0.920 |
| RXRB | 4.0e-01 | 0.920 |
| MYL12A | 4.0e-01 | 0.920 |
| FBXL6 | 4.0e-01 | 0.920 |
| MBNL1 | 4.0e-01 | 0.920 |
| SARS2 | 4.0e-01 | 0.920 |
| GNA12 | 4.0e-01 | 0.920 |
| ADAM17 | 4.0e-01 | 0.920 |
| NGFR | 4.0e-01 | 0.920 |
| HSD17B12 | 4.0e-01 | 0.920 |
| HIST1H2AC | 4.0e-01 | 0.920 |
| TXNDC15 | 4.0e-01 | 0.920 |
| USP16 | 4.0e-01 | 0.920 |
| PDCD2 | 4.0e-01 | 0.920 |
| PGBD1 | 4.0e-01 | 0.920 |
| GAPDH | 4.0e-01 | 0.920 |
| CTDP1 | 4.0e-01 | 0.920 |
| C14orf2 | 4.0e-01 | 0.920 |
| AACS | 4.0e-01 | 0.920 |
| FAM122B | 4.0e-01 | 0.920 |
| WDR59 | 4.0e-01 | 0.920 |
| ST7L | 4.0e-01 | 0.920 |
| IFITM3 | 4.1e-01 | 0.920 |
| GTPBP3 | 4.1e-01 | 0.920 |
| CDC7 | 4.1e-01 | 0.920 |
| ATP5G3 | 4.1e-01 | 0.920 |
| ALKBH8 | 4.1e-01 | 0.920 |
| XIAP | 4.1e-01 | 0.920 |
| L3MBTL2 | 4.1e-01 | 0.920 |
| C19orf54 | 4.1e-01 | 0.920 |
| SLC30A6 | 4.1e-01 | 0.920 |
| DERL1 | 4.1e-01 | 0.920 |
| C1orf106 | 4.1e-01 | 0.920 |
| ETFDH | 4.1e-01 | 0.920 |
| DUSP14 | 4.1e-01 | 0.920 |
| DUS2 | 4.1e-01 | 0.920 |
| TTC14 | 4.1e-01 | 0.920 |
| ZNF830 | 4.1e-01 | 0.920 |
| SBK1 | 4.1e-01 | 0.920 |
| PSMB1 | 4.1e-01 | 0.920 |
| NDUFS2 | 4.1e-01 | 0.920 |
| TSC1 | 4.1e-01 | 0.920 |
| BUD13 | 4.1e-01 | 0.920 |
| SRP72 | 4.1e-01 | 0.920 |
| SART3 | 4.1e-01 | 0.920 |
| THOC1 | 4.1e-01 | 0.920 |
| KIAA1549 | 4.1e-01 | 0.920 |
| TBL1X | 4.1e-01 | 0.920 |
| AMOT | 4.1e-01 | 0.920 |
| PGLS | 4.1e-01 | 0.920 |
| STX4 | 4.1e-01 | 0.920 |
| EIF4B | 4.1e-01 | 0.920 |
| AUNIP | 4.1e-01 | 0.920 |
| QSER1 | 4.1e-01 | 0.920 |
| TFIP11 | 4.1e-01 | 0.920 |
| MCM4 | 4.1e-01 | 0.920 |
| EIF2D | 4.1e-01 | 0.920 |
| TEAD3 | 4.1e-01 | 0.920 |
| SPG21 | 4.1e-01 | 0.920 |
| SDR39U1 | 4.1e-01 | 0.920 |
| PNO1 | 4.1e-01 | 0.920 |
| ASMTL | 4.1e-01 | 0.920 |
| SDHAF1 | 4.1e-01 | 0.920 |
| ZNF83 | 4.1e-01 | 0.920 |
| DR1 | 4.1e-01 | 0.920 |
| DSEL | 4.1e-01 | 0.920 |
| UBE2F | 4.1e-01 | 0.920 |
| KBTBD4 | 4.1e-01 | 0.920 |
| VSIG10 | 4.1e-01 | 0.920 |
| HYAL2 | 4.1e-01 | 0.920 |
| C6orf203 | 4.1e-01 | 0.920 |
| PWWP2A | 4.1e-01 | 0.920 |
| CCNB1 | 4.1e-01 | 0.920 |
| HOXB2 | 4.1e-01 | 0.920 |
| TFPT | 4.1e-01 | 0.920 |
| C19orf79 | 4.1e-01 | 0.920 |
| LITAF | 4.1e-01 | 0.920 |
| WNT7A | 4.1e-01 | 0.920 |
| DIS3L2 | 4.1e-01 | 0.920 |
| RRP15 | 4.1e-01 | 0.920 |
| SYNRG | 4.1e-01 | 0.920 |
| CLN8 | 4.1e-01 | 0.920 |
| DCUN1D4 | 4.1e-01 | 0.920 |
| ORMDL1 | 4.1e-01 | 0.920 |
| JPX | 4.1e-01 | 0.920 |
| RPL37 | 4.1e-01 | 0.920 |
| STAG2 | 4.1e-01 | 0.920 |
| CCNT1 | 4.1e-01 | 0.920 |
| PGM2 | 4.1e-01 | 0.920 |
| LONRF1 | 4.1e-01 | 0.920 |
| CSPP1 | 4.1e-01 | 0.920 |
| SMC4 | 4.1e-01 | 0.920 |
| FAM19A5 | 4.1e-01 | 0.920 |
| DGCR6L | 4.1e-01 | 0.920 |
| CSRNP3 | 4.1e-01 | 0.920 |
| DUSP6 | 4.1e-01 | 0.920 |
| UBE2M | 4.1e-01 | 0.920 |
| GINS4 | 4.1e-01 | 0.920 |
| HEBP2 | 4.1e-01 | 0.920 |
| SARAF | 4.1e-01 | 0.920 |
| SMYD2 | 4.1e-01 | 0.920 |
| LMBR1L | 4.1e-01 | 0.920 |
| PPIB | 4.1e-01 | 0.920 |
| LONP2 | 4.1e-01 | 0.920 |
| RP11-752G15.7 | 4.1e-01 | 0.920 |
| ARV1 | 4.1e-01 | 0.920 |
| UHRF1 | 4.2e-01 | 0.920 |
| KLF12 | 4.2e-01 | 0.920 |
| BIN1 | 4.2e-01 | 0.920 |
| EFNA4 | 4.2e-01 | 0.920 |
| TSGA10 | 4.2e-01 | 0.920 |
| SUV39H1 | 4.2e-01 | 0.920 |
| LYRM4 | 4.2e-01 | 0.920 |
| WIPI2 | 4.2e-01 | 0.920 |
| PMS1 | 4.2e-01 | 0.920 |
| OTUD5 | 4.2e-01 | 0.920 |
| RP11-522B15.3 | 4.2e-01 | 0.920 |
| UPF3B | 4.2e-01 | 0.920 |
| NUBPL | 4.2e-01 | 0.920 |
| ELMOD2 | 4.2e-01 | 0.920 |
| ELK4 | 4.2e-01 | 0.920 |
| MAP1S | 4.2e-01 | 0.920 |
| PFKM | 4.2e-01 | 0.920 |
| TSSC1 | 4.2e-01 | 0.920 |
| SNRPE | 4.2e-01 | 0.920 |
| NAA10 | 4.2e-01 | 0.920 |
| AGTPBP1 | 4.2e-01 | 0.920 |
| C1orf131 | 4.2e-01 | 0.920 |
| EPN1 | 4.2e-01 | 0.920 |
| DERL2 | 4.2e-01 | 0.920 |
| ARID1B | 4.2e-01 | 0.920 |
| PGM2L1 | 4.2e-01 | 0.920 |
| DOLPP1 | 4.2e-01 | 0.920 |
| HSPA9 | 4.2e-01 | 0.920 |
| RFX7 | 4.2e-01 | 0.920 |
| FLVCR1 | 4.2e-01 | 0.920 |
| PPT1 | 4.2e-01 | 0.920 |
| CDH2 | 4.2e-01 | 0.920 |
| PSPC1 | 4.2e-01 | 0.920 |
| POLR1C | 4.2e-01 | 0.920 |
| C9orf3 | 4.2e-01 | 0.920 |
| USP4 | 4.2e-01 | 0.920 |
| YIF1B | 4.2e-01 | 0.920 |
| RHOC | 4.2e-01 | 0.920 |
| SYT17 | 4.2e-01 | 0.920 |
| TBC1D10B | 4.2e-01 | 0.920 |
| MASTL | 4.2e-01 | 0.920 |
| SBNO1 | 4.2e-01 | 0.920 |
| EIF4G1 | 4.2e-01 | 0.920 |
| FTL | 4.2e-01 | 0.920 |
| NUP43 | 4.2e-01 | 0.920 |
| FAM161A | 4.2e-01 | 0.920 |
| CXXC4 | 4.2e-01 | 0.930 |
| ACAD9 | 4.2e-01 | 0.930 |
| GTF2I | 4.2e-01 | 0.930 |
| ORAOV1 | 4.2e-01 | 0.930 |
| WBP4 | 4.2e-01 | 0.930 |
| ERGIC3 | 4.2e-01 | 0.930 |
| PSMD5 | 4.2e-01 | 0.930 |
| SMARCA5 | 4.2e-01 | 0.930 |
| MLLT4 | 4.2e-01 | 0.930 |
| NCOA5 | 4.2e-01 | 0.930 |
| ATXN2 | 4.2e-01 | 0.930 |
| PHC3 | 4.2e-01 | 0.930 |
| CREBZF | 4.3e-01 | 0.930 |
| UCHL3 | 4.3e-01 | 0.930 |
| ACYP1 | 4.3e-01 | 0.930 |
| N4BP2 | 4.3e-01 | 0.930 |
| TROAP | 4.3e-01 | 0.930 |
| PRKAG1 | 4.3e-01 | 0.930 |
| THOC5 | 4.3e-01 | 0.930 |
| ZBTB25 | 4.3e-01 | 0.930 |
| CTDSP1 | 4.3e-01 | 0.930 |
| TOPBP1 | 4.3e-01 | 0.930 |
| TMEM231 | 4.3e-01 | 0.930 |
| CCDC184 | 4.3e-01 | 0.930 |
| TAF13 | 4.3e-01 | 0.930 |
| BMPR1B | 4.3e-01 | 0.930 |
| FBXW5 | 4.3e-01 | 0.930 |
| NKTR | 4.3e-01 | 0.930 |
| RCHY1 | 4.3e-01 | 0.930 |
| LARS | 4.3e-01 | 0.930 |
| AKAP8L | 4.3e-01 | 0.930 |
| ZFC3H1 | 4.3e-01 | 0.930 |
| ABCB7 | 4.3e-01 | 0.930 |
| TMSB10 | 4.3e-01 | 0.930 |
| PRAG1 | 4.3e-01 | 0.930 |
| PCDH19 | 4.3e-01 | 0.930 |
| LYRM5 | 4.3e-01 | 0.930 |
| LACTB2 | 4.3e-01 | 0.930 |
| FADS2 | 4.3e-01 | 0.930 |
| LTBP4 | 4.3e-01 | 0.930 |
| ZNF420 | 4.3e-01 | 0.930 |
| SFT2D3 | 4.3e-01 | 0.930 |
| COPS5 | 4.3e-01 | 0.930 |
| SNHG14 | 4.3e-01 | 0.930 |
| TFE3 | 4.3e-01 | 0.930 |
| RP9 | 4.3e-01 | 0.930 |
| NAA25 | 4.3e-01 | 0.930 |
| GSTK1 | 4.3e-01 | 0.930 |
| COPG2 | 4.3e-01 | 0.930 |
| CIAPIN1 | 4.3e-01 | 0.930 |
| UGGT1 | 4.3e-01 | 0.930 |
| FAM96A | 4.3e-01 | 0.930 |
| SLC35D2 | 4.3e-01 | 0.930 |
| TAZ | 4.3e-01 | 0.930 |
| SPTAN1 | 4.3e-01 | 0.930 |
| CERS5 | 4.3e-01 | 0.930 |
| ANAPC13 | 4.3e-01 | 0.930 |
| HSPD1 | 4.3e-01 | 0.930 |
| RTN4 | 4.3e-01 | 0.930 |
| MTIF3 | 4.3e-01 | 0.930 |
| CTC-539A10.4 | 4.3e-01 | 0.930 |
| PRKAA2 | 4.3e-01 | 0.930 |
| TBPL1 | 4.3e-01 | 0.930 |
| TPM1 | 4.3e-01 | 0.930 |
| SETD9 | 4.3e-01 | 0.930 |
| PRDX5 | 4.3e-01 | 0.930 |
| LIPA | 4.3e-01 | 0.930 |
| SSBP3 | 4.3e-01 | 0.930 |
| TUBGCP3 | 4.3e-01 | 0.930 |
| KBTBD6 | 4.3e-01 | 0.930 |
| KBTBD7 | 4.3e-01 | 0.930 |
| FDXR | 4.3e-01 | 0.930 |
| CYB5D2 | 4.3e-01 | 0.930 |
| RP3-329A5.4 | 4.3e-01 | 0.930 |
| FAM63B | 4.3e-01 | 0.930 |
| STRAP | 4.3e-01 | 0.930 |
| CRYBB2 | 4.3e-01 | 0.930 |
| ADD1 | 4.3e-01 | 0.930 |
| THEM6 | 4.3e-01 | 0.930 |
| CASP7 | 4.3e-01 | 0.930 |
| TMEM141 | 4.4e-01 | 0.930 |
| SRPK2 | 4.4e-01 | 0.930 |
| MED30 | 4.4e-01 | 0.930 |
| ELP6 | 4.4e-01 | 0.930 |
| UBE2B | 4.4e-01 | 0.930 |
| PPP1R16A | 4.4e-01 | 0.930 |
| PHTF1 | 4.4e-01 | 0.930 |
| CUL5 | 4.4e-01 | 0.930 |
| SPECC1L | 4.4e-01 | 0.930 |
| TRO | 4.4e-01 | 0.930 |
| SEPW1 | 4.4e-01 | 0.930 |
| PSMB4 | 4.4e-01 | 0.930 |
| JAK1 | 4.4e-01 | 0.930 |
| BPHL | 4.4e-01 | 0.930 |
| LSM6 | 4.4e-01 | 0.930 |
| ATF4 | 4.4e-01 | 0.930 |
| IGF1R | 4.4e-01 | 0.930 |
| MGAT2 | 4.4e-01 | 0.930 |
| ERLIN2 | 4.4e-01 | 0.930 |
| MAGED2 | 4.4e-01 | 0.930 |
| AP3S1 | 4.4e-01 | 0.930 |
| PRRC2B | 4.4e-01 | 0.930 |
| MTMR14 | 4.4e-01 | 0.930 |
| TTC28 | 4.4e-01 | 0.930 |
| DENR | 4.4e-01 | 0.930 |
| ABCE1 | 4.4e-01 | 0.930 |
| RAB39B | 4.4e-01 | 0.930 |
| DRAXIN | 4.4e-01 | 0.930 |
| NOC3L | 4.4e-01 | 0.930 |
| PODXL2 | 4.4e-01 | 0.930 |
| TMEM50A | 4.4e-01 | 0.930 |
| BCKDK | 4.4e-01 | 0.930 |
| SIMC1 | 4.4e-01 | 0.930 |
| C7orf50 | 4.4e-01 | 0.930 |
| CLUH | 4.4e-01 | 0.930 |
| PRMT2 | 4.4e-01 | 0.930 |
| RAB2A | 4.4e-01 | 0.930 |
| C9orf142 | 4.4e-01 | 0.930 |
| AURKA | 4.4e-01 | 0.930 |
| KRT8 | 4.4e-01 | 0.930 |
| TRDMT1 | 4.4e-01 | 0.930 |
| KATNBL1 | 4.4e-01 | 0.930 |
| ASB6 | 4.4e-01 | 0.930 |
| TSPAN17 | 4.4e-01 | 0.930 |
| SLC35E1 | 4.4e-01 | 0.930 |
| CRNDE | 4.4e-01 | 0.930 |
| ZNF260 | 4.4e-01 | 0.930 |
| NSD2 | 4.4e-01 | 0.930 |
| VPS35 | 4.4e-01 | 0.930 |
| SLC2A8 | 4.4e-01 | 0.930 |
| ANXA1 | 4.4e-01 | 0.930 |
| ZNF322 | 4.4e-01 | 0.930 |
| PAK1 | 4.4e-01 | 0.930 |
| ACTR3B | 4.4e-01 | 0.930 |
| ZNF710 | 4.4e-01 | 0.930 |
| THRA | 4.4e-01 | 0.930 |
| KMT5B | 4.4e-01 | 0.930 |
| MYO1C | 4.4e-01 | 0.930 |
| MRPL2 | 4.4e-01 | 0.930 |
| SRRM1 | 4.4e-01 | 0.930 |
| PMPCB | 4.4e-01 | 0.930 |
| EIF1AX | 4.4e-01 | 0.930 |
| ARNT | 4.4e-01 | 0.930 |
| ERBIN | 4.4e-01 | 0.930 |
| RBM22 | 4.4e-01 | 0.930 |
| FOCAD | 4.4e-01 | 0.930 |
| ZMYM5 | 4.4e-01 | 0.930 |
| HPCAL1 | 4.4e-01 | 0.930 |
| SBF1 | 4.4e-01 | 0.930 |
| RWDD2B | 4.4e-01 | 0.930 |
| UBE3A | 4.4e-01 | 0.930 |
| GADD45A | 4.4e-01 | 0.930 |
| ZCRB1 | 4.4e-01 | 0.930 |
| IDI1 | 4.4e-01 | 0.930 |
| TTC27 | 4.4e-01 | 0.930 |
| LIMK1 | 4.4e-01 | 0.930 |
| MAP2K3 | 4.4e-01 | 0.930 |
| ESCO1 | 4.4e-01 | 0.930 |
| DIAPH3 | 4.4e-01 | 0.930 |
| SBDS | 4.4e-01 | 0.930 |
| OPN3 | 4.4e-01 | 0.930 |
| RPRD1B | 4.4e-01 | 0.930 |
| MRPS7 | 4.4e-01 | 0.930 |
| PNN | 4.4e-01 | 0.930 |
| SERPING1 | 4.4e-01 | 0.930 |
| COMMD7 | 4.4e-01 | 0.930 |
| TSPYL2 | 4.5e-01 | 0.930 |
| TIMM22 | 4.5e-01 | 0.930 |
| SART1 | 4.5e-01 | 0.930 |
| RPL15 | 4.5e-01 | 0.930 |
| RP11-111M22.3 | 4.5e-01 | 0.930 |
| WBSCR16 | 4.5e-01 | 0.930 |
| ATPAF2 | 4.5e-01 | 0.930 |
| AGK | 4.5e-01 | 0.930 |
| BET1L | 4.5e-01 | 0.930 |
| ARHGEF9 | 4.5e-01 | 0.930 |
| PPP1R35 | 4.5e-01 | 0.930 |
| ROCK2 | 4.5e-01 | 0.930 |
| SEMA5B | 4.5e-01 | 0.930 |
| MUM1 | 4.5e-01 | 0.930 |
| PSKH1 | 4.5e-01 | 0.930 |
| RP11-15H20.6 | 4.5e-01 | 0.930 |
| PLEKHG4B | 4.5e-01 | 0.930 |
| SIRT6 | 4.5e-01 | 0.930 |
| TBC1D15 | 4.5e-01 | 0.930 |
| AEN | 4.5e-01 | 0.930 |
| BCAP29 | 4.5e-01 | 0.930 |
| CDC42SE1 | 4.5e-01 | 0.930 |
| DHX40 | 4.5e-01 | 0.930 |
| XPC | 4.5e-01 | 0.930 |
| YPEL1 | 4.5e-01 | 0.930 |
| PPP2CB | 4.5e-01 | 0.930 |
| TUBE1 | 4.5e-01 | 0.930 |
| JOSD2 | 4.5e-01 | 0.930 |
| ZNF561 | 4.5e-01 | 0.930 |
| CERS2 | 4.5e-01 | 0.930 |
| ZNF771 | 4.5e-01 | 0.930 |
| PRPF39 | 4.5e-01 | 0.930 |
| PCBP4 | 4.5e-01 | 0.930 |
| CASD1 | 4.5e-01 | 0.930 |
| EIF3D | 4.5e-01 | 0.930 |
| CYP2R1 | 4.5e-01 | 0.930 |
| UNKL | 4.5e-01 | 0.930 |
| NUDT1 | 4.5e-01 | 0.930 |
| PHOSPHO2 | 4.5e-01 | 0.930 |
| SESN2 | 4.5e-01 | 0.930 |
| BAD | 4.5e-01 | 0.930 |
| AKTIP | 4.5e-01 | 0.930 |
| ATG4B | 4.5e-01 | 0.930 |
| ACYP2 | 4.5e-01 | 0.930 |
| ORC4 | 4.5e-01 | 0.940 |
| MAK16 | 4.5e-01 | 0.940 |
| SAYSD1 | 4.5e-01 | 0.940 |
| MFGE8 | 4.5e-01 | 0.940 |
| ATP6AP1 | 4.5e-01 | 0.940 |
| SERGEF | 4.5e-01 | 0.940 |
| NOL12 | 4.5e-01 | 0.940 |
| COG2 | 4.5e-01 | 0.940 |
| ITPK1 | 4.5e-01 | 0.940 |
| ZDHHC6 | 4.5e-01 | 0.940 |
| SP3 | 4.5e-01 | 0.940 |
| MPLKIP | 4.5e-01 | 0.940 |
| PPP1R8 | 4.5e-01 | 0.940 |
| TMEM263 | 4.5e-01 | 0.940 |
| AMZ2 | 4.5e-01 | 0.940 |
| PIP5K2B | 4.5e-01 | 0.940 |
| RP5-1129J21.3 | 4.5e-01 | 0.940 |
| HMGB1 | 4.5e-01 | 0.940 |
| NTRK2 | 4.5e-01 | 0.940 |
| SV2A | 4.5e-01 | 0.940 |
| PDE12 | 4.5e-01 | 0.940 |
| DYNC1LI2 | 4.5e-01 | 0.940 |
| FAM181A | 4.5e-01 | 0.940 |
| TTYH3 | 4.5e-01 | 0.940 |
| CAMSAP2 | 4.5e-01 | 0.940 |
| AKR1B1 | 4.5e-01 | 0.940 |
| PIK3C2A | 4.5e-01 | 0.940 |
| NUCB1 | 4.5e-01 | 0.940 |
| MOB2 | 4.6e-01 | 0.940 |
| GSTP1 | 4.6e-01 | 0.940 |
| UBE2D2 | 4.6e-01 | 0.940 |
| ZNF428 | 4.6e-01 | 0.940 |
| MRPL36 | 4.6e-01 | 0.940 |
| MIIP | 4.6e-01 | 0.940 |
| GLO1 | 4.6e-01 | 0.940 |
| TAMM41 | 4.6e-01 | 0.940 |
| BAIAP2-AS1 | 4.6e-01 | 0.940 |
| TMEM245 | 4.6e-01 | 0.940 |
| STAU2 | 4.6e-01 | 0.940 |
| ACOT7 | 4.6e-01 | 0.940 |
| COQ7 | 4.6e-01 | 0.940 |
| RP11-186B7.4 | 4.6e-01 | 0.940 |
| TMEM68 | 4.6e-01 | 0.940 |
| ZBTB18 | 4.6e-01 | 0.940 |
| CRTC3 | 4.6e-01 | 0.940 |
| AKIP1 | 4.6e-01 | 0.940 |
| SPIN2B | 4.6e-01 | 0.940 |
| TOX | 4.6e-01 | 0.940 |
| ATG13 | 4.6e-01 | 0.940 |
| NCOA1 | 4.6e-01 | 0.940 |
| ATPAF1 | 4.6e-01 | 0.940 |
| CDCA2 | 4.6e-01 | 0.940 |
| RRP8 | 4.6e-01 | 0.940 |
| ATP2C1 | 4.6e-01 | 0.940 |
| ZSWIM6 | 4.6e-01 | 0.940 |
| NR2F1 | 4.6e-01 | 0.940 |
| TMEM67 | 4.6e-01 | 0.940 |
| PTGES2 | 4.6e-01 | 0.940 |
| CRIPT | 4.6e-01 | 0.940 |
| PPID | 4.6e-01 | 0.940 |
| SLC16A14 | 4.6e-01 | 0.940 |
| ELOB | 4.6e-01 | 0.940 |
| DCTN4 | 4.6e-01 | 0.940 |
| PCCA | 4.6e-01 | 0.940 |
| ZC3H7A | 4.6e-01 | 0.940 |
| SOCS7 | 4.6e-01 | 0.940 |
| APPBP2 | 4.6e-01 | 0.940 |
| STXBP1 | 4.6e-01 | 0.940 |
| SHC1 | 4.6e-01 | 0.940 |
| DMPK | 4.6e-01 | 0.940 |
| CMC1 | 4.6e-01 | 0.940 |
| TCTN3 | 4.6e-01 | 0.940 |
| CCND2 | 4.6e-01 | 0.940 |
| GMFB | 4.6e-01 | 0.940 |
| PUDP | 4.6e-01 | 0.940 |
| CXXC5 | 4.6e-01 | 0.940 |
| RXRA | 4.6e-01 | 0.940 |
| BRE | 4.6e-01 | 0.940 |
| VMA21 | 4.6e-01 | 0.940 |
| POMP | 4.6e-01 | 0.940 |
| ATF7IP2 | 4.6e-01 | 0.940 |
| SUB1 | 4.6e-01 | 0.940 |
| PPIL3 | 4.6e-01 | 0.940 |
| KRAS | 4.6e-01 | 0.940 |
| RNF44 | 4.7e-01 | 0.940 |
| HMGCS1 | 4.7e-01 | 0.940 |
| RRAGD | 4.7e-01 | 0.940 |
| DNM2 | 4.7e-01 | 0.940 |
| CIB1 | 4.7e-01 | 0.940 |
| PRMT8 | 4.7e-01 | 0.940 |
| MORN4 | 4.7e-01 | 0.940 |
| NKIRAS2 | 4.7e-01 | 0.940 |
| HNRNPH1 | 4.7e-01 | 0.940 |
| GPS1 | 4.7e-01 | 0.940 |
| STARD4 | 4.7e-01 | 0.940 |
| CTGF | 4.7e-01 | 0.940 |
| ACAP3 | 4.7e-01 | 0.940 |
| DCAF15 | 4.7e-01 | 0.940 |
| C1orf159 | 4.7e-01 | 0.940 |
| MRPL9 | 4.7e-01 | 0.940 |
| CASKIN2 | 4.7e-01 | 0.940 |
| PDE1B | 4.7e-01 | 0.940 |
| GNA13 | 4.7e-01 | 0.940 |
| PREB | 4.7e-01 | 0.940 |
| PRKX | 4.7e-01 | 0.940 |
| GABARAPL2 | 4.7e-01 | 0.940 |
| COPG1 | 4.7e-01 | 0.940 |
| ZNF213 | 4.7e-01 | 0.940 |
| IFT122 | 4.7e-01 | 0.940 |
| SON | 4.7e-01 | 0.940 |
| ASNSD1 | 4.7e-01 | 0.940 |
| SLC52A2 | 4.7e-01 | 0.940 |
| STX6 | 4.7e-01 | 0.940 |
| AKIRIN1 | 4.7e-01 | 0.940 |
| ITGB8 | 4.7e-01 | 0.940 |
| ZBTB48 | 4.7e-01 | 0.940 |
| AIDA | 4.7e-01 | 0.940 |
| KLHDC4 | 4.7e-01 | 0.940 |
| NUPL2 | 4.7e-01 | 0.940 |
| GFRA1 | 4.7e-01 | 0.940 |
| POLR2D | 4.7e-01 | 0.940 |
| GATB | 4.7e-01 | 0.940 |
| C11orf57 | 4.7e-01 | 0.940 |
| VCP | 4.7e-01 | 0.940 |
| APBB2 | 4.7e-01 | 0.940 |
| NSD3 | 4.7e-01 | 0.950 |
| SREBF2 | 4.7e-01 | 0.950 |
| ZNF212 | 4.7e-01 | 0.950 |
| CADM1 | 4.7e-01 | 0.950 |
| RNF40 | 4.7e-01 | 0.950 |
| RSL24D1 | 4.7e-01 | 0.950 |
| LRRFIP2 | 4.7e-01 | 0.950 |
| FAF2 | 4.7e-01 | 0.950 |
| NDUFB11 | 4.7e-01 | 0.950 |
| CDC40 | 4.7e-01 | 0.950 |
| SNX13 | 4.7e-01 | 0.950 |
| CDADC1 | 4.7e-01 | 0.950 |
| CD200 | 4.7e-01 | 0.950 |
| AGPS | 4.7e-01 | 0.950 |
| MED16 | 4.7e-01 | 0.950 |
| POLR3F | 4.7e-01 | 0.950 |
| EEF2KMT | 4.7e-01 | 0.950 |
| MARS | 4.7e-01 | 0.950 |
| VCPKMT | 4.7e-01 | 0.950 |
| PRMT6 | 4.7e-01 | 0.950 |
| PIK3R3 | 4.7e-01 | 0.950 |
| ELOA | 4.7e-01 | 0.950 |
| SLC22A23 | 4.7e-01 | 0.950 |
| TM7SF2 | 4.7e-01 | 0.950 |
| CAMSAP1 | 4.7e-01 | 0.950 |
| ZNF598 | 4.7e-01 | 0.950 |
| TBC1D12 | 4.7e-01 | 0.950 |
| PLD3 | 4.7e-01 | 0.950 |
| TMEM169 | 4.7e-01 | 0.950 |
| DDX39A | 4.7e-01 | 0.950 |
| ETHE1 | 4.8e-01 | 0.950 |
| PARD3B | 4.8e-01 | 0.950 |
| KMT2C | 4.8e-01 | 0.950 |
| SCO1 | 4.8e-01 | 0.950 |
| SMYD3 | 4.8e-01 | 0.950 |
| RBM23 | 4.8e-01 | 0.950 |
| NEDD8 | 4.8e-01 | 0.950 |
| LMNB1 | 4.8e-01 | 0.950 |
| ETNK1 | 4.8e-01 | 0.950 |
| RPL26 | 4.8e-01 | 0.950 |
| MRGBP | 4.8e-01 | 0.950 |
| MAZ | 4.8e-01 | 0.950 |
| BUD31 | 4.8e-01 | 0.950 |
| ZBED4 | 4.8e-01 | 0.950 |
| PSAP | 4.8e-01 | 0.950 |
| MAP2K5 | 4.8e-01 | 0.950 |
| ATXN2L | 4.8e-01 | 0.950 |
| KLHL12 | 4.8e-01 | 0.950 |
| FTSJ3 | 4.8e-01 | 0.950 |
| IRF2BPL | 4.8e-01 | 0.950 |
| TMEM184C | 4.8e-01 | 0.950 |
| GNG12 | 4.8e-01 | 0.950 |
| DEPDC1B | 4.8e-01 | 0.950 |
| COX10 | 4.8e-01 | 0.950 |
| SNRNP48 | 4.8e-01 | 0.950 |
| DYRK1A | 4.8e-01 | 0.950 |
| WWOX | 4.8e-01 | 0.950 |
| IQCB1 | 4.8e-01 | 0.950 |
| ZNF106 | 4.8e-01 | 0.950 |
| CREB1 | 4.8e-01 | 0.950 |
| DCAF11 | 4.8e-01 | 0.950 |
| RPS13 | 4.8e-01 | 0.950 |
| VPS33B | 4.8e-01 | 0.950 |
| M6PR | 4.8e-01 | 0.950 |
| ZHX1 | 4.8e-01 | 0.950 |
| KDM5B | 4.8e-01 | 0.950 |
| GPANK1 | 4.8e-01 | 0.950 |
| CLP1 | 4.8e-01 | 0.950 |
| SPSB3 | 4.8e-01 | 0.950 |
| FGFR3 | 4.8e-01 | 0.950 |
| WDR82 | 4.8e-01 | 0.950 |
| WDR41 | 4.8e-01 | 0.950 |
| ZBED3 | 4.8e-01 | 0.950 |
| IDH3B | 4.8e-01 | 0.950 |
| MAP4 | 4.8e-01 | 0.950 |
| NIT2 | 4.8e-01 | 0.950 |
| NEFM | 4.8e-01 | 0.950 |
| LPAR2 | 4.8e-01 | 0.950 |
| SHMT2 | 4.8e-01 | 0.950 |
| CDK8 | 4.8e-01 | 0.950 |
| COQ6 | 4.8e-01 | 0.950 |
| SEPT2 | 4.8e-01 | 0.950 |
| ST5 | 4.8e-01 | 0.950 |
| RNF146 | 4.8e-01 | 0.950 |
| VPS4A | 4.8e-01 | 0.950 |
| HIBCH | 4.8e-01 | 0.950 |
| HMGCR | 4.8e-01 | 0.950 |
| TMEM189 | 4.8e-01 | 0.950 |
| C16orf91 | 4.8e-01 | 0.950 |
| PNKD | 4.8e-01 | 0.950 |
| ATF2 | 4.8e-01 | 0.950 |
| HNRNPL | 4.8e-01 | 0.950 |
| COG3 | 4.8e-01 | 0.950 |
| GNG11 | 4.8e-01 | 0.950 |
| CES2 | 4.8e-01 | 0.950 |
| ING1 | 4.8e-01 | 0.950 |
| RB1 | 4.8e-01 | 0.950 |
| TMEM14C | 4.8e-01 | 0.950 |
| ALDH4A1 | 4.8e-01 | 0.950 |
| HGH1 | 4.8e-01 | 0.950 |
| SERPINE2 | 4.8e-01 | 0.950 |
| FAM210B | 4.8e-01 | 0.950 |
| PHF20 | 4.8e-01 | 0.950 |
| LAMA1 | 4.8e-01 | 0.950 |
| UBR7 | 4.9e-01 | 0.950 |
| PNRC2 | 4.9e-01 | 0.950 |
| NBN | 4.9e-01 | 0.950 |
| DZIP1 | 4.9e-01 | 0.950 |
| RPS27L | 4.9e-01 | 0.950 |
| C21orf62 | 4.9e-01 | 0.950 |
| CDK5 | 4.9e-01 | 0.950 |
| OPTN | 4.9e-01 | 0.950 |
| CLINT1 | 4.9e-01 | 0.950 |
| TYW1 | 4.9e-01 | 0.950 |
| PSMB5 | 4.9e-01 | 0.950 |
| FH | 4.9e-01 | 0.950 |
| FAM104B | 4.9e-01 | 0.950 |
| UBA5 | 4.9e-01 | 0.950 |
| HNRNPLL | 4.9e-01 | 0.950 |
| MAFB | 4.9e-01 | 0.950 |
| REPIN1 | 4.9e-01 | 0.950 |
| SNX5 | 4.9e-01 | 0.950 |
| MCPH1 | 4.9e-01 | 0.950 |
| TRIP6 | 4.9e-01 | 0.950 |
| CORO1A | 4.9e-01 | 0.950 |
| WDR92 | 4.9e-01 | 0.950 |
| MAML2 | 4.9e-01 | 0.950 |
| RBX1 | 4.9e-01 | 0.950 |
| NPAS3 | 4.9e-01 | 0.950 |
| PHLDA3 | 4.9e-01 | 0.950 |
| BACE1 | 4.9e-01 | 0.950 |
| RBM18 | 4.9e-01 | 0.950 |
| PELO | 4.9e-01 | 0.950 |
| EXOSC7 | 4.9e-01 | 0.950 |
| MXI1 | 4.9e-01 | 0.950 |
| KDM2B | 4.9e-01 | 0.950 |
| UQCRQ | 4.9e-01 | 0.950 |
| MRPS33 | 4.9e-01 | 0.950 |
| SEC63 | 4.9e-01 | 0.950 |
| RPP25L | 4.9e-01 | 0.950 |
| FAM136A | 4.9e-01 | 0.950 |
| CRYZ | 4.9e-01 | 0.950 |
| CETN3 | 4.9e-01 | 0.950 |
| HMG20A | 4.9e-01 | 0.950 |
| BICD2 | 4.9e-01 | 0.950 |
| RP11-357C3.3 | 4.9e-01 | 0.950 |
| BCKDHB | 4.9e-01 | 0.950 |
| CYB5R1 | 4.9e-01 | 0.950 |
| BICD1 | 4.9e-01 | 0.950 |
| TMEM115 | 4.9e-01 | 0.950 |
| ANKZF1 | 4.9e-01 | 0.950 |
| PHACTR4 | 4.9e-01 | 0.950 |
| KPNA6 | 4.9e-01 | 0.950 |
| C15orf40 | 4.9e-01 | 0.950 |
| ULK3 | 4.9e-01 | 0.950 |
| PARK7 | 4.9e-01 | 0.950 |
| CLSTN1 | 4.9e-01 | 0.950 |
| HIST3H2A | 4.9e-01 | 0.950 |
| TBK1 | 4.9e-01 | 0.950 |
| ANP32B | 4.9e-01 | 0.950 |
| SWI5 | 4.9e-01 | 0.950 |
| GNB5 | 4.9e-01 | 0.950 |
| CFDP1 | 4.9e-01 | 0.950 |
| TADA3 | 4.9e-01 | 0.950 |
| ABHD5 | 4.9e-01 | 0.950 |
| ZDHHC3 | 4.9e-01 | 0.950 |
| DDHD2 | 4.9e-01 | 0.950 |
| C1orf52 | 4.9e-01 | 0.950 |
| USP5 | 4.9e-01 | 0.950 |
| APIP | 4.9e-01 | 0.950 |
| FAM229B | 4.9e-01 | 0.950 |
| CRABP1 | 4.9e-01 | 0.950 |
| MAPK8IP1 | 4.9e-01 | 0.950 |
| PARL | 4.9e-01 | 0.950 |
| SLC25A26 | 4.9e-01 | 0.950 |
| MTERF1 | 4.9e-01 | 0.950 |
| BTBD7 | 4.9e-01 | 0.950 |
| ZC2HC1A | 4.9e-01 | 0.950 |
| WTIP | 4.9e-01 | 0.950 |
| SPCS1 | 4.9e-01 | 0.950 |
| PALMD | 4.9e-01 | 0.950 |
| COQ3 | 4.9e-01 | 0.950 |
| SAFB | 4.9e-01 | 0.950 |
| RP11-32K4.2 | 4.9e-01 | 0.950 |
| PYGL | 4.9e-01 | 0.950 |
| HECTD1 | 4.9e-01 | 0.950 |
| ALG14 | 4.9e-01 | 0.950 |
| TMEM19 | 4.9e-01 | 0.950 |
| TUBGCP2 | 4.9e-01 | 0.950 |
| PARP6 | 4.9e-01 | 0.950 |
| STK3 | 4.9e-01 | 0.950 |
| THAP5 | 4.9e-01 | 0.950 |
| SEC24B | 4.9e-01 | 0.950 |
| AKR1A1 | 4.9e-01 | 0.950 |
| POLR3E | 4.9e-01 | 0.950 |
| HOMER2 | 4.9e-01 | 0.950 |
| RPL7A | 4.9e-01 | 0.950 |
| SLC39A10 | 4.9e-01 | 0.950 |
| EIF2AK4 | 4.9e-01 | 0.950 |
| SORT1 | 4.9e-01 | 0.950 |
| PEX16 | 4.9e-01 | 0.950 |
| KLHL8 | 5.0e-01 | 0.950 |
| AK4 | 5.0e-01 | 0.950 |
| SEM1 | 5.0e-01 | 0.950 |
| SRD5A3 | 5.0e-01 | 0.950 |
| MRPL1 | 5.0e-01 | 0.950 |
| NOP14 | 5.0e-01 | 0.950 |
| ID1 | 5.0e-01 | 0.950 |
| DDX52 | 5.0e-01 | 0.950 |
| PLEKHJ1 | 5.0e-01 | 0.950 |
| SLF1 | 5.0e-01 | 0.950 |
| SLC35B1 | 5.0e-01 | 0.950 |
| RALGDS | 5.0e-01 | 0.950 |
| CTB-175P5.4 | 5.0e-01 | 0.950 |
| TMA16 | 5.0e-01 | 0.950 |
| KMT2B | 5.0e-01 | 0.950 |
| TMEM14B | 5.0e-01 | 0.950 |
| SOWAHC | 5.0e-01 | 0.950 |
| EPB41L4A-AS1 | 5.0e-01 | 0.950 |
| RAB28 | 5.0e-01 | 0.950 |
| NCBP1 | 5.0e-01 | 0.950 |
| CTDSP2 | 5.0e-01 | 0.950 |
| ECH1 | 5.0e-01 | 0.950 |
| RPL4 | 5.0e-01 | 0.950 |
| ZMAT3 | 5.0e-01 | 0.950 |
| PSMC6 | 5.0e-01 | 0.950 |
| SYAP1 | 5.0e-01 | 0.950 |
| OCIAD1 | 5.0e-01 | 0.950 |
| KRIT1 | 5.0e-01 | 0.950 |
| METTL2B | 5.0e-01 | 0.950 |
| RNFT1 | 5.0e-01 | 0.950 |
| SHF | 5.0e-01 | 0.950 |
| SPOP | 5.0e-01 | 0.950 |
| FBN2 | 5.0e-01 | 0.950 |
| EMP2 | 5.0e-01 | 0.950 |
| MLYCD | 5.0e-01 | 0.950 |
| HDHD3 | 5.0e-01 | 0.950 |
| NIPSNAP1 | 5.0e-01 | 0.950 |
| TBCD | 5.0e-01 | 0.950 |
| EIF2B3 | 5.0e-01 | 0.950 |
| PDHB | 5.0e-01 | 0.950 |
| URI1 | 5.0e-01 | 0.950 |
| RNASEH1 | 5.0e-01 | 0.950 |
| PLAGL2 | 5.0e-01 | 0.950 |
| RNMTL1 | 5.0e-01 | 0.950 |
| CRISPLD1 | 5.0e-01 | 0.950 |
| PARG | 5.0e-01 | 0.950 |
| UBE2V2 | 5.0e-01 | 0.950 |
| OIP5-AS1 | 5.0e-01 | 0.950 |
| THOC7 | 5.0e-01 | 0.950 |
| TJAP1 | 5.0e-01 | 0.950 |
| RPS6KA6 | 5.0e-01 | 0.950 |
| RBBP9 | 5.0e-01 | 0.950 |
| PHF10 | 5.0e-01 | 0.950 |
| ZNF317 | 5.0e-01 | 0.950 |
| EDF1 | 5.0e-01 | 0.950 |
| OSGIN1 | 5.0e-01 | 0.950 |
| BET1 | 5.0e-01 | 0.950 |
| CTDNEP1 | 5.0e-01 | 0.950 |
| SDCBP | 5.0e-01 | 0.950 |
| RPL27 | 5.0e-01 | 0.950 |
| COIL | 5.0e-01 | 0.950 |
| NACC1 | 5.0e-01 | 0.950 |
| SMC3 | 5.0e-01 | 0.950 |
| UBN1 | 5.0e-01 | 0.950 |
| C18orf21 | 5.0e-01 | 0.950 |
| ACAT1 | 5.0e-01 | 0.950 |
| MT-ND2 | 5.0e-01 | 0.950 |
| ARL4A | 5.0e-01 | 0.950 |
| ALDH7A1 | 5.0e-01 | 0.950 |
| GMCL1 | 5.0e-01 | 0.950 |
| EPM2AIP1 | 5.0e-01 | 0.950 |
| PAN3 | 5.0e-01 | 0.950 |
| FUNDC2 | 5.0e-01 | 0.950 |
| PVR | 5.0e-01 | 0.950 |
| COQ2 | 5.0e-01 | 0.950 |
| ACBD6 | 5.0e-01 | 0.950 |
| ARID3A | 5.0e-01 | 0.950 |
| ARCN1 | 5.0e-01 | 0.950 |
| UBE2D4 | 5.0e-01 | 0.950 |
| HABP4 | 5.0e-01 | 0.950 |
| DNAJC19 | 5.0e-01 | 0.950 |
| ANKRD40 | 5.0e-01 | 0.950 |
| SNX14 | 5.0e-01 | 0.950 |
| TFB1M | 5.1e-01 | 0.950 |
| OGG1 | 5.1e-01 | 0.950 |
| KIAA0141 | 5.1e-01 | 0.950 |
| ARID4A | 5.1e-01 | 0.950 |
| ALDH16A1 | 5.1e-01 | 0.950 |
| ZFAND3 | 5.1e-01 | 0.950 |
| C1orf198 | 5.1e-01 | 0.950 |
| SDAD1 | 5.1e-01 | 0.950 |
| GPR89A | 5.1e-01 | 0.950 |
| HCFC1 | 5.1e-01 | 0.950 |
| PTBP3 | 5.1e-01 | 0.950 |
| DYNC1H1 | 5.1e-01 | 0.950 |
| RSRC2 | 5.1e-01 | 0.950 |
| ABI2 | 5.1e-01 | 0.950 |
| UFC1 | 5.1e-01 | 0.950 |
| TMTC4 | 5.1e-01 | 0.950 |
| ECI1 | 5.1e-01 | 0.950 |
| UBE4A | 5.1e-01 | 0.950 |
| CRY1 | 5.1e-01 | 0.950 |
| HOOK2 | 5.1e-01 | 0.950 |
| LYPLAL1 | 5.1e-01 | 0.950 |
| CXADR | 5.1e-01 | 0.950 |
| EIF2B4 | 5.1e-01 | 0.950 |
| HES6 | 5.1e-01 | 0.950 |
| SUMF1 | 5.1e-01 | 0.950 |
| MRPL16 | 5.1e-01 | 0.950 |
| NOB1 | 5.1e-01 | 0.950 |
| CDK5RAP1 | 5.1e-01 | 0.950 |
| NLRP1 | 5.1e-01 | 0.950 |
| EIF3H | 5.1e-01 | 0.950 |
| CCDC71L | 5.1e-01 | 0.950 |
| MRPL49 | 5.1e-01 | 0.950 |
| ZCCHC11 | 5.1e-01 | 0.950 |
| PFKL | 5.1e-01 | 0.950 |
| PSMB2 | 5.1e-01 | 0.950 |
| FUOM | 5.1e-01 | 0.950 |
| DHX29 | 5.1e-01 | 0.950 |
| TRAF6 | 5.1e-01 | 0.950 |
| ANXA2 | 5.1e-01 | 0.950 |
| ARFGAP3 | 5.1e-01 | 0.950 |
| LTA4H | 5.1e-01 | 0.950 |
| NDUFA3 | 5.1e-01 | 0.950 |
| RASSF8 | 5.1e-01 | 0.950 |
| ENDOV | 5.1e-01 | 0.950 |
| MMP2 | 5.1e-01 | 0.950 |
| POLR2I | 5.1e-01 | 0.950 |
| SNRPB2 | 5.1e-01 | 0.950 |
| ATP6V0A1 | 5.1e-01 | 0.950 |
| GORAB | 5.1e-01 | 0.950 |
| NDUFB2 | 5.1e-01 | 0.950 |
| POLR2J | 5.1e-01 | 0.950 |
| ZNF37A | 5.1e-01 | 0.950 |
| TSTD3 | 5.1e-01 | 0.950 |
| SOGA1 | 5.1e-01 | 0.950 |
| FOXM1 | 5.1e-01 | 0.950 |
| CNOT6L | 5.1e-01 | 0.950 |
| C4orf46 | 5.1e-01 | 0.950 |
| STK38 | 5.1e-01 | 0.950 |
| CITED2 | 5.1e-01 | 0.950 |
| TEX9 | 5.1e-01 | 0.950 |
| OXNAD1 | 5.1e-01 | 0.950 |
| GDF11 | 5.1e-01 | 0.950 |
| PSMB3 | 5.1e-01 | 0.950 |
| LRCH3 | 5.1e-01 | 0.950 |
| ASB1 | 5.1e-01 | 0.950 |
| AK6 | 5.1e-01 | 0.950 |
| NOP10 | 5.1e-01 | 0.950 |
| CTD-2119F7.2 | 5.1e-01 | 0.950 |
| TMEM120A | 5.1e-01 | 0.950 |
| SNRPD3 | 5.1e-01 | 0.950 |
| PFDN5 | 5.1e-01 | 0.950 |
| SPIRE1 | 5.1e-01 | 0.950 |
| SRF | 5.1e-01 | 0.950 |
| KRT18 | 5.1e-01 | 0.950 |
| RPS28 | 5.1e-01 | 0.950 |
| MTFMT | 5.1e-01 | 0.950 |
| MAP3K2 | 5.1e-01 | 0.950 |
| SERTAD2 | 5.1e-01 | 0.950 |
| CENPN | 5.1e-01 | 0.950 |
| PRKAR1A | 5.1e-01 | 0.950 |
| AMD1 | 5.1e-01 | 0.950 |
| SNAP47 | 5.1e-01 | 0.950 |
| GPC2 | 5.1e-01 | 0.950 |
| POMT2 | 5.1e-01 | 0.950 |
| SLC25A36 | 5.1e-01 | 0.950 |
| SULF2 | 5.1e-01 | 0.950 |
| RNASEH2B | 5.1e-01 | 0.950 |
| SSR4 | 5.1e-01 | 0.950 |
| PCM1 | 5.2e-01 | 0.950 |
| LGMN | 5.2e-01 | 0.950 |
| ID2 | 5.2e-01 | 0.950 |
| ABHD13 | 5.2e-01 | 0.950 |
| EMC8 | 5.2e-01 | 0.950 |
| TCOF1 | 5.2e-01 | 0.950 |
| TRA2B | 5.2e-01 | 0.950 |
| CCHCR1 | 5.2e-01 | 0.950 |
| NDUFA13 | 5.2e-01 | 0.950 |
| PCID2 | 5.2e-01 | 0.950 |
| NCAPG2 | 5.2e-01 | 0.950 |
| ABR | 5.2e-01 | 0.950 |
| TRIM2 | 5.2e-01 | 0.950 |
| KLHDC8B | 5.2e-01 | 0.950 |
| ANKIB1 | 5.2e-01 | 0.950 |
| ZMPSTE24 | 5.2e-01 | 0.950 |
| DHX36 | 5.2e-01 | 0.950 |
| DEGS1 | 5.2e-01 | 0.950 |
| C6orf47 | 5.2e-01 | 0.950 |
| NADK | 5.2e-01 | 0.950 |
| PDHA1 | 5.2e-01 | 0.950 |
| GALNT1 | 5.2e-01 | 0.950 |
| ZNF12 | 5.2e-01 | 0.950 |
| SMOC1 | 5.2e-01 | 0.950 |
| RNF166 | 5.2e-01 | 0.950 |
| RSPRY1 | 5.2e-01 | 0.950 |
| CTNNB1 | 5.2e-01 | 0.950 |
| TPP2 | 5.2e-01 | 0.950 |
| MCFD2 | 5.2e-01 | 0.950 |
| NUP107 | 5.2e-01 | 0.950 |
| NSD1 | 5.2e-01 | 0.950 |
| CDC5L | 5.2e-01 | 0.950 |
| DYRK2 | 5.2e-01 | 0.950 |
| CNN2 | 5.2e-01 | 0.950 |
| MRPL44 | 5.2e-01 | 0.950 |
| TATDN1 | 5.2e-01 | 0.950 |
| AATF | 5.2e-01 | 0.950 |
| TOP2B | 5.2e-01 | 0.950 |
| MARCH9 | 5.2e-01 | 0.950 |
| B4GALT2 | 5.2e-01 | 0.950 |
| C16orf88 | 5.2e-01 | 0.950 |
| CACNB3 | 5.2e-01 | 0.950 |
| TRMU | 5.2e-01 | 0.950 |
| PSAT1 | 5.2e-01 | 0.950 |
| STOML1 | 5.2e-01 | 0.950 |
| ACVR1B | 5.2e-01 | 0.950 |
| PIAS4 | 5.2e-01 | 0.950 |
| TMEM198 | 5.2e-01 | 0.950 |
| RAP2A | 5.2e-01 | 0.950 |
| PYCR1 | 5.2e-01 | 0.950 |
| SPOCK1 | 5.2e-01 | 0.950 |
| PDHX | 5.2e-01 | 0.950 |
| DNAJC1 | 5.2e-01 | 0.950 |
| SECISBP2 | 5.2e-01 | 0.950 |
| ENTPD6 | 5.2e-01 | 0.950 |
| UBALD1 | 5.2e-01 | 0.950 |
| TMEM242 | 5.2e-01 | 0.950 |
| ING4 | 5.2e-01 | 0.950 |
| SMARCD1 | 5.2e-01 | 0.950 |
| ARHGDIA | 5.2e-01 | 0.950 |
| PRIM1 | 5.2e-01 | 0.950 |
| LAS1L | 5.2e-01 | 0.950 |
| CAPN1 | 5.2e-01 | 0.950 |
| FAM192A | 5.2e-01 | 0.950 |
| TMEM25 | 5.2e-01 | 0.950 |
| CDC27 | 5.2e-01 | 0.950 |
| C6orf48 | 5.2e-01 | 0.950 |
| DNAJB11 | 5.2e-01 | 0.950 |
| PIK3C2B | 5.2e-01 | 0.950 |
| SMAD5 | 5.2e-01 | 0.950 |
| MED4 | 5.2e-01 | 0.950 |
| RRBP1 | 5.2e-01 | 0.950 |
| DPY30 | 5.2e-01 | 0.950 |
| HNRNPDL | 5.2e-01 | 0.950 |
| TSNAX | 5.2e-01 | 0.950 |
| MOGS | 5.2e-01 | 0.950 |
| CLIC1 | 5.2e-01 | 0.950 |
| CAMLG | 5.2e-01 | 0.950 |
| C8orf82 | 5.2e-01 | 0.950 |
| LPCAT1 | 5.2e-01 | 0.950 |
| C12orf48 | 5.3e-01 | 0.950 |
| ZNF544 | 5.3e-01 | 0.950 |
| DSCR3 | 5.3e-01 | 0.950 |
| RBM48 | 5.3e-01 | 0.950 |
| THOC2 | 5.3e-01 | 0.950 |
| GNG10 | 5.3e-01 | 0.950 |
| NAT10 | 5.3e-01 | 0.950 |
| FSD1 | 5.3e-01 | 0.950 |
| SYT1 | 5.3e-01 | 0.950 |
| TAF1 | 5.3e-01 | 0.950 |
| SDC1 | 5.3e-01 | 0.950 |
| FAXC | 5.3e-01 | 0.950 |
| PARVA | 5.3e-01 | 0.950 |
| CHD1L | 5.3e-01 | 0.950 |
| LOXL1 | 5.3e-01 | 0.950 |
| ACAA2 | 5.3e-01 | 0.950 |
| TEFM | 5.3e-01 | 0.950 |
| RGS12 | 5.3e-01 | 0.950 |
| NDST1 | 5.3e-01 | 0.950 |
| TIMM10B | 5.3e-01 | 0.950 |
| CCDC160 | 5.3e-01 | 0.950 |
| FIGN | 5.3e-01 | 0.950 |
| TFCP2 | 5.3e-01 | 0.950 |
| C1orf112 | 5.3e-01 | 0.950 |
| LINC00467 | 5.3e-01 | 0.950 |
| CTR9 | 5.3e-01 | 0.950 |
| CDCA3 | 5.3e-01 | 0.950 |
| GMPPB | 5.3e-01 | 0.950 |
| HDLBP | 5.3e-01 | 0.950 |
| XPO7 | 5.3e-01 | 0.950 |
| ZWILCH | 5.3e-01 | 0.950 |
| TTC1 | 5.3e-01 | 0.950 |
| RPP14 | 5.3e-01 | 0.950 |
| FAM118B | 5.3e-01 | 0.950 |
| GCLM | 5.3e-01 | 0.950 |
| OXLD1 | 5.3e-01 | 0.950 |
| WBSCR22 | 5.3e-01 | 0.950 |
| ALMS1 | 5.3e-01 | 0.950 |
| FBXL5 | 5.3e-01 | 0.950 |
| ANKRD39 | 5.3e-01 | 0.950 |
| FN3KRP | 5.3e-01 | 0.950 |
| HS6ST1 | 5.3e-01 | 0.950 |
| ALDH6A1 | 5.3e-01 | 0.950 |
| GALNT16 | 5.3e-01 | 0.950 |
| MPV17 | 5.3e-01 | 0.950 |
| C18orf32 | 5.3e-01 | 0.950 |
| PSME2 | 5.3e-01 | 0.950 |
| EVA1B | 5.3e-01 | 0.950 |
| CDKN1C | 5.3e-01 | 0.950 |
| METTL16 | 5.3e-01 | 0.950 |
| C20orf96 | 5.3e-01 | 0.950 |
| RANBP2 | 5.3e-01 | 0.950 |
| FHL2 | 5.3e-01 | 0.950 |
| PLEC | 5.3e-01 | 0.950 |
| DUSP11 | 5.3e-01 | 0.950 |
| NGDN | 5.3e-01 | 0.950 |
| PACRGL | 5.3e-01 | 0.950 |
| USP44 | 5.3e-01 | 0.950 |
| RFX5 | 5.3e-01 | 0.950 |
| FAM193A | 5.3e-01 | 0.950 |
| LIX1L | 5.3e-01 | 0.950 |
| LEPROTL1 | 5.3e-01 | 0.950 |
| GNAI1 | 5.3e-01 | 0.950 |
| FOXP4 | 5.3e-01 | 0.950 |
| AFG3L2 | 5.3e-01 | 0.950 |
| UNC119 | 5.3e-01 | 0.950 |
| RPS2 | 5.3e-01 | 0.950 |
| NIF3L1 | 5.3e-01 | 0.950 |
| FDX1 | 5.3e-01 | 0.950 |
| GTF2H5 | 5.3e-01 | 0.950 |
| CEP85L | 5.3e-01 | 0.950 |
| LAMTOR1 | 5.3e-01 | 0.950 |
| PDXK | 5.3e-01 | 0.950 |
| CSNK1E | 5.3e-01 | 0.950 |
| DDX56 | 5.3e-01 | 0.950 |
| FMR1 | 5.3e-01 | 0.950 |
| CDKN2D | 5.3e-01 | 0.950 |
| KAZN | 5.3e-01 | 0.950 |
| GPRC5B | 5.3e-01 | 0.950 |
| BCL7C | 5.3e-01 | 0.950 |
| TUBA1A | 5.3e-01 | 0.950 |
| IFT74 | 5.3e-01 | 0.950 |
| KIAA1257 | 5.3e-01 | 0.950 |
| AFTPH | 5.3e-01 | 0.950 |
| HNRNPUL2 | 5.3e-01 | 0.950 |
| TMEM18 | 5.3e-01 | 0.950 |
| TIMM10 | 5.3e-01 | 0.950 |
| CDKN2AIPNL | 5.3e-01 | 0.950 |
| ZNF43 | 5.4e-01 | 0.950 |
| GDAP1L1 | 5.4e-01 | 0.950 |
| SERINC2 | 5.4e-01 | 0.950 |
| SUPT5H | 5.4e-01 | 0.950 |
| SDK2 | 5.4e-01 | 0.950 |
| VPS28 | 5.4e-01 | 0.950 |
| LSG1 | 5.4e-01 | 0.950 |
| ESRRA | 5.4e-01 | 0.950 |
| BMS1 | 5.4e-01 | 0.950 |
| ARFGAP2 | 5.4e-01 | 0.950 |
| SASS6 | 5.4e-01 | 0.950 |
| ABCF3 | 5.4e-01 | 0.950 |
| COMMD10 | 5.4e-01 | 0.950 |
| RNF20 | 5.4e-01 | 0.950 |
| UBAC2 | 5.4e-01 | 0.950 |
| RPS20 | 5.4e-01 | 0.950 |
| MN1 | 5.4e-01 | 0.950 |
| SMDT1 | 5.4e-01 | 0.950 |
| FZD2 | 5.4e-01 | 0.950 |
| COA7 | 5.4e-01 | 0.950 |
| CBX3 | 5.4e-01 | 0.950 |
| EIF3E | 5.4e-01 | 0.950 |
| DOK5 | 5.4e-01 | 0.950 |
| WHAMM | 5.4e-01 | 0.950 |
| LOXL2 | 5.4e-01 | 0.950 |
| SRRD | 5.4e-01 | 0.950 |
| TMEM161A | 5.4e-01 | 0.950 |
| C14orf37 | 5.4e-01 | 0.950 |
| AKIRIN2 | 5.4e-01 | 0.950 |
| MAP2K1 | 5.4e-01 | 0.950 |
| CDK5RAP2 | 5.4e-01 | 0.950 |
| KCNG1 | 5.4e-01 | 0.950 |
| MVB12A | 5.4e-01 | 0.950 |
| SETD2 | 5.4e-01 | 0.950 |
| ZNF623 | 5.4e-01 | 0.950 |
| CDC25B | 5.4e-01 | 0.950 |
| NECTIN1 | 5.4e-01 | 0.950 |
| COTL1 | 5.4e-01 | 0.950 |
| NDUFS6 | 5.4e-01 | 0.950 |
| CD47 | 5.4e-01 | 0.950 |
| CCDC59 | 5.4e-01 | 0.950 |
| ZCCHC10 | 5.4e-01 | 0.950 |
| ARF4 | 5.4e-01 | 0.950 |
| POLA1 | 5.4e-01 | 0.950 |
| CNTFR | 5.4e-01 | 0.950 |
| DLD | 5.4e-01 | 0.950 |
| TMEM93 | 5.4e-01 | 0.950 |
| LARP6 | 5.4e-01 | 0.950 |
| SLC19A1 | 5.4e-01 | 0.950 |
| CCDC136 | 5.4e-01 | 0.950 |
| SIGIRR | 5.4e-01 | 0.950 |
| NKD1 | 5.4e-01 | 0.950 |
| POU2F1 | 5.4e-01 | 0.950 |
| SLC9A3R1 | 5.4e-01 | 0.950 |
| NUF2 | 5.4e-01 | 0.950 |
| STAT3 | 5.4e-01 | 0.950 |
| MPHOSPH8 | 5.4e-01 | 0.950 |
| KIF5C | 5.4e-01 | 0.950 |
| ABLIM1 | 5.4e-01 | 0.950 |
| ANKRD13C | 5.4e-01 | 0.950 |
| CHCHD8 | 5.4e-01 | 0.950 |
| PAIP1 | 5.4e-01 | 0.950 |
| NEO1 | 5.4e-01 | 0.950 |
| BRCA1 | 5.4e-01 | 0.950 |
| TBC1D14 | 5.4e-01 | 0.950 |
| FRMD4B | 5.4e-01 | 0.950 |
| KCTD17 | 5.4e-01 | 0.950 |
| UROS | 5.4e-01 | 0.950 |
| PPP4R2 | 5.4e-01 | 0.950 |
| CTTNBP2NL | 5.4e-01 | 0.950 |
| BRD1 | 5.4e-01 | 0.950 |
| FAM135A | 5.4e-01 | 0.950 |
| TMEM216 | 5.5e-01 | 0.950 |
| YWHAE | 5.5e-01 | 0.950 |
| CTD-2192D1.1 | 5.5e-01 | 0.950 |
| ADNP2 | 5.5e-01 | 0.950 |
| RSRP1 | 5.5e-01 | 0.950 |
| PET117 | 5.5e-01 | 0.950 |
| HDAC3 | 5.5e-01 | 0.950 |
| DCAF6 | 5.5e-01 | 0.950 |
| UAP1 | 5.5e-01 | 0.950 |
| CTSL | 5.5e-01 | 0.950 |
| DPY19L4 | 5.5e-01 | 0.950 |
| HMMR | 5.5e-01 | 0.950 |
| PHF23 | 5.5e-01 | 0.950 |
| NOC2L | 5.5e-01 | 0.950 |
| SRGAP2C | 5.5e-01 | 0.950 |
| IDH3A | 5.5e-01 | 0.950 |
| ZNHIT1 | 5.5e-01 | 0.950 |
| FAM193B | 5.5e-01 | 0.950 |
| TOMM7 | 5.5e-01 | 0.950 |
| NCDN | 5.5e-01 | 0.950 |
| C11orf95 | 5.5e-01 | 0.950 |
| KAT7 | 5.5e-01 | 0.950 |
| COMMD9 | 5.5e-01 | 0.950 |
| GNPNAT1 | 5.5e-01 | 0.950 |
| HES1 | 5.5e-01 | 0.950 |
| ARID4B | 5.5e-01 | 0.950 |
| E2F4 | 5.5e-01 | 0.950 |
| TAB2 | 5.5e-01 | 0.950 |
| CCDC14 | 5.5e-01 | 0.950 |
| ZMYM3 | 5.5e-01 | 0.950 |
| NUMBL | 5.5e-01 | 0.950 |
| CSTF2 | 5.5e-01 | 0.950 |
| PPHLN1 | 5.5e-01 | 0.950 |
| REXO4 | 5.5e-01 | 0.950 |
| FAM207A | 5.5e-01 | 0.950 |
| IFI27L1 | 5.5e-01 | 0.950 |
| CS | 5.5e-01 | 0.950 |
| DHODH | 5.5e-01 | 0.950 |
| THAP4 | 5.5e-01 | 0.950 |
| CPOX | 5.5e-01 | 0.950 |
| RASAL2 | 5.5e-01 | 0.950 |
| ARHGEF12 | 5.5e-01 | 0.950 |
| SRPK1 | 5.5e-01 | 0.950 |
| RTKN2 | 5.5e-01 | 0.950 |
| RAC3 | 5.5e-01 | 0.950 |
| DGCR2 | 5.5e-01 | 0.950 |
| STAT1 | 5.5e-01 | 0.950 |
| CD63 | 5.5e-01 | 0.950 |
| PARD6A | 5.5e-01 | 0.950 |
| ALDH9A1 | 5.5e-01 | 0.950 |
| DALRD3 | 5.5e-01 | 0.950 |
| PPIL2 | 5.5e-01 | 0.950 |
| SEC61G | 5.5e-01 | 0.950 |
| GXYLT1 | 5.5e-01 | 0.950 |
| BAALC | 5.5e-01 | 0.950 |
| MDC1 | 5.5e-01 | 0.950 |
| CLIP3 | 5.5e-01 | 0.950 |
| RFK | 5.5e-01 | 0.950 |
| E2F3 | 5.5e-01 | 0.950 |
| WARS2 | 5.5e-01 | 0.950 |
| IGF2BP2 | 5.5e-01 | 0.950 |
| MICU2 | 5.5e-01 | 0.950 |
| CA2 | 5.5e-01 | 0.950 |
| RSF1 | 5.5e-01 | 0.950 |
| TSSC4 | 5.5e-01 | 0.950 |
| RNF216 | 5.5e-01 | 0.950 |
| LSAMP | 5.5e-01 | 0.950 |
| ZKSCAN5 | 5.5e-01 | 0.950 |
| MAP2K2 | 5.5e-01 | 0.950 |
| PTOV1 | 5.5e-01 | 0.950 |
| PLXNA2 | 5.5e-01 | 0.950 |
| CSTF3 | 5.5e-01 | 0.950 |
| BEX2 | 5.5e-01 | 0.950 |
| CDC16 | 5.5e-01 | 0.950 |
| FHL1 | 5.5e-01 | 0.950 |
| METTL13 | 5.5e-01 | 0.950 |
| TMEM39B | 5.5e-01 | 0.950 |
| RBMS1 | 5.5e-01 | 0.950 |
| NES | 5.5e-01 | 0.950 |
| KLHL7 | 5.6e-01 | 0.950 |
| SLC38A1 | 5.6e-01 | 0.950 |
| PGM1 | 5.6e-01 | 0.950 |
| GGH | 5.6e-01 | 0.950 |
| CYB561D2 | 5.6e-01 | 0.950 |
| THAP7 | 5.6e-01 | 0.950 |
| PLEKHA8 | 5.6e-01 | 0.950 |
| GPN2 | 5.6e-01 | 0.950 |
| TMEM126B | 5.6e-01 | 0.950 |
| IP6K2 | 5.6e-01 | 0.950 |
| FAH | 5.6e-01 | 0.950 |
| BEX1 | 5.6e-01 | 0.950 |
| ALG8 | 5.6e-01 | 0.950 |
| NR6A1 | 5.6e-01 | 0.950 |
| CAPN10 | 5.6e-01 | 0.950 |
| RUFY3 | 5.6e-01 | 0.950 |
| B4GALNT4 | 5.6e-01 | 0.950 |
| PIGQ | 5.6e-01 | 0.950 |
| PYURF | 5.6e-01 | 0.950 |
| TRIP11 | 5.6e-01 | 0.950 |
| YBX3 | 5.6e-01 | 0.950 |
| OGFR | 5.6e-01 | 0.950 |
| UACA | 5.6e-01 | 0.950 |
| C5orf15 | 5.6e-01 | 0.950 |
| SLC35F1 | 5.6e-01 | 0.950 |
| GTF3C3 | 5.6e-01 | 0.950 |
| SNW1 | 5.6e-01 | 0.950 |
| TPST1 | 5.6e-01 | 0.950 |
| ATP5H | 5.6e-01 | 0.950 |
| CYR61 | 5.6e-01 | 0.950 |
| CXorf56 | 5.6e-01 | 0.950 |
| TAB1 | 5.6e-01 | 0.950 |
| ZNF324 | 5.6e-01 | 0.950 |
| NQO2 | 5.6e-01 | 0.950 |
| STK26 | 5.6e-01 | 0.950 |
| ANKRD1 | 5.6e-01 | 0.950 |
| CRIP2 | 5.6e-01 | 0.950 |
| UQCR10 | 5.6e-01 | 0.950 |
| NDUFC1 | 5.6e-01 | 0.950 |
| DBX2 | 5.6e-01 | 0.950 |
| HIP1 | 5.6e-01 | 0.950 |
| CHID1 | 5.6e-01 | 0.950 |
| TLE1 | 5.6e-01 | 0.950 |
| CUEDC2 | 5.6e-01 | 0.950 |
| POM121C | 5.6e-01 | 0.950 |
| ACAD8 | 5.6e-01 | 0.950 |
| CREB5 | 5.6e-01 | 0.960 |
| PPM1K | 5.6e-01 | 0.960 |
| CAPZA2 | 5.6e-01 | 0.960 |
| OTUD4 | 5.6e-01 | 0.960 |
| SETD3 | 5.6e-01 | 0.960 |
| UBL5 | 5.6e-01 | 0.960 |
| MDK | 5.6e-01 | 0.960 |
| UBXN11 | 5.6e-01 | 0.960 |
| DECR2 | 5.6e-01 | 0.960 |
| ZC3H4 | 5.6e-01 | 0.960 |
| MCM8 | 5.6e-01 | 0.960 |
| POLR3D | 5.6e-01 | 0.960 |
| PACSIN2 | 5.6e-01 | 0.960 |
| NCOR2 | 5.6e-01 | 0.960 |
| HEATR6 | 5.6e-01 | 0.960 |
| TGOLN2 | 5.6e-01 | 0.960 |
| YTHDC1 | 5.6e-01 | 0.960 |
| FBXW7 | 5.6e-01 | 0.960 |
| NEU1 | 5.6e-01 | 0.960 |
| IRF2BP2 | 5.6e-01 | 0.960 |
| AKAP12 | 5.6e-01 | 0.960 |
| DYNC2H1 | 5.6e-01 | 0.960 |
| RRM2B | 5.7e-01 | 0.960 |
| TM2D3 | 5.7e-01 | 0.960 |
| TCF4 | 5.7e-01 | 0.960 |
| PTPRZ1 | 5.7e-01 | 0.960 |
| FJX1 | 5.7e-01 | 0.960 |
| IFT27 | 5.7e-01 | 0.960 |
| YTHDF2 | 5.7e-01 | 0.960 |
| CHMP4B | 5.7e-01 | 0.960 |
| VAX2 | 5.7e-01 | 0.960 |
| KIFAP3 | 5.7e-01 | 0.960 |
| OLMALINC | 5.7e-01 | 0.960 |
| ZNF625 | 5.7e-01 | 0.960 |
| TOX3 | 5.7e-01 | 0.960 |
| MCM10 | 5.7e-01 | 0.960 |
| CDKN3 | 5.7e-01 | 0.960 |
| TRIM16 | 5.7e-01 | 0.960 |
| GEMIN2 | 5.7e-01 | 0.960 |
| USP36 | 5.7e-01 | 0.960 |
| NR2C1 | 5.7e-01 | 0.960 |
| PFDN1 | 5.7e-01 | 0.960 |
| TRIO | 5.7e-01 | 0.960 |
| UBE2N | 5.7e-01 | 0.960 |
| ORAI2 | 5.7e-01 | 0.960 |
| GLTSCR2 | 5.7e-01 | 0.960 |
| DDI2 | 5.7e-01 | 0.960 |
| AP2B1 | 5.7e-01 | 0.960 |
| OSCP1 | 5.7e-01 | 0.960 |
| GCHFR | 5.7e-01 | 0.960 |
| GPX4 | 5.7e-01 | 0.960 |
| TRIB3 | 5.7e-01 | 0.960 |
| UNC13B | 5.7e-01 | 0.960 |
| HNRNPA1L2 | 5.7e-01 | 0.960 |
| DIDO1 | 5.7e-01 | 0.960 |
| IDE | 5.7e-01 | 0.960 |
| ATP5G1 | 5.7e-01 | 0.960 |
| NRBP1 | 5.7e-01 | 0.960 |
| LAMTOR2 | 5.7e-01 | 0.960 |
| SEC22B | 5.7e-01 | 0.960 |
| KBTBD2 | 5.7e-01 | 0.960 |
| GGNBP2 | 5.7e-01 | 0.960 |
| INO80C | 5.7e-01 | 0.960 |
| MIB1 | 5.7e-01 | 0.960 |
| ZFYVE16 | 5.7e-01 | 0.960 |
| TRIM8 | 5.7e-01 | 0.960 |
| XPO4 | 5.7e-01 | 0.960 |
| MIS18BP1 | 5.7e-01 | 0.960 |
| SCAP | 5.7e-01 | 0.960 |
| MSRB1 | 5.7e-01 | 0.960 |
| SAMD4B | 5.7e-01 | 0.960 |
| RP11-14N7.2 | 5.7e-01 | 0.960 |
| CDK2AP2 | 5.7e-01 | 0.960 |
| E2F5 | 5.7e-01 | 0.960 |
| CTSC | 5.7e-01 | 0.960 |
| SCAF11 | 5.7e-01 | 0.960 |
| HSD17B10 | 5.7e-01 | 0.960 |
| ZNF511 | 5.7e-01 | 0.960 |
| TSPYL4 | 5.7e-01 | 0.960 |
| C15orf57 | 5.7e-01 | 0.960 |
| PAWR | 5.7e-01 | 0.960 |
| PLEKHO1 | 5.7e-01 | 0.960 |
| SFR1 | 5.7e-01 | 0.960 |
| SMC2 | 5.7e-01 | 0.960 |
| ZNF682 | 5.7e-01 | 0.960 |
| GDPD2 | 5.7e-01 | 0.960 |
| CCNYL1 | 5.7e-01 | 0.960 |
| PLK2 | 5.7e-01 | 0.960 |
| EMC10 | 5.7e-01 | 0.960 |
| GBE1 | 5.7e-01 | 0.960 |
| VAT1 | 5.7e-01 | 0.960 |
| TCAF1 | 5.8e-01 | 0.960 |
| CENPO | 5.8e-01 | 0.960 |
| D2HGDH | 5.8e-01 | 0.960 |
| HLA-C | 5.8e-01 | 0.960 |
| SAC3D1 | 5.8e-01 | 0.960 |
| ZNF408 | 5.8e-01 | 0.960 |
| PPME1 | 5.8e-01 | 0.960 |
| MIF4GD | 5.8e-01 | 0.960 |
| TUBB3 | 5.8e-01 | 0.960 |
| TIPIN | 5.8e-01 | 0.960 |
| CLTA | 5.8e-01 | 0.960 |
| XAB2 | 5.8e-01 | 0.960 |
| TMBIM6 | 5.8e-01 | 0.960 |
| UPP1 | 5.8e-01 | 0.960 |
| ALKBH7 | 5.8e-01 | 0.960 |
| TUFM | 5.8e-01 | 0.960 |
| RBM33 | 5.8e-01 | 0.960 |
| HAUS8 | 5.8e-01 | 0.960 |
| TTF1 | 5.8e-01 | 0.960 |
| PITPNM1 | 5.8e-01 | 0.960 |
| NEDD4L | 5.8e-01 | 0.960 |
| MPHOSPH9 | 5.8e-01 | 0.960 |
| ZNF462 | 5.8e-01 | 0.960 |
| FAM73B | 5.8e-01 | 0.960 |
| PSMB7 | 5.8e-01 | 0.960 |
| MRPL19 | 5.8e-01 | 0.960 |
| TMEM223 | 5.8e-01 | 0.960 |
| FNBP1 | 5.8e-01 | 0.960 |
| API5 | 5.8e-01 | 0.960 |
| DYNLL1 | 5.8e-01 | 0.960 |
| SPHK1 | 5.8e-01 | 0.960 |
| CNIH4 | 5.8e-01 | 0.960 |
| EIF4E | 5.8e-01 | 0.960 |
| EXOSC6 | 5.8e-01 | 0.960 |
| RARA | 5.8e-01 | 0.960 |
| DLGAP4 | 5.8e-01 | 0.960 |
| CNOT4 | 5.8e-01 | 0.960 |
| CSK | 5.8e-01 | 0.960 |
| PPFIA3 | 5.8e-01 | 0.960 |
| DPPA4 | 5.8e-01 | 0.960 |
| METTL15 | 5.8e-01 | 0.960 |
| C5orf38 | 5.8e-01 | 0.960 |
| MDM1 | 5.8e-01 | 0.960 |
| PROSC | 5.8e-01 | 0.960 |
| RPL18 | 5.8e-01 | 0.960 |
| RPL32 | 5.8e-01 | 0.960 |
| ARHGEF2 | 5.8e-01 | 0.960 |
| BBC3 | 5.8e-01 | 0.960 |
| TRAIP | 5.8e-01 | 0.960 |
| GOLIM4 | 5.8e-01 | 0.960 |
| AURKAIP1 | 5.8e-01 | 0.960 |
| IQGAP2 | 5.8e-01 | 0.960 |
| PIK3C3 | 5.8e-01 | 0.960 |
| NOL10 | 5.8e-01 | 0.960 |
| TSC22D4 | 5.8e-01 | 0.960 |
| AIMP2 | 5.8e-01 | 0.960 |
| ZNF668 | 5.8e-01 | 0.960 |
| COMMD8 | 5.8e-01 | 0.960 |
| COPS2 | 5.8e-01 | 0.960 |
| CBLL1 | 5.8e-01 | 0.960 |
| CINP | 5.8e-01 | 0.960 |
| TEX264 | 5.8e-01 | 0.960 |
| DARS | 5.8e-01 | 0.960 |
| C2orf76 | 5.8e-01 | 0.960 |
| PRR14L | 5.8e-01 | 0.960 |
| MAP3K13 | 5.8e-01 | 0.960 |
| SMIM14 | 5.8e-01 | 0.960 |
| RHOQ | 5.8e-01 | 0.960 |
| HYOU1 | 5.8e-01 | 0.960 |
| DLGAP1 | 5.8e-01 | 0.960 |
| POFUT1 | 5.8e-01 | 0.960 |
| SFPQ | 5.8e-01 | 0.960 |
| RNF38 | 5.8e-01 | 0.960 |
| GKAP1 | 5.8e-01 | 0.960 |
| MPC2 | 5.8e-01 | 0.960 |
| COASY | 5.8e-01 | 0.960 |
| GPM6B | 5.8e-01 | 0.960 |
| KMT5C | 5.8e-01 | 0.960 |
| PSMD12 | 5.8e-01 | 0.960 |
| SLMAP | 5.8e-01 | 0.960 |
| NELFA | 5.8e-01 | 0.960 |
| CMPK1 | 5.8e-01 | 0.960 |
| MYO9B | 5.8e-01 | 0.960 |
| RPUSD2 | 5.8e-01 | 0.960 |
| SNX2 | 5.8e-01 | 0.960 |
| PTPN12 | 5.8e-01 | 0.960 |
| ZNF784 | 5.8e-01 | 0.960 |
| ANAPC7 | 5.8e-01 | 0.960 |
| PI4KB | 5.8e-01 | 0.960 |
| LDOC1L | 5.9e-01 | 0.960 |
| NUDT8 | 5.9e-01 | 0.960 |
| SMIM19 | 5.9e-01 | 0.960 |
| ABHD17B | 5.9e-01 | 0.960 |
| ZNF629 | 5.9e-01 | 0.960 |
| LYRM7 | 5.9e-01 | 0.960 |
| TMEM129 | 5.9e-01 | 0.960 |
| YIPF1 | 5.9e-01 | 0.960 |
| GALNT11 | 5.9e-01 | 0.960 |
| PURB | 5.9e-01 | 0.960 |
| MTPAP | 5.9e-01 | 0.960 |
| GRIPAP1 | 5.9e-01 | 0.960 |
| GCFC2 | 5.9e-01 | 0.960 |
| C11orf49 | 5.9e-01 | 0.960 |
| PELP1 | 5.9e-01 | 0.960 |
| SOCS3 | 5.9e-01 | 0.960 |
| CSNK2B | 5.9e-01 | 0.960 |
| MAPRE2 | 5.9e-01 | 0.960 |
| FAM13A | 5.9e-01 | 0.960 |
| SLC30A9 | 5.9e-01 | 0.960 |
| ACBD3 | 5.9e-01 | 0.960 |
| R3HCC1 | 5.9e-01 | 0.960 |
| FOSL1 | 5.9e-01 | 0.960 |
| NDUFS8 | 5.9e-01 | 0.960 |
| KRI1 | 5.9e-01 | 0.960 |
| CTTN | 5.9e-01 | 0.960 |
| NOP56 | 5.9e-01 | 0.960 |
| PTBP1 | 5.9e-01 | 0.960 |
| NAPG | 5.9e-01 | 0.960 |
| CREBL2 | 5.9e-01 | 0.960 |
| NAA60 | 5.9e-01 | 0.960 |
| TFDP2 | 5.9e-01 | 0.960 |
| OSBPL1A | 5.9e-01 | 0.960 |
| RER1 | 5.9e-01 | 0.960 |
| MGST3 | 5.9e-01 | 0.960 |
| EML1 | 5.9e-01 | 0.960 |
| ARPC5 | 5.9e-01 | 0.960 |
| EXOSC5 | 5.9e-01 | 0.960 |
| NT5C3B | 5.9e-01 | 0.960 |
| RARS2 | 5.9e-01 | 0.960 |
| SDHD | 5.9e-01 | 0.960 |
| FAM98A | 5.9e-01 | 0.960 |
| CYP20A1 | 5.9e-01 | 0.960 |
| UBAC1 | 5.9e-01 | 0.960 |
| NDUFAF6 | 5.9e-01 | 0.960 |
| PLOD3 | 5.9e-01 | 0.960 |
| GPC6 | 5.9e-01 | 0.960 |
| EHMT2 | 5.9e-01 | 0.960 |
| MRPL48 | 5.9e-01 | 0.960 |
| MARCH6 | 5.9e-01 | 0.960 |
| UBE2D3 | 5.9e-01 | 0.960 |
| MBD1 | 5.9e-01 | 0.960 |
| AGL | 5.9e-01 | 0.960 |
| ATXN3 | 5.9e-01 | 0.960 |
| PPP1CA | 5.9e-01 | 0.960 |
| RBP1 | 5.9e-01 | 0.960 |
| LEF1 | 5.9e-01 | 0.960 |
| POLE2 | 5.9e-01 | 0.960 |
| C9orf69 | 5.9e-01 | 0.960 |
| LZTS2 | 5.9e-01 | 0.960 |
| PHPT1 | 5.9e-01 | 0.960 |
| CPSF1 | 5.9e-01 | 0.960 |
| TMEM132A | 5.9e-01 | 0.960 |
| GPC4 | 5.9e-01 | 0.960 |
| TRRAP | 5.9e-01 | 0.960 |
| CLCN6 | 5.9e-01 | 0.960 |
| PRKCI | 5.9e-01 | 0.960 |
| DTD2 | 5.9e-01 | 0.960 |
| OSBP | 5.9e-01 | 0.960 |
| PTBP2 | 5.9e-01 | 0.960 |
| GSTM4 | 5.9e-01 | 0.960 |
| FERMT2 | 5.9e-01 | 0.960 |
| CAD | 5.9e-01 | 0.960 |
| WDR45B | 5.9e-01 | 0.960 |
| IFI6 | 5.9e-01 | 0.960 |
| DCAF16 | 5.9e-01 | 0.960 |
| RAPGEF2 | 6.0e-01 | 0.960 |
| NIFK | 6.0e-01 | 0.960 |
| GALE | 6.0e-01 | 0.960 |
| TIA1 | 6.0e-01 | 0.960 |
| SLC16A9 | 6.0e-01 | 0.960 |
| RPAP1 | 6.0e-01 | 0.960 |
| AASS | 6.0e-01 | 0.960 |
| GULP1 | 6.0e-01 | 0.960 |
| RANBP6 | 6.0e-01 | 0.960 |
| CHERP | 6.0e-01 | 0.960 |
| GATC | 6.0e-01 | 0.960 |
| MFSD5 | 6.0e-01 | 0.960 |
| TBC1D17 | 6.0e-01 | 0.960 |
| PSD | 6.0e-01 | 0.960 |
| TCF7 | 6.0e-01 | 0.960 |
| XBP1 | 6.0e-01 | 0.960 |
| PMVK | 6.0e-01 | 0.960 |
| VIMP | 6.0e-01 | 0.960 |
| ZNF609 | 6.0e-01 | 0.960 |
| TRNAU1AP | 6.0e-01 | 0.960 |
| PPP2R4 | 6.0e-01 | 0.960 |
| RPL11 | 6.0e-01 | 0.960 |
| FKBP2 | 6.0e-01 | 0.960 |
| NRP2 | 6.0e-01 | 0.960 |
| ATF7 | 6.0e-01 | 0.960 |
| LRRC47 | 6.0e-01 | 0.960 |
| CREBBP | 6.0e-01 | 0.960 |
| PDDC1 | 6.0e-01 | 0.960 |
| OSBPL11 | 6.0e-01 | 0.960 |
| PSMD3 | 6.0e-01 | 0.960 |
| MEN1 | 6.0e-01 | 0.960 |
| EFR3B | 6.0e-01 | 0.960 |
| NUP210 | 6.0e-01 | 0.960 |
| TGIF2 | 6.0e-01 | 0.960 |
| CCDC74A | 6.0e-01 | 0.960 |
| CBFB | 6.0e-01 | 0.960 |
| NUBP2 | 6.0e-01 | 0.960 |
| NGLY1 | 6.0e-01 | 0.960 |
| BPNT1 | 6.0e-01 | 0.960 |
| IMMP2L | 6.0e-01 | 0.960 |
| HSPA4 | 6.0e-01 | 0.960 |
| EMSY | 6.0e-01 | 0.960 |
| PINK1 | 6.0e-01 | 0.960 |
| SLC2A4RG | 6.0e-01 | 0.960 |
| CCDC56 | 6.0e-01 | 0.960 |
| LSM10 | 6.0e-01 | 0.960 |
| ANKRD13A | 6.0e-01 | 0.960 |
| ROBO1 | 6.0e-01 | 0.960 |
| ELFN1 | 6.0e-01 | 0.960 |
| TOP1MT | 6.0e-01 | 0.960 |
| ZSCAN16 | 6.0e-01 | 0.960 |
| QKI | 6.0e-01 | 0.960 |
| MFSD11 | 6.0e-01 | 0.960 |
| RFX1 | 6.0e-01 | 0.960 |
| TRMT10A | 6.0e-01 | 0.960 |
| TRUB2 | 6.0e-01 | 0.960 |
| RP11-539L10.3 | 6.0e-01 | 0.960 |
| HBS1L | 6.0e-01 | 0.960 |
| DCP1A | 6.0e-01 | 0.960 |
| NAA38 | 6.0e-01 | 0.960 |
| KDSR | 6.0e-01 | 0.960 |
| CEP170 | 6.0e-01 | 0.960 |
| SF3B5 | 6.0e-01 | 0.960 |
| TMEM9B | 6.0e-01 | 0.960 |
| DPM3 | 6.0e-01 | 0.960 |
| ARL5B | 6.0e-01 | 0.960 |
| NKAP | 6.0e-01 | 0.960 |
| ZNF91 | 6.0e-01 | 0.960 |
| KCTD5 | 6.0e-01 | 0.960 |
| MIA3 | 6.0e-01 | 0.960 |
| LRRC57 | 6.0e-01 | 0.960 |
| UBQLN2 | 6.1e-01 | 0.960 |
| DLL3 | 6.1e-01 | 0.960 |
| NUMA1 | 6.1e-01 | 0.960 |
| FST | 6.1e-01 | 0.960 |
| MRPL55 | 6.1e-01 | 0.960 |
| RPL36 | 6.1e-01 | 0.960 |
| POMGNT1 | 6.1e-01 | 0.960 |
| MFHAS1 | 6.1e-01 | 0.960 |
| SNX24 | 6.1e-01 | 0.960 |
| EFNB2 | 6.1e-01 | 0.960 |
| PTMS | 6.1e-01 | 0.960 |
| NOSIP | 6.1e-01 | 0.960 |
| PCYOX1 | 6.1e-01 | 0.960 |
| COL4A3BP | 6.1e-01 | 0.960 |
| MAP2K6 | 6.1e-01 | 0.960 |
| SCAND1 | 6.1e-01 | 0.960 |
| C1D | 6.1e-01 | 0.960 |
| FASTKD5 | 6.1e-01 | 0.960 |
| CHIC2 | 6.1e-01 | 0.960 |
| FASTKD2 | 6.1e-01 | 0.960 |
| ABCD4 | 6.1e-01 | 0.960 |
| DTNB | 6.1e-01 | 0.960 |
| RRP1 | 6.1e-01 | 0.960 |
| RCOR2 | 6.1e-01 | 0.960 |
| SRPRA | 6.1e-01 | 0.960 |
| DDX21 | 6.1e-01 | 0.960 |
| PAK3 | 6.1e-01 | 0.960 |
| PPP2R1B | 6.1e-01 | 0.960 |
| DHX33 | 6.1e-01 | 0.960 |
| SRSF8 | 6.1e-01 | 0.960 |
| RUSC1 | 6.1e-01 | 0.960 |
| GNL3L | 6.1e-01 | 0.960 |
| MAD2L1BP | 6.1e-01 | 0.960 |
| RIC8A | 6.1e-01 | 0.960 |
| SLC25A4 | 6.1e-01 | 0.960 |
| COMMD1 | 6.1e-01 | 0.960 |
| CHMP2A | 6.1e-01 | 0.960 |
| TUBB4B | 6.1e-01 | 0.960 |
| AGAP1 | 6.1e-01 | 0.960 |
| PARD3 | 6.1e-01 | 0.960 |
| TUBD1 | 6.1e-01 | 0.960 |
| MRPL52 | 6.1e-01 | 0.960 |
| WDR62 | 6.1e-01 | 0.960 |
| HMGN1 | 6.1e-01 | 0.960 |
| SP2 | 6.1e-01 | 0.960 |
| GUF1 | 6.1e-01 | 0.960 |
| SLC7A6OS | 6.1e-01 | 0.960 |
| ABHD10 | 6.1e-01 | 0.960 |
| LGALS3 | 6.1e-01 | 0.960 |
| RBBP4 | 6.1e-01 | 0.960 |
| SRFBP1 | 6.1e-01 | 0.960 |
| FAM120AOS | 6.1e-01 | 0.960 |
| SRPRB | 6.1e-01 | 0.960 |
| WTAP | 6.1e-01 | 0.960 |
| BTBD2 | 6.1e-01 | 0.960 |
| PHC2 | 6.1e-01 | 0.960 |
| SLTM | 6.1e-01 | 0.960 |
| ARL5A | 6.1e-01 | 0.960 |
| CELF2 | 6.1e-01 | 0.960 |
| CLCN3 | 6.1e-01 | 0.960 |
| ZNF226 | 6.1e-01 | 0.960 |
| PAK2 | 6.1e-01 | 0.960 |
| SLC16A1 | 6.1e-01 | 0.960 |
| CCDC12 | 6.1e-01 | 0.960 |
| EDC3 | 6.1e-01 | 0.960 |
| MED27 | 6.1e-01 | 0.960 |
| TMEM55A | 6.1e-01 | 0.960 |
| RCC2 | 6.1e-01 | 0.960 |
| NME2 | 6.1e-01 | 0.960 |
| ZFP64 | 6.1e-01 | 0.960 |
| FAM50A | 6.1e-01 | 0.960 |
| NDUFA12 | 6.1e-01 | 0.960 |
| C17orf97 | 6.1e-01 | 0.960 |
| NDUFB3 | 6.1e-01 | 0.960 |
| TANGO2 | 6.1e-01 | 0.960 |
| EZH2 | 6.1e-01 | 0.960 |
| FAM171A1 | 6.1e-01 | 0.960 |
| ZFX | 6.1e-01 | 0.960 |
| CST3 | 6.1e-01 | 0.960 |
| ZNF608 | 6.1e-01 | 0.960 |
| PDE9A | 6.1e-01 | 0.960 |
| TRAF3IP1 | 6.1e-01 | 0.960 |
| NDUFS1 | 6.1e-01 | 0.960 |
| ADGRV1 | 6.1e-01 | 0.960 |
| COG8 | 6.1e-01 | 0.960 |
| ETV4 | 6.1e-01 | 0.960 |
| KLHL5 | 6.1e-01 | 0.960 |
| NAXE | 6.1e-01 | 0.960 |
| C21orf58 | 6.1e-01 | 0.960 |
| DGKZ | 6.2e-01 | 0.960 |
| DAZAP2 | 6.2e-01 | 0.960 |
| SNX19 | 6.2e-01 | 0.960 |
| UBE2D1 | 6.2e-01 | 0.960 |
| UROD | 6.2e-01 | 0.960 |
| ANXA7 | 6.2e-01 | 0.960 |
| HADHB | 6.2e-01 | 0.960 |
| RACGAP1 | 6.2e-01 | 0.960 |
| TRAPPC2B | 6.2e-01 | 0.960 |
| FAM49B | 6.2e-01 | 0.960 |
| UCK2 | 6.2e-01 | 0.960 |
| HSPA13 | 6.2e-01 | 0.960 |
| TAF1B | 6.2e-01 | 0.960 |
| SCP2 | 6.2e-01 | 0.960 |
| TCEAL9 | 6.2e-01 | 0.960 |
| WRAP53 | 6.2e-01 | 0.960 |
| ASAP1 | 6.2e-01 | 0.960 |
| MINPP1 | 6.2e-01 | 0.960 |
| RPAIN | 6.2e-01 | 0.960 |
| CRMP1 | 6.2e-01 | 0.960 |
| RPL14 | 6.2e-01 | 0.960 |
| MAD2L1 | 6.2e-01 | 0.960 |
| BLOC1S6 | 6.2e-01 | 0.960 |
| TUBA1C | 6.2e-01 | 0.960 |
| LAMP1 | 6.2e-01 | 0.960 |
| CISD3 | 6.2e-01 | 0.960 |
| CFL2 | 6.2e-01 | 0.960 |
| FANCA | 6.2e-01 | 0.960 |
| ZNF92 | 6.2e-01 | 0.960 |
| SMIM4 | 6.2e-01 | 0.960 |
| IL13RA2 | 6.2e-01 | 0.960 |
| STARD10 | 6.2e-01 | 0.960 |
| ZNF281 | 6.2e-01 | 0.960 |
| MAP3K11 | 6.2e-01 | 0.960 |
| AAAS | 6.2e-01 | 0.960 |
| RCN1 | 6.2e-01 | 0.960 |
| MIPEP | 6.2e-01 | 0.960 |
| MXD4 | 6.2e-01 | 0.960 |
| ZBTB22 | 6.2e-01 | 0.960 |
| DIO3 | 6.2e-01 | 0.960 |
| PPP4R1 | 6.2e-01 | 0.960 |
| RPL19 | 6.2e-01 | 0.960 |
| CTU1 | 6.2e-01 | 0.960 |
| MARK2 | 6.2e-01 | 0.960 |
| COPE | 6.2e-01 | 0.960 |
| RABEP2 | 6.2e-01 | 0.960 |
| SMIM20 | 6.2e-01 | 0.960 |
| MPHOSPH10 | 6.2e-01 | 0.960 |
| ARMC7 | 6.2e-01 | 0.960 |
| SMIM15 | 6.2e-01 | 0.960 |
| DTX4 | 6.2e-01 | 0.960 |
| SCFD1 | 6.2e-01 | 0.960 |
| CHMP6 | 6.2e-01 | 0.960 |
| TXNL1 | 6.2e-01 | 0.960 |
| AAGAB | 6.2e-01 | 0.960 |
| GNL2 | 6.2e-01 | 0.960 |
| PCDH18 | 6.2e-01 | 0.960 |
| TOMM22 | 6.2e-01 | 0.960 |
| TAF1C | 6.2e-01 | 0.960 |
| SF3B6 | 6.2e-01 | 0.960 |
| SGPL1 | 6.2e-01 | 0.960 |
| C1QBP | 6.2e-01 | 0.960 |
| DIXDC1 | 6.2e-01 | 0.960 |
| WWC2 | 6.2e-01 | 0.960 |
| ZNF706 | 6.2e-01 | 0.960 |
| THUMPD3 | 6.2e-01 | 0.960 |
| ZFR | 6.2e-01 | 0.960 |
| MTHFD1L | 6.2e-01 | 0.960 |
| PLEKHB2 | 6.2e-01 | 0.960 |
| NECTIN3 | 6.2e-01 | 0.960 |
| SUPT20H | 6.2e-01 | 0.960 |
| SEPT6 | 6.2e-01 | 0.960 |
| GPSM1 | 6.2e-01 | 0.960 |
| EWSR1 | 6.2e-01 | 0.960 |
| DCX | 6.2e-01 | 0.960 |
| RAD17 | 6.2e-01 | 0.960 |
| RAB8B | 6.2e-01 | 0.960 |
| ATP5B | 6.2e-01 | 0.960 |
| RPS24 | 6.2e-01 | 0.960 |
| ORAI1 | 6.2e-01 | 0.960 |
| ANAPC10 | 6.2e-01 | 0.960 |
| KATNAL1 | 6.2e-01 | 0.960 |
| FUT8 | 6.2e-01 | 0.960 |
| XRRA1 | 6.2e-01 | 0.960 |
| FKBP7 | 6.2e-01 | 0.960 |
| MRPS35 | 6.3e-01 | 0.960 |
| MYO6 | 6.3e-01 | 0.960 |
| CLTB | 6.3e-01 | 0.960 |
| MGST1 | 6.3e-01 | 0.960 |
| GYS1 | 6.3e-01 | 0.960 |
| MSI2 | 6.3e-01 | 0.960 |
| FAM221A | 6.3e-01 | 0.960 |
| ARL1 | 6.3e-01 | 0.960 |
| LRRC75A-AS1 | 6.3e-01 | 0.960 |
| COX4I1 | 6.3e-01 | 0.960 |
| RLF | 6.3e-01 | 0.960 |
| CDC25C | 6.3e-01 | 0.960 |
| TIMM13 | 6.3e-01 | 0.960 |
| XRCC2 | 6.3e-01 | 0.960 |
| SOD1 | 6.3e-01 | 0.960 |
| ARL14EP | 6.3e-01 | 0.960 |
| RLIM | 6.3e-01 | 0.960 |
| SFMBT1 | 6.3e-01 | 0.960 |
| SMU1 | 6.3e-01 | 0.960 |
| CSNK2A1 | 6.3e-01 | 0.960 |
| CCNJL | 6.3e-01 | 0.960 |
| SLC25A39 | 6.3e-01 | 0.960 |
| IPO7 | 6.3e-01 | 0.960 |
| ANKMY2 | 6.3e-01 | 0.960 |
| SESN3 | 6.3e-01 | 0.960 |
| ARL2BP | 6.3e-01 | 0.960 |
| ENOSF1 | 6.3e-01 | 0.960 |
| TXNDC17 | 6.3e-01 | 0.960 |
| ACSL4 | 6.3e-01 | 0.960 |
| BASP1 | 6.3e-01 | 0.960 |
| TP53BP2 | 6.3e-01 | 0.960 |
| SCRN2 | 6.3e-01 | 0.960 |
| SVIL | 6.3e-01 | 0.960 |
| PLCD1 | 6.3e-01 | 0.960 |
| TPPP3 | 6.3e-01 | 0.960 |
| ZBTB39 | 6.3e-01 | 0.960 |
| CCDC92 | 6.3e-01 | 0.960 |
| AKT2 | 6.3e-01 | 0.960 |
| VMP1 | 6.3e-01 | 0.960 |
| DCLK1 | 6.3e-01 | 0.960 |
| TP53TG1 | 6.3e-01 | 0.960 |
| SSR2 | 6.3e-01 | 0.960 |
| KCMF1 | 6.3e-01 | 0.960 |
| TRAM2 | 6.3e-01 | 0.960 |
| NUP85 | 6.3e-01 | 0.960 |
| SMPD4 | 6.3e-01 | 0.960 |
| DPYSL2 | 6.3e-01 | 0.960 |
| ACTB | 6.3e-01 | 0.960 |
| C8orf59 | 6.3e-01 | 0.960 |
| IMP4 | 6.3e-01 | 0.960 |
| PTP4A2 | 6.3e-01 | 0.960 |
| PGAM1 | 6.3e-01 | 0.960 |
| COX18 | 6.3e-01 | 0.960 |
| SESN1 | 6.3e-01 | 0.960 |
| NOL9 | 6.3e-01 | 0.960 |
| MYBBP1A | 6.3e-01 | 0.960 |
| CAPZB | 6.3e-01 | 0.960 |
| NBEAL1 | 6.3e-01 | 0.960 |
| RBFOX2 | 6.3e-01 | 0.960 |
| RRP7A | 6.3e-01 | 0.960 |
| HSF1 | 6.3e-01 | 0.960 |
| ZNF638 | 6.3e-01 | 0.960 |
| LIG3 | 6.3e-01 | 0.960 |
| CTSD | 6.3e-01 | 0.960 |
| MUS81 | 6.3e-01 | 0.960 |
| ZNF519 | 6.3e-01 | 0.960 |
| PDPR | 6.3e-01 | 0.960 |
| HSDL1 | 6.3e-01 | 0.960 |
| RPF1 | 6.3e-01 | 0.960 |
| XRCC5 | 6.3e-01 | 0.960 |
| AURKB | 6.3e-01 | 0.960 |
| C11orf71 | 6.3e-01 | 0.960 |
| ADRM1 | 6.3e-01 | 0.960 |
| ATP6V1D | 6.3e-01 | 0.960 |
| ATF7IP | 6.3e-01 | 0.960 |
| AC136007.2 | 6.3e-01 | 0.960 |
| FAAP100 | 6.4e-01 | 0.960 |
| CYB561A3 | 6.4e-01 | 0.960 |
| ZNF358 | 6.4e-01 | 0.960 |
| PAPD5 | 6.4e-01 | 0.960 |
| SYTL2 | 6.4e-01 | 0.960 |
| TRMT5 | 6.4e-01 | 0.960 |
| YIPF5 | 6.4e-01 | 0.960 |
| LIAS | 6.4e-01 | 0.960 |
| EPB41 | 6.4e-01 | 0.960 |
| MAP2 | 6.4e-01 | 0.960 |
| PMM1 | 6.4e-01 | 0.960 |
| ISCA2 | 6.4e-01 | 0.960 |
| BNIP2 | 6.4e-01 | 0.960 |
| NDRG3 | 6.4e-01 | 0.960 |
| SRSF4 | 6.4e-01 | 0.960 |
| SIPA1L2 | 6.4e-01 | 0.960 |
| STRADA | 6.4e-01 | 0.960 |
| NSA2 | 6.4e-01 | 0.960 |
| ATP13A1 | 6.4e-01 | 0.960 |
| WASF2 | 6.4e-01 | 0.960 |
| ZNF689 | 6.4e-01 | 0.960 |
| HMCES | 6.4e-01 | 0.960 |
| ZNF503 | 6.4e-01 | 0.960 |
| ERF | 6.4e-01 | 0.960 |
| GHITM | 6.4e-01 | 0.960 |
| VAV2 | 6.4e-01 | 0.960 |
| FKBP10 | 6.4e-01 | 0.960 |
| ERI1 | 6.4e-01 | 0.960 |
| CREM | 6.4e-01 | 0.960 |
| PLCG1 | 6.4e-01 | 0.960 |
| AMDHD2 | 6.4e-01 | 0.960 |
| POLR2C | 6.4e-01 | 0.960 |
| CDC42 | 6.4e-01 | 0.960 |
| AASDH | 6.4e-01 | 0.960 |
| C17orf89 | 6.4e-01 | 0.960 |
| DNTTIP1 | 6.4e-01 | 0.960 |
| PRKD2 | 6.4e-01 | 0.960 |
| FBXL3 | 6.4e-01 | 0.960 |
| DGCR6 | 6.4e-01 | 0.960 |
| UBE2L6 | 6.4e-01 | 0.960 |
| RPS8 | 6.4e-01 | 0.960 |
| CASC3 | 6.4e-01 | 0.960 |
| SUMO3 | 6.4e-01 | 0.960 |
| VAPB | 6.4e-01 | 0.960 |
| SMAD4 | 6.4e-01 | 0.960 |
| AMMECR1 | 6.4e-01 | 0.960 |
| COG1 | 6.4e-01 | 0.960 |
| AZI2 | 6.4e-01 | 0.960 |
| ADCK4 | 6.4e-01 | 0.960 |
| DNAJC11 | 6.4e-01 | 0.960 |
| SLC4A1AP | 6.4e-01 | 0.960 |
| DNAJA1 | 6.4e-01 | 0.970 |
| MMGT1 | 6.4e-01 | 0.970 |
| GSTCD | 6.4e-01 | 0.970 |
| P4HB | 6.4e-01 | 0.970 |
| C1GALT1C1 | 6.4e-01 | 0.970 |
| DDOST | 6.4e-01 | 0.970 |
| GPATCH1 | 6.4e-01 | 0.970 |
| SLU7 | 6.4e-01 | 0.970 |
| CASP3 | 6.5e-01 | 0.970 |
| TBL1XR1 | 6.5e-01 | 0.970 |
| LONP1 | 6.5e-01 | 0.970 |
| HNRNPUL1 | 6.5e-01 | 0.970 |
| STX16 | 6.5e-01 | 0.970 |
| SPSB4 | 6.5e-01 | 0.970 |
| PDAP1 | 6.5e-01 | 0.970 |
| SEPT9 | 6.5e-01 | 0.970 |
| TRMT6 | 6.5e-01 | 0.970 |
| PPP6C | 6.5e-01 | 0.970 |
| CNTLN | 6.5e-01 | 0.970 |
| DCAKD | 6.5e-01 | 0.970 |
| ZCCHC7 | 6.5e-01 | 0.970 |
| GNAL | 6.5e-01 | 0.970 |
| STX12 | 6.5e-01 | 0.970 |
| SGSM3 | 6.5e-01 | 0.970 |
| FBXW9 | 6.5e-01 | 0.970 |
| PEX19 | 6.5e-01 | 0.970 |
| NUDCD2 | 6.5e-01 | 0.970 |
| GANAB | 6.5e-01 | 0.970 |
| AAR2 | 6.5e-01 | 0.970 |
| DHX9 | 6.5e-01 | 0.970 |
| SH3YL1 | 6.5e-01 | 0.970 |
| FNTB | 6.5e-01 | 0.970 |
| USP6NL | 6.5e-01 | 0.970 |
| NAP1L1 | 6.5e-01 | 0.970 |
| PRDX3 | 6.5e-01 | 0.970 |
| PIGL | 6.5e-01 | 0.970 |
| BLCAP | 6.5e-01 | 0.970 |
| CTD-2568A17.3 | 6.5e-01 | 0.970 |
| MT-CO1 | 6.5e-01 | 0.970 |
| RPS15 | 6.5e-01 | 0.970 |
| ING2 | 6.5e-01 | 0.970 |
| AGGF1 | 6.5e-01 | 0.970 |
| KIF4A | 6.5e-01 | 0.970 |
| WAPL | 6.5e-01 | 0.970 |
| IBTK | 6.5e-01 | 0.970 |
| MYH9 | 6.5e-01 | 0.970 |
| FAT1 | 6.5e-01 | 0.970 |
| SDSL | 6.5e-01 | 0.970 |
| CENPW | 6.5e-01 | 0.970 |
| SCAMP2 | 6.5e-01 | 0.970 |
| SAP30 | 6.5e-01 | 0.970 |
| SUZ12 | 6.5e-01 | 0.970 |
| GPX8 | 6.5e-01 | 0.970 |
| ANKS6 | 6.5e-01 | 0.970 |
| MFSD14B | 6.5e-01 | 0.970 |
| TRAPPC2L | 6.5e-01 | 0.970 |
| VAMP3 | 6.5e-01 | 0.970 |
| DNPEP | 6.5e-01 | 0.970 |
| CKAP2L | 6.5e-01 | 0.970 |
| EFNB1 | 6.5e-01 | 0.970 |
| TTC38 | 6.5e-01 | 0.970 |
| TUSC3 | 6.5e-01 | 0.970 |
| MAGEH1 | 6.5e-01 | 0.970 |
| MOB1A | 6.5e-01 | 0.970 |
| NUP153 | 6.5e-01 | 0.970 |
| MTF2 | 6.5e-01 | 0.970 |
| C5orf24 | 6.5e-01 | 0.970 |
| FGFR1OP2 | 6.5e-01 | 0.970 |
| FAM195A | 6.5e-01 | 0.970 |
| GINS1 | 6.5e-01 | 0.970 |
| LRRC42 | 6.5e-01 | 0.970 |
| PDCD6IP | 6.5e-01 | 0.970 |
| SRRT | 6.5e-01 | 0.970 |
| CNOT3 | 6.5e-01 | 0.970 |
| KIAA1524 | 6.5e-01 | 0.970 |
| DUSP22 | 6.5e-01 | 0.970 |
| HMGN3 | 6.5e-01 | 0.970 |
| DYNLL2 | 6.5e-01 | 0.970 |
| APOPT1 | 6.5e-01 | 0.970 |
| HAUS2 | 6.5e-01 | 0.970 |
| EIF3I | 6.5e-01 | 0.970 |
| PIAS1 | 6.6e-01 | 0.970 |
| DYRK4 | 6.6e-01 | 0.970 |
| RPLP0 | 6.6e-01 | 0.970 |
| RPS6KA3 | 6.6e-01 | 0.970 |
| DCAF10 | 6.6e-01 | 0.970 |
| C16orf13 | 6.6e-01 | 0.970 |
| PALLD | 6.6e-01 | 0.970 |
| MFAP2 | 6.6e-01 | 0.970 |
| PAAF1 | 6.6e-01 | 0.970 |
| LARP1 | 6.6e-01 | 0.970 |
| RAB34 | 6.6e-01 | 0.970 |
| TMEM39A | 6.6e-01 | 0.970 |
| GEMIN6 | 6.6e-01 | 0.970 |
| HAUS5 | 6.6e-01 | 0.970 |
| FGF13 | 6.6e-01 | 0.970 |
| MYH10 | 6.6e-01 | 0.970 |
| ARF6 | 6.6e-01 | 0.970 |
| KCTD15 | 6.6e-01 | 0.970 |
| GADD45GIP1 | 6.6e-01 | 0.970 |
| MRPS23 | 6.6e-01 | 0.970 |
| DNAJC16 | 6.6e-01 | 0.970 |
| ELAVL3 | 6.6e-01 | 0.970 |
| DPM2 | 6.6e-01 | 0.970 |
| CAST | 6.6e-01 | 0.970 |
| HS2ST1 | 6.6e-01 | 0.970 |
| TEX261 | 6.6e-01 | 0.970 |
| PRKCZ | 6.6e-01 | 0.970 |
| CEP70 | 6.6e-01 | 0.970 |
| ATRAID | 6.6e-01 | 0.970 |
| RPS11 | 6.6e-01 | 0.970 |
| BCR | 6.6e-01 | 0.970 |
| PHKB | 6.6e-01 | 0.970 |
| ZMYM4 | 6.6e-01 | 0.970 |
| USP47 | 6.6e-01 | 0.970 |
| BEST1 | 6.6e-01 | 0.970 |
| POLR2H | 6.6e-01 | 0.970 |
| JADE1 | 6.6e-01 | 0.970 |
| REEP2 | 6.6e-01 | 0.970 |
| MGEA5 | 6.6e-01 | 0.970 |
| HAUS4 | 6.6e-01 | 0.970 |
| UBE2O | 6.6e-01 | 0.970 |
| IARS2 | 6.6e-01 | 0.970 |
| RBM25 | 6.6e-01 | 0.970 |
| MICALL1 | 6.6e-01 | 0.970 |
| C19orf60 | 6.6e-01 | 0.970 |
| PDIA4 | 6.6e-01 | 0.970 |
| RPS6KB1 | 6.6e-01 | 0.970 |
| PITPNC1 | 6.6e-01 | 0.970 |
| RBM28 | 6.6e-01 | 0.970 |
| POLK | 6.6e-01 | 0.970 |
| NUDT6 | 6.6e-01 | 0.970 |
| SEPT7 | 6.6e-01 | 0.970 |
| PDZD8 | 6.6e-01 | 0.970 |
| TBC1D5 | 6.6e-01 | 0.970 |
| UCHL1 | 6.6e-01 | 0.970 |
| NMT2 | 6.6e-01 | 0.970 |
| KIAA2026 | 6.6e-01 | 0.970 |
| GLA | 6.6e-01 | 0.970 |
| C5orf30 | 6.6e-01 | 0.970 |
| RBM27 | 6.6e-01 | 0.970 |
| MED14 | 6.6e-01 | 0.970 |
| VGF | 6.6e-01 | 0.970 |
| SNHG6 | 6.6e-01 | 0.970 |
| TBCB | 6.6e-01 | 0.970 |
| MEG3 | 6.6e-01 | 0.970 |
| ZNF232 | 6.6e-01 | 0.970 |
| MAFF | 6.6e-01 | 0.970 |
| AP4M1 | 6.6e-01 | 0.970 |
| SGF29 | 6.6e-01 | 0.970 |
| FGFR2 | 6.6e-01 | 0.970 |
| TTC33 | 6.6e-01 | 0.970 |
| CCNT2 | 6.6e-01 | 0.970 |
| SH3GLB2 | 6.6e-01 | 0.970 |
| ATG5 | 6.6e-01 | 0.970 |
| NEFL | 6.6e-01 | 0.970 |
| ACOX3 | 6.6e-01 | 0.970 |
| NREP | 6.7e-01 | 0.970 |
| TMEM99 | 6.7e-01 | 0.970 |
| FYTTD1 | 6.7e-01 | 0.970 |
| RAB1A | 6.7e-01 | 0.970 |
| PEG10 | 6.7e-01 | 0.970 |
| TMEM147 | 6.7e-01 | 0.970 |
| NUDCD3 | 6.7e-01 | 0.970 |
| BMI1 | 6.7e-01 | 0.970 |
| SNHG1 | 6.7e-01 | 0.970 |
| PHC1 | 6.7e-01 | 0.970 |
| SYNGR2 | 6.7e-01 | 0.970 |
| NUP133 | 6.7e-01 | 0.970 |
| LSM1 | 6.7e-01 | 0.970 |
| GNG5 | 6.7e-01 | 0.970 |
| ZNF532 | 6.7e-01 | 0.970 |
| PRADC1 | 6.7e-01 | 0.970 |
| UBE2Q1 | 6.7e-01 | 0.970 |
| CKS1B | 6.7e-01 | 0.970 |
| XRN2 | 6.7e-01 | 0.970 |
| BTBD10 | 6.7e-01 | 0.970 |
| DCAF13 | 6.7e-01 | 0.970 |
| CCDC157 | 6.7e-01 | 0.970 |
| NCSTN | 6.7e-01 | 0.970 |
| CLOCK | 6.7e-01 | 0.970 |
| MTPN | 6.7e-01 | 0.970 |
| FAM210A | 6.7e-01 | 0.970 |
| CKS2 | 6.7e-01 | 0.970 |
| NACC2 | 6.7e-01 | 0.970 |
| LRP3 | 6.7e-01 | 0.970 |
| ALG2 | 6.7e-01 | 0.970 |
| ZNF627 | 6.7e-01 | 0.970 |
| ST7 | 6.7e-01 | 0.970 |
| GSKIP | 6.7e-01 | 0.970 |
| MECR | 6.7e-01 | 0.970 |
| RABL2A | 6.7e-01 | 0.970 |
| DNAJC10 | 6.7e-01 | 0.970 |
| C12orf45 | 6.7e-01 | 0.970 |
| IQCK | 6.7e-01 | 0.970 |
| DROSHA | 6.7e-01 | 0.970 |
| RC3H1 | 6.7e-01 | 0.970 |
| EXOC1 | 6.7e-01 | 0.970 |
| MEF2A | 6.7e-01 | 0.970 |
| DDX31 | 6.7e-01 | 0.970 |
| MRPS17 | 6.7e-01 | 0.970 |
| KDM2A | 6.7e-01 | 0.970 |
| RPL5 | 6.7e-01 | 0.970 |
| SDHB | 6.7e-01 | 0.970 |
| KCTD2 | 6.7e-01 | 0.970 |
| TLE3 | 6.7e-01 | 0.970 |
| SETD1A | 6.7e-01 | 0.970 |
| THUMPD2 | 6.7e-01 | 0.970 |
| SMS | 6.7e-01 | 0.970 |
| ME2 | 6.7e-01 | 0.970 |
| GABARAP | 6.7e-01 | 0.970 |
| CPNE1 | 6.7e-01 | 0.970 |
| TBC1D9B | 6.7e-01 | 0.970 |
| SIRT7 | 6.7e-01 | 0.970 |
| ASCC3 | 6.7e-01 | 0.970 |
| MRPS30 | 6.7e-01 | 0.970 |
| GTPBP4 | 6.7e-01 | 0.970 |
| RAB1B | 6.7e-01 | 0.970 |
| CCDC112 | 6.7e-01 | 0.970 |
| BRI3 | 6.7e-01 | 0.970 |
| CCDC6 | 6.7e-01 | 0.970 |
| FAM206A | 6.7e-01 | 0.970 |
| ASB3 | 6.7e-01 | 0.970 |
| ZNF148 | 6.7e-01 | 0.970 |
| UBN2 | 6.7e-01 | 0.970 |
| CSRP2 | 6.7e-01 | 0.970 |
| CASP6 | 6.8e-01 | 0.970 |
| MBD5 | 6.8e-01 | 0.970 |
| TET1 | 6.8e-01 | 0.970 |
| TSPAN5 | 6.8e-01 | 0.970 |
| NEIL2 | 6.8e-01 | 0.970 |
| ARF3 | 6.8e-01 | 0.970 |
| SLC35C1 | 6.8e-01 | 0.970 |
| SUMF2 | 6.8e-01 | 0.970 |
| MLF1 | 6.8e-01 | 0.970 |
| MSN | 6.8e-01 | 0.970 |
| ZBTB16 | 6.8e-01 | 0.970 |
| POLRMT | 6.8e-01 | 0.970 |
| MUT | 6.8e-01 | 0.970 |
| ARHGAP21 | 6.8e-01 | 0.970 |
| PCYT2 | 6.8e-01 | 0.970 |
| MRPL20 | 6.8e-01 | 0.970 |
| CCDC85C | 6.8e-01 | 0.970 |
| VPS11 | 6.8e-01 | 0.970 |
| FDX1L | 6.8e-01 | 0.970 |
| RND3 | 6.8e-01 | 0.970 |
| PHAX | 6.8e-01 | 0.970 |
| TMEM256 | 6.8e-01 | 0.970 |
| SLC39A7 | 6.8e-01 | 0.970 |
| GDE1 | 6.8e-01 | 0.970 |
| TMED10 | 6.8e-01 | 0.970 |
| PFDN6 | 6.8e-01 | 0.970 |
| NAB1 | 6.8e-01 | 0.970 |
| TRAPPC4 | 6.8e-01 | 0.970 |
| TPST2 | 6.8e-01 | 0.970 |
| TRPC4AP | 6.8e-01 | 0.970 |
| ARL8B | 6.8e-01 | 0.970 |
| GPHN | 6.8e-01 | 0.970 |
| NOTCH1 | 6.8e-01 | 0.970 |
| SAR1B | 6.8e-01 | 0.970 |
| COG6 | 6.8e-01 | 0.970 |
| AMFR | 6.8e-01 | 0.970 |
| PPCDC | 6.8e-01 | 0.970 |
| TAF7 | 6.8e-01 | 0.970 |
| GTF3C5 | 6.8e-01 | 0.970 |
| LAMP2 | 6.8e-01 | 0.970 |
| CTD-2135J3.2 | 6.8e-01 | 0.970 |
| ZNF775 | 6.8e-01 | 0.970 |
| IQSEC1 | 6.8e-01 | 0.970 |
| HMGXB4 | 6.8e-01 | 0.970 |
| FAM86C1 | 6.8e-01 | 0.970 |
| DRG1 | 6.8e-01 | 0.970 |
| POLG2 | 6.8e-01 | 0.970 |
| NTAN1 | 6.8e-01 | 0.970 |
| PSMC3 | 6.8e-01 | 0.970 |
| ADGRG1 | 6.8e-01 | 0.970 |
| ZNF592 | 6.8e-01 | 0.970 |
| MIER1 | 6.8e-01 | 0.970 |
| ZNF593 | 6.8e-01 | 0.970 |
| DHRS4L2 | 6.8e-01 | 0.970 |
| NIP7 | 6.8e-01 | 0.970 |
| TUBB2A | 6.8e-01 | 0.970 |
| SRP54 | 6.8e-01 | 0.970 |
| ARID3B | 6.8e-01 | 0.970 |
| KDM4A | 6.8e-01 | 0.970 |
| SSBP4 | 6.8e-01 | 0.970 |
| NIPSNAP3A | 6.8e-01 | 0.970 |
| DYNLRB1 | 6.8e-01 | 0.970 |
| NAMPT | 6.8e-01 | 0.970 |
| EP300 | 6.8e-01 | 0.970 |
| EXD3 | 6.8e-01 | 0.970 |
| FAM109A | 6.8e-01 | 0.970 |
| WDR61 | 6.8e-01 | 0.970 |
| PQBP1 | 6.8e-01 | 0.970 |
| FKBP8 | 6.8e-01 | 0.970 |
| MT-ND5 | 6.8e-01 | 0.970 |
| FAM214B | 6.8e-01 | 0.970 |
| GRAMD1A | 6.9e-01 | 0.970 |
| LYRM1 | 6.9e-01 | 0.970 |
| CCNH | 6.9e-01 | 0.970 |
| PCNP | 6.9e-01 | 0.970 |
| ACO1 | 6.9e-01 | 0.970 |
| TAGLN3 | 6.9e-01 | 0.980 |
| FAM134A | 6.9e-01 | 0.980 |
| ZC3H13 | 6.9e-01 | 0.980 |
| ARIH2 | 6.9e-01 | 0.980 |
| GNG3 | 6.9e-01 | 0.980 |
| AFMID | 6.9e-01 | 0.980 |
| BBS2 | 6.9e-01 | 0.980 |
| CNN1 | 6.9e-01 | 0.980 |
| PSEN2 | 6.9e-01 | 0.980 |
| TMEM9 | 6.9e-01 | 0.980 |
| RNF11 | 6.9e-01 | 0.980 |
| ZFYVE19 | 6.9e-01 | 0.980 |
| HIGD2A | 6.9e-01 | 0.980 |
| SNX3 | 6.9e-01 | 0.980 |
| VCL | 6.9e-01 | 0.980 |
| GDI1 | 6.9e-01 | 0.980 |
| ZSCAN21 | 6.9e-01 | 0.980 |
| ITPA | 6.9e-01 | 0.980 |
| KLC1 | 6.9e-01 | 0.980 |
| BCHE | 6.9e-01 | 0.980 |
| SYS1 | 6.9e-01 | 0.980 |
| ZNF302 | 6.9e-01 | 0.980 |
| CHD4 | 6.9e-01 | 0.980 |
| COPZ1 | 6.9e-01 | 0.980 |
| ALYREF | 6.9e-01 | 0.980 |
| QSOX1 | 6.9e-01 | 0.980 |
| PLXNB2 | 6.9e-01 | 0.980 |
| TPRG1L | 6.9e-01 | 0.980 |
| ASL | 6.9e-01 | 0.980 |
| ZFP36L2 | 6.9e-01 | 0.980 |
| ZMIZ2 | 6.9e-01 | 0.980 |
| NECAB3 | 6.9e-01 | 0.980 |
| TIMP3 | 6.9e-01 | 0.980 |
| HKR1 | 6.9e-01 | 0.980 |
| C1orf50 | 6.9e-01 | 0.980 |
| ICA1 | 6.9e-01 | 0.980 |
| FADD | 6.9e-01 | 0.980 |
| EGLN1 | 6.9e-01 | 0.980 |
| GTF2H3 | 6.9e-01 | 0.980 |
| IP6K1 | 6.9e-01 | 0.980 |
| HN1 | 6.9e-01 | 0.980 |
| HERC2 | 6.9e-01 | 0.980 |
| RABEPK | 6.9e-01 | 0.980 |
| ATP5S | 6.9e-01 | 0.980 |
| CDC42SE2 | 6.9e-01 | 0.980 |
| ARPC4 | 6.9e-01 | 0.980 |
| HERPUD2 | 6.9e-01 | 0.980 |
| TMEM116 | 6.9e-01 | 0.980 |
| SNRNP35 | 6.9e-01 | 0.980 |
| CCDC82 | 6.9e-01 | 0.980 |
| EFCAB14 | 6.9e-01 | 0.980 |
| TUSC2 | 6.9e-01 | 0.980 |
| ABCB8 | 6.9e-01 | 0.980 |
| FAHD1 | 6.9e-01 | 0.980 |
| PSMD11 | 6.9e-01 | 0.980 |
| PDPN | 6.9e-01 | 0.980 |
| CCDC80 | 6.9e-01 | 0.980 |
| TIMM8A | 6.9e-01 | 0.980 |
| PLAGL1 | 6.9e-01 | 0.980 |
| ECSIT | 6.9e-01 | 0.980 |
| FTSJ1 | 6.9e-01 | 0.980 |
| C1orf122 | 6.9e-01 | 0.980 |
| FAM127B | 6.9e-01 | 0.980 |
| CBWD5 | 6.9e-01 | 0.980 |
| ENY2 | 6.9e-01 | 0.980 |
| TSC22D3 | 6.9e-01 | 0.980 |
| SLC35A4 | 6.9e-01 | 0.980 |
| ANAPC1 | 6.9e-01 | 0.980 |
| PUM2 | 7.0e-01 | 0.980 |
| SNTG1 | 7.0e-01 | 0.980 |
| MPDZ | 7.0e-01 | 0.980 |
| SLC36A4 | 7.0e-01 | 0.980 |
| DMAP1 | 7.0e-01 | 0.980 |
| RPF2 | 7.0e-01 | 0.980 |
| TPM3 | 7.0e-01 | 0.980 |
| ATP5SL | 7.0e-01 | 0.980 |
| GPM6A | 7.0e-01 | 0.980 |
| HSDL2 | 7.0e-01 | 0.980 |
| AKT3 | 7.0e-01 | 0.980 |
| DHX32 | 7.0e-01 | 0.980 |
| TMEM230 | 7.0e-01 | 0.980 |
| BMPR1A | 7.0e-01 | 0.980 |
| GAS1 | 7.0e-01 | 0.980 |
| THBS3 | 7.0e-01 | 0.980 |
| FKBP1A | 7.0e-01 | 0.980 |
| MVK | 7.0e-01 | 0.980 |
| PCGF3 | 7.0e-01 | 0.980 |
| PHYKPL | 7.0e-01 | 0.980 |
| C4orf3 | 7.0e-01 | 0.980 |
| POLR2L | 7.0e-01 | 0.980 |
| ANKRD49 | 7.0e-01 | 0.980 |
| SEC23B | 7.0e-01 | 0.980 |
| TTC26 | 7.0e-01 | 0.980 |
| THOP1 | 7.0e-01 | 0.980 |
| MYNN | 7.0e-01 | 0.980 |
| CMTM6 | 7.0e-01 | 0.980 |
| ADSS | 7.0e-01 | 0.980 |
| ZNF445 | 7.0e-01 | 0.980 |
| PPP1R14C | 7.0e-01 | 0.980 |
| TRMO | 7.0e-01 | 0.980 |
| CSNK2A2 | 7.0e-01 | 0.980 |
| DPY19L3 | 7.0e-01 | 0.980 |
| ID4 | 7.0e-01 | 0.980 |
| DPAGT1 | 7.0e-01 | 0.980 |
| SF3B3 | 7.0e-01 | 0.980 |
| MORC2 | 7.0e-01 | 0.980 |
| RIF1 | 7.0e-01 | 0.980 |
| DDX20 | 7.0e-01 | 0.980 |
| CENPF | 7.0e-01 | 0.980 |
| LEMD2 | 7.0e-01 | 0.980 |
| FUCA2 | 7.0e-01 | 0.980 |
| VPS37C | 7.0e-01 | 0.980 |
| SLC43A2 | 7.0e-01 | 0.980 |
| RTFDC1 | 7.0e-01 | 0.980 |
| ZC3H15 | 7.0e-01 | 0.980 |
| HSBP1 | 7.0e-01 | 0.980 |
| SSSCA1 | 7.0e-01 | 0.980 |
| ATXN7L3 | 7.0e-01 | 0.980 |
| TMEM164 | 7.0e-01 | 0.980 |
| MGME1 | 7.0e-01 | 0.980 |
| TSC22D1 | 7.0e-01 | 0.980 |
| ITGB1 | 7.0e-01 | 0.980 |
| C1orf43 | 7.0e-01 | 0.980 |
| EIF4A2 | 7.0e-01 | 0.980 |
| BBS4 | 7.0e-01 | 0.980 |
| DLEU1 | 7.0e-01 | 0.980 |
| SUCLG2 | 7.0e-01 | 0.980 |
| UBL3 | 7.0e-01 | 0.980 |
| RP11-382A20.3 | 7.0e-01 | 0.980 |
| ELAC2 | 7.0e-01 | 0.980 |
| EED | 7.0e-01 | 0.980 |
| SPRTN | 7.0e-01 | 0.980 |
| TBCK | 7.1e-01 | 0.980 |
| CMTR1 | 7.1e-01 | 0.980 |
| RAB7A | 7.1e-01 | 0.980 |
| CASC5 | 7.1e-01 | 0.980 |
| CREB3 | 7.1e-01 | 0.980 |
| JRK | 7.1e-01 | 0.980 |
| SEC24C | 7.1e-01 | 0.980 |
| MT-ATP6 | 7.1e-01 | 0.980 |
| UNC50 | 7.1e-01 | 0.980 |
| CHMP2B | 7.1e-01 | 0.980 |
| PATL1 | 7.1e-01 | 0.980 |
| TRIOBP | 7.1e-01 | 0.980 |
| ARPC2 | 7.1e-01 | 0.980 |
| TWISTNB | 7.1e-01 | 0.980 |
| MTFR1L | 7.1e-01 | 0.980 |
| FAM65A | 7.1e-01 | 0.980 |
| EMC2 | 7.1e-01 | 0.980 |
| USP40 | 7.1e-01 | 0.980 |
| FOXJ1 | 7.1e-01 | 0.980 |
| VPS37D | 7.1e-01 | 0.980 |
| TTC8 | 7.1e-01 | 0.980 |
| CALCOCO2 | 7.1e-01 | 0.980 |
| MYADM | 7.1e-01 | 0.980 |
| SUGT1 | 7.1e-01 | 0.980 |
| MOAP1 | 7.1e-01 | 0.980 |
| CDC42BPA | 7.1e-01 | 0.980 |
| CUEDC1 | 7.1e-01 | 0.980 |
| RPUSD4 | 7.1e-01 | 0.980 |
| ZNF622 | 7.1e-01 | 0.980 |
| CUX1 | 7.1e-01 | 0.980 |
| IPO13 | 7.1e-01 | 0.980 |
| DCXR | 7.1e-01 | 0.980 |
| SNAP29 | 7.1e-01 | 0.980 |
| DESI2 | 7.1e-01 | 0.980 |
| MGAT1 | 7.1e-01 | 0.980 |
| SLC29A1 | 7.1e-01 | 0.980 |
| GEMIN5 | 7.1e-01 | 0.980 |
| PRPF4B | 7.1e-01 | 0.980 |
| MCU | 7.1e-01 | 0.980 |
| PEX11B | 7.1e-01 | 0.980 |
| ABHD14A | 7.1e-01 | 0.980 |
| LRPAP1 | 7.1e-01 | 0.980 |
| MNAT1 | 7.1e-01 | 0.980 |
| POGZ | 7.1e-01 | 0.980 |
| ZMYND8 | 7.1e-01 | 0.980 |
| INA | 7.1e-01 | 0.980 |
| ZNF71 | 7.1e-01 | 0.980 |
| ZNF766 | 7.1e-01 | 0.980 |
| TWSG1 | 7.1e-01 | 0.980 |
| POLR2B | 7.1e-01 | 0.980 |
| SCLT1 | 7.1e-01 | 0.980 |
| RBPMS | 7.1e-01 | 0.980 |
| C2orf44 | 7.1e-01 | 0.980 |
| ECHDC1 | 7.1e-01 | 0.980 |
| NEXN | 7.1e-01 | 0.980 |
| RGP1 | 7.1e-01 | 0.980 |
| TOMM70 | 7.1e-01 | 0.980 |
| C19orf10 | 7.1e-01 | 0.980 |
| CTDSPL | 7.1e-01 | 0.980 |
| TMF1 | 7.1e-01 | 0.980 |
| CAPNS1 | 7.1e-01 | 0.980 |
| MRE11A | 7.1e-01 | 0.980 |
| SPTY2D1 | 7.1e-01 | 0.980 |
| SOD2 | 7.1e-01 | 0.980 |
| CLUAP1 | 7.1e-01 | 0.980 |
| GINM1 | 7.1e-01 | 0.980 |
| RYBP | 7.1e-01 | 0.980 |
| TARDBP | 7.1e-01 | 0.980 |
| NFYC | 7.1e-01 | 0.980 |
| RNF19A | 7.1e-01 | 0.980 |
| PRKAB1 | 7.1e-01 | 0.980 |
| BAG3 | 7.1e-01 | 0.980 |
| ELMO1 | 7.1e-01 | 0.980 |
| YIPF3 | 7.1e-01 | 0.980 |
| SSR1 | 7.1e-01 | 0.980 |
| FOXK1 | 7.1e-01 | 0.980 |
| FHOD3 | 7.1e-01 | 0.980 |
| CLCC1 | 7.1e-01 | 0.980 |
| SH3BGRL | 7.2e-01 | 0.980 |
| HNRNPC | 7.2e-01 | 0.980 |
| SAP30BP | 7.2e-01 | 0.980 |
| MIR99AHG | 7.2e-01 | 0.980 |
| TMEM123 | 7.2e-01 | 0.980 |
| GABRB3 | 7.2e-01 | 0.980 |
| TPM2 | 7.2e-01 | 0.980 |
| CHURC1 | 7.2e-01 | 0.980 |
| CCNDBP1 | 7.2e-01 | 0.980 |
| HLTF | 7.2e-01 | 0.980 |
| MYL9 | 7.2e-01 | 0.980 |
| S100A13 | 7.2e-01 | 0.980 |
| CHD2 | 7.2e-01 | 0.980 |
| SMARCD3 | 7.2e-01 | 0.980 |
| RNF25 | 7.2e-01 | 0.980 |
| MRPL39 | 7.2e-01 | 0.980 |
| AFF3 | 7.2e-01 | 0.980 |
| IFT52 | 7.2e-01 | 0.980 |
| RHOG | 7.2e-01 | 0.980 |
| ACLY | 7.2e-01 | 0.980 |
| AC005154.6 | 7.2e-01 | 0.980 |
| HNRNPH3 | 7.2e-01 | 0.980 |
| ADIPOR2 | 7.2e-01 | 0.980 |
| EP400 | 7.2e-01 | 0.980 |
| CHCHD5 | 7.2e-01 | 0.980 |
| SMG7 | 7.2e-01 | 0.980 |
| TMX2 | 7.2e-01 | 0.980 |
| ZNF197 | 7.2e-01 | 0.980 |
| RPL22 | 7.2e-01 | 0.980 |
| FAM120A | 7.2e-01 | 0.980 |
| INTS11 | 7.2e-01 | 0.980 |
| C17orf75 | 7.2e-01 | 0.980 |
| KIFC3 | 7.2e-01 | 0.980 |
| DPH6 | 7.2e-01 | 0.980 |
| TMOD3 | 7.2e-01 | 0.980 |
| FOXP1 | 7.2e-01 | 0.980 |
| AP3D1 | 7.2e-01 | 0.980 |
| TMEM248 | 7.2e-01 | 0.980 |
| ATOX1 | 7.2e-01 | 0.980 |
| DAP | 7.2e-01 | 0.980 |
| RABIF | 7.2e-01 | 0.980 |
| CDV3 | 7.2e-01 | 0.980 |
| IVNS1ABP | 7.2e-01 | 0.980 |
| DNAJC30 | 7.2e-01 | 0.980 |
| MSMO1 | 7.2e-01 | 0.980 |
| LRRC4 | 7.2e-01 | 0.980 |
| ATP6V1E1 | 7.2e-01 | 0.980 |
| EMG1 | 7.2e-01 | 0.980 |
| APEH | 7.2e-01 | 0.980 |
| AGBL5 | 7.2e-01 | 0.980 |
| LYRM2 | 7.2e-01 | 0.980 |
| STIP1 | 7.2e-01 | 0.980 |
| ZNF236 | 7.2e-01 | 0.980 |
| FAM175B | 7.2e-01 | 0.980 |
| MRPL43 | 7.2e-01 | 0.980 |
| JKAMP | 7.2e-01 | 0.980 |
| FTSJ2 | 7.2e-01 | 0.980 |
| CUTA | 7.2e-01 | 0.980 |
| EIF2AK1 | 7.2e-01 | 0.980 |
| P3H4 | 7.2e-01 | 0.980 |
| ATPIF1 | 7.2e-01 | 0.980 |
| SUGP2 | 7.2e-01 | 0.980 |
| MT-ND4L | 7.2e-01 | 0.980 |
| ACVR2B | 7.2e-01 | 0.980 |
| HIST1H1C | 7.2e-01 | 0.980 |
| TIMP2 | 7.2e-01 | 0.980 |
| VPS39 | 7.2e-01 | 0.980 |
| SEH1L | 7.2e-01 | 0.980 |
| DEDD | 7.2e-01 | 0.980 |
| FBXO17 | 7.2e-01 | 0.980 |
| UTRN | 7.2e-01 | 0.980 |
| TSC22D2 | 7.2e-01 | 0.980 |
| TOR1A | 7.2e-01 | 0.980 |
| ZNF584 | 7.2e-01 | 0.980 |
| DHRSX | 7.2e-01 | 0.980 |
| TMEM150A | 7.2e-01 | 0.980 |
| CDCA8 | 7.2e-01 | 0.980 |
| SRR | 7.2e-01 | 0.980 |
| AIP | 7.2e-01 | 0.980 |
| AIMP1 | 7.2e-01 | 0.980 |
| RAB29 | 7.2e-01 | 0.980 |
| PCK2 | 7.3e-01 | 0.980 |
| DHRS7B | 7.3e-01 | 0.980 |
| DEPDC1 | 7.3e-01 | 0.980 |
| RBM34 | 7.3e-01 | 0.980 |
| UBXN1 | 7.3e-01 | 0.980 |
| RPL39L | 7.3e-01 | 0.980 |
| INTS7 | 7.3e-01 | 0.980 |
| DDX55 | 7.3e-01 | 0.980 |
| LMAN1 | 7.3e-01 | 0.980 |
| C5orf45 | 7.3e-01 | 0.980 |
| EXOC5 | 7.3e-01 | 0.980 |
| DAPK3 | 7.3e-01 | 0.980 |
| ERLIN1 | 7.3e-01 | 0.980 |
| FEZ1 | 7.3e-01 | 0.980 |
| MGRN1 | 7.3e-01 | 0.980 |
| FIGNL2 | 7.3e-01 | 0.980 |
| PMPCA | 7.3e-01 | 0.980 |
| C9orf85 | 7.3e-01 | 0.980 |
| CAMK2D | 7.3e-01 | 0.980 |
| IPPK | 7.3e-01 | 0.980 |
| TTC19 | 7.3e-01 | 0.980 |
| RP11-566K11.6 | 7.3e-01 | 0.980 |
| GINS3 | 7.3e-01 | 0.980 |
| INPP5E | 7.3e-01 | 0.980 |
| LIMCH1 | 7.3e-01 | 0.980 |
| CCDC18 | 7.3e-01 | 0.980 |
| POLL | 7.3e-01 | 0.980 |
| CHCHD1 | 7.3e-01 | 0.980 |
| TMEM2 | 7.3e-01 | 0.980 |
| DDB1 | 7.3e-01 | 0.980 |
| CCDC130 | 7.3e-01 | 0.980 |
| C1orf21 | 7.3e-01 | 0.980 |
| ELOVL1 | 7.3e-01 | 0.980 |
| SPATS2L | 7.3e-01 | 0.980 |
| METTL17 | 7.3e-01 | 0.980 |
| ABCF2 | 7.3e-01 | 0.980 |
| ZNHIT2 | 7.3e-01 | 0.980 |
| C14orf1 | 7.3e-01 | 0.980 |
| CBR1 | 7.3e-01 | 0.980 |
| MACROD1 | 7.3e-01 | 0.980 |
| RPS9 | 7.3e-01 | 0.980 |
| IFNAR2 | 7.3e-01 | 0.980 |
| METTL12 | 7.3e-01 | 0.980 |
| FAM20B | 7.3e-01 | 0.980 |
| ZNF431 | 7.3e-01 | 0.980 |
| RBSN | 7.3e-01 | 0.980 |
| ANOS1 | 7.3e-01 | 0.980 |
| RPL23 | 7.3e-01 | 0.980 |
| LAPTM4B | 7.3e-01 | 0.980 |
| GORASP1 | 7.3e-01 | 0.980 |
| GTF2E2 | 7.3e-01 | 0.980 |
| ANKRD36C | 7.3e-01 | 0.980 |
| RAP1GDS1 | 7.3e-01 | 0.980 |
| CACTIN | 7.3e-01 | 0.980 |
| CCDC43 | 7.3e-01 | 0.980 |
| MLST8 | 7.3e-01 | 0.980 |
| SCNM1 | 7.3e-01 | 0.980 |
| BTN2A1 | 7.3e-01 | 0.980 |
| TRIP13 | 7.3e-01 | 0.980 |
| DOCK7 | 7.3e-01 | 0.980 |
| NOL6 | 7.3e-01 | 0.980 |
| MRPS5 | 7.3e-01 | 0.980 |
| TJP1 | 7.3e-01 | 0.980 |
| FANCE | 7.3e-01 | 0.980 |
| MECP2 | 7.3e-01 | 0.980 |
| SMC1A | 7.3e-01 | 0.980 |
| NAT8L | 7.3e-01 | 0.980 |
| CCT5 | 7.3e-01 | 0.980 |
| SALL2 | 7.3e-01 | 0.980 |
| LRFN4 | 7.3e-01 | 0.980 |
| BLVRB | 7.3e-01 | 0.980 |
| KAT6A | 7.3e-01 | 0.980 |
| APBA2 | 7.3e-01 | 0.980 |
| TTL | 7.3e-01 | 0.980 |
| RPL18A | 7.3e-01 | 0.980 |
| KIF2A | 7.3e-01 | 0.980 |
| PPFIBP1 | 7.3e-01 | 0.980 |
| NOL8 | 7.3e-01 | 0.980 |
| RPLP2 | 7.3e-01 | 0.980 |
| EPS8 | 7.3e-01 | 0.980 |
| DPF2 | 7.3e-01 | 0.980 |
| ORC3 | 7.3e-01 | 0.980 |
| KDM3B | 7.3e-01 | 0.980 |
| ZNF496 | 7.3e-01 | 0.980 |
| NRGN | 7.4e-01 | 0.980 |
| C14orf179 | 7.4e-01 | 0.980 |
| SC5D | 7.4e-01 | 0.980 |
| AK1 | 7.4e-01 | 0.980 |
| SYPL1 | 7.4e-01 | 0.980 |
| CCDC58 | 7.4e-01 | 0.980 |
| BCDIN3D | 7.4e-01 | 0.980 |
| RNF121 | 7.4e-01 | 0.980 |
| CHD8 | 7.4e-01 | 0.980 |
| POLDIP3 | 7.4e-01 | 0.980 |
| TTBK2 | 7.4e-01 | 0.980 |
| ACTR1A | 7.4e-01 | 0.980 |
| DNAJC3 | 7.4e-01 | 0.980 |
| PSMC2 | 7.4e-01 | 0.980 |
| DLL1 | 7.4e-01 | 0.980 |
| CFAP97 | 7.4e-01 | 0.980 |
| CBX6 | 7.4e-01 | 0.980 |
| HMGB3 | 7.4e-01 | 0.980 |
| TAF15 | 7.4e-01 | 0.980 |
| C19orf70 | 7.4e-01 | 0.980 |
| USP13 | 7.4e-01 | 0.980 |
| CPTP | 7.4e-01 | 0.980 |
| ADH5 | 7.4e-01 | 0.980 |
| VKORC1 | 7.4e-01 | 0.980 |
| SLF2 | 7.4e-01 | 0.980 |
| CRAT | 7.4e-01 | 0.980 |
| DUSP12 | 7.4e-01 | 0.980 |
| USP7 | 7.4e-01 | 0.980 |
| GREB1 | 7.4e-01 | 0.980 |
| ASXL2 | 7.4e-01 | 0.980 |
| SETD4 | 7.4e-01 | 0.980 |
| SLC35B2 | 7.4e-01 | 0.980 |
| CRTC1 | 7.4e-01 | 0.980 |
| MTMR2 | 7.4e-01 | 0.980 |
| UBA1 | 7.4e-01 | 0.980 |
| C9orf78 | 7.4e-01 | 0.980 |
| MTMR4 | 7.4e-01 | 0.980 |
| C8orf58 | 7.4e-01 | 0.980 |
| PML | 7.4e-01 | 0.980 |
| RANBP9 | 7.4e-01 | 0.980 |
| FYN | 7.4e-01 | 0.980 |
| UBA6 | 7.4e-01 | 0.980 |
| NCAPH2 | 7.4e-01 | 0.980 |
| GPR108 | 7.4e-01 | 0.980 |
| PDCD4 | 7.4e-01 | 0.980 |
| NF2 | 7.4e-01 | 0.980 |
| GSTM3 | 7.4e-01 | 0.980 |
| ARFRP1 | 7.4e-01 | 0.980 |
| ZNF90 | 7.4e-01 | 0.980 |
| RPS25 | 7.4e-01 | 0.980 |
| PCCB | 7.4e-01 | 0.980 |
| SLC11A2 | 7.4e-01 | 0.980 |
| ZNF827 | 7.4e-01 | 0.980 |
| CSNK1D | 7.4e-01 | 0.980 |
| ADPGK | 7.4e-01 | 0.980 |
| E2F6 | 7.4e-01 | 0.980 |
| TRAPPC2 | 7.4e-01 | 0.980 |
| HSP90AA1 | 7.4e-01 | 0.980 |
| NSUN2 | 7.4e-01 | 0.980 |
| DCTN1 | 7.4e-01 | 0.980 |
| UBA3 | 7.4e-01 | 0.980 |
| PRH1 | 7.4e-01 | 0.980 |
| CAB39L | 7.4e-01 | 0.980 |
| KCNQ2 | 7.4e-01 | 0.980 |
| DNAJA3 | 7.4e-01 | 0.980 |
| WEE1 | 7.4e-01 | 0.980 |
| UQCC1 | 7.4e-01 | 0.980 |
| COPS8 | 7.4e-01 | 0.980 |
| RITA1 | 7.4e-01 | 0.980 |
| NOVA2 | 7.4e-01 | 0.980 |
| GFM2 | 7.4e-01 | 0.980 |
| HEATR1 | 7.4e-01 | 0.980 |
| NDUFB4 | 7.4e-01 | 0.980 |
| ATF5 | 7.4e-01 | 0.980 |
| PIAS2 | 7.4e-01 | 0.980 |
| TACC1 | 7.4e-01 | 0.980 |
| RGMB | 7.4e-01 | 0.980 |
| NUP37 | 7.4e-01 | 0.980 |
| POLE4 | 7.4e-01 | 0.980 |
| MOSPD1 | 7.4e-01 | 0.980 |
| ANKRD6 | 7.4e-01 | 0.980 |
| GOLM1 | 7.4e-01 | 0.980 |
| TST | 7.4e-01 | 0.980 |
| LCMT2 | 7.5e-01 | 0.980 |
| PAF1 | 7.5e-01 | 0.980 |
| ABI1 | 7.5e-01 | 0.980 |
| IDH3G | 7.5e-01 | 0.980 |
| MFN2 | 7.5e-01 | 0.980 |
| DDX18 | 7.5e-01 | 0.980 |
| ZFP91 | 7.5e-01 | 0.980 |
| SLC30A7 | 7.5e-01 | 0.980 |
| AGPAT4 | 7.5e-01 | 0.980 |
| MMS19 | 7.5e-01 | 0.980 |
| ACACA | 7.5e-01 | 0.980 |
| BLVRA | 7.5e-01 | 0.980 |
| MRFAP1L1 | 7.5e-01 | 0.980 |
| CASC4 | 7.5e-01 | 0.980 |
| OGFOD1 | 7.5e-01 | 0.980 |
| ZNF26 | 7.5e-01 | 0.980 |
| PLA2G12A | 7.5e-01 | 0.980 |
| TTYH1 | 7.5e-01 | 0.980 |
| SECISBP2L | 7.5e-01 | 0.980 |
| AMOTL2 | 7.5e-01 | 0.980 |
| DIMT1 | 7.5e-01 | 0.980 |
| FAF1 | 7.5e-01 | 0.980 |
| SERPINH1 | 7.5e-01 | 0.980 |
| SAR1A | 7.5e-01 | 0.980 |
| DDX24 | 7.5e-01 | 0.980 |
| NME3 | 7.5e-01 | 0.980 |
| GNPDA2 | 7.5e-01 | 0.980 |
| GGPS1 | 7.5e-01 | 0.980 |
| ZNF200 | 7.5e-01 | 0.980 |
| SLC12A4 | 7.5e-01 | 0.980 |
| C7orf55 | 7.5e-01 | 0.980 |
| OAT | 7.5e-01 | 0.980 |
| VPRBP | 7.5e-01 | 0.980 |
| DIS3L | 7.5e-01 | 0.980 |
| SCMH1 | 7.5e-01 | 0.980 |
| TMEM192 | 7.5e-01 | 0.980 |
| NCAPD3 | 7.5e-01 | 0.980 |
| PCNX4 | 7.5e-01 | 0.980 |
| KCTD9 | 7.5e-01 | 0.980 |
| QRSL1 | 7.5e-01 | 0.980 |
| SLC5A3 | 7.5e-01 | 0.980 |
| DBN1 | 7.5e-01 | 0.980 |
| ATP6V1G1 | 7.5e-01 | 0.980 |
| SPCS3 | 7.5e-01 | 0.980 |
| MFF | 7.5e-01 | 0.980 |
| ANP32E | 7.5e-01 | 0.980 |
| MZT2A | 7.5e-01 | 0.980 |
| SLC25A29 | 7.5e-01 | 0.980 |
| DUSP18 | 7.5e-01 | 0.980 |
| PPP2R5A | 7.5e-01 | 0.980 |
| ATP1A1 | 7.5e-01 | 0.980 |
| POLR1E | 7.5e-01 | 0.980 |
| TMEM185A | 7.5e-01 | 0.980 |
| SPECC1 | 7.5e-01 | 0.980 |
| HACD1 | 7.5e-01 | 0.980 |
| TPT1-AS1 | 7.5e-01 | 0.980 |
| DDX41 | 7.5e-01 | 0.980 |
| POLDIP2 | 7.5e-01 | 0.980 |
| MRPS18B | 7.5e-01 | 0.980 |
| PRKACA | 7.5e-01 | 0.980 |
| ERBB4 | 7.5e-01 | 0.980 |
| CBR4 | 7.5e-01 | 0.980 |
| GAK | 7.5e-01 | 0.980 |
| UBE2E3 | 7.5e-01 | 0.980 |
| GJC1 | 7.5e-01 | 0.980 |
| GPS2 | 7.5e-01 | 0.980 |
| CCZ1 | 7.5e-01 | 0.980 |
| RNASET2 | 7.5e-01 | 0.980 |
| NUAK1 | 7.5e-01 | 0.980 |
| SRI | 7.6e-01 | 0.980 |
| PRKCSH | 7.6e-01 | 0.980 |
| ANKRA2 | 7.6e-01 | 0.980 |
| DUSP4 | 7.6e-01 | 0.980 |
| TMEM234 | 7.6e-01 | 0.980 |
| DNAJB2 | 7.6e-01 | 0.980 |
| GIGYF2 | 7.6e-01 | 0.980 |
| KHDRBS3 | 7.6e-01 | 0.980 |
| TWF2 | 7.6e-01 | 0.980 |
| HMGN5 | 7.6e-01 | 0.980 |
| RPL27A | 7.6e-01 | 0.980 |
| VPS36 | 7.6e-01 | 0.980 |
| NUAK2 | 7.6e-01 | 0.980 |
| CCNC | 7.6e-01 | 0.980 |
| SEPT1 | 7.6e-01 | 0.980 |
| LRP10 | 7.6e-01 | 0.980 |
| MCUR1 | 7.6e-01 | 0.980 |
| JARID2 | 7.6e-01 | 0.980 |
| RNF175 | 7.6e-01 | 0.980 |
| MEST | 7.6e-01 | 0.980 |
| NPC2 | 7.6e-01 | 0.980 |
| LCOR | 7.6e-01 | 0.980 |
| HINT3 | 7.6e-01 | 0.980 |
| BRD2 | 7.6e-01 | 0.980 |
| PSRC1 | 7.6e-01 | 0.980 |
| MKI67 | 7.6e-01 | 0.980 |
| MYCBP | 7.6e-01 | 0.980 |
| FNTA | 7.6e-01 | 0.980 |
| C4orf27 | 7.6e-01 | 0.980 |
| C11orf1 | 7.6e-01 | 0.980 |
| EIF2B2 | 7.6e-01 | 0.980 |
| SUDS3 | 7.6e-01 | 0.980 |
| MYO1B | 7.6e-01 | 0.980 |
| MINOS1 | 7.6e-01 | 0.980 |
| DAG1 | 7.6e-01 | 0.980 |
| L2HGDH | 7.6e-01 | 0.980 |
| GOLGA8A | 7.6e-01 | 0.980 |
| NCKAP5 | 7.6e-01 | 0.980 |
| NPM3 | 7.6e-01 | 0.980 |
| ACIN1 | 7.6e-01 | 0.980 |
| SH3BGRL3 | 7.6e-01 | 0.980 |
| KTI12 | 7.6e-01 | 0.980 |
| CHD9 | 7.6e-01 | 0.980 |
| CHMP7 | 7.6e-01 | 0.980 |
| RBMX | 7.6e-01 | 0.980 |
| VTI1B | 7.6e-01 | 0.980 |
| VBP1 | 7.6e-01 | 0.980 |
| ZFP14 | 7.6e-01 | 0.980 |
| SRA1 | 7.6e-01 | 0.980 |
| TMEM14A | 7.6e-01 | 0.980 |
| RIMKLB | 7.6e-01 | 0.980 |
| TMEM45A | 7.6e-01 | 0.980 |
| ARF1 | 7.6e-01 | 0.980 |
| RNF170 | 7.6e-01 | 0.980 |
| RPS3A | 7.6e-01 | 0.980 |
| FBRSL1 | 7.6e-01 | 0.980 |
| SEC11C | 7.6e-01 | 0.980 |
| DOCK1 | 7.6e-01 | 0.980 |
| MSTO1 | 7.6e-01 | 0.980 |
| FAM89B | 7.6e-01 | 0.980 |
| EEF1AKMT4 | 7.6e-01 | 0.980 |
| GCN1 | 7.6e-01 | 0.980 |
| CHD1 | 7.6e-01 | 0.980 |
| EIF2S2 | 7.6e-01 | 0.980 |
| AKT1S1 | 7.6e-01 | 0.980 |
| DYNLT1 | 7.6e-01 | 0.980 |
| CEP55 | 7.6e-01 | 0.980 |
| EPHA4 | 7.6e-01 | 0.980 |
| HOOK3 | 7.6e-01 | 0.980 |
| CORO1B | 7.6e-01 | 0.980 |
| TAOK1 | 7.6e-01 | 0.980 |
| ERRFI1 | 7.6e-01 | 0.980 |
| MPST | 7.6e-01 | 0.980 |
| TOP3B | 7.6e-01 | 0.980 |
| ZFYVE27 | 7.6e-01 | 0.980 |
| HHLA3 | 7.6e-01 | 0.980 |
| LINC00476 | 7.7e-01 | 0.980 |
| HSPA4L | 7.7e-01 | 0.980 |
| CH17-118O6.2 | 7.7e-01 | 0.980 |
| TRAPPC12 | 7.7e-01 | 0.980 |
| CTB-89H12.4 | 7.7e-01 | 0.980 |
| ZNF300 | 7.7e-01 | 0.980 |
| LPAR4 | 7.7e-01 | 0.980 |
| C14orf142 | 7.7e-01 | 0.980 |
| TMEM107 | 7.7e-01 | 0.980 |
| OPA1 | 7.7e-01 | 0.980 |
| RPL39 | 7.7e-01 | 0.980 |
| PCNT | 7.7e-01 | 0.980 |
| TMED1 | 7.7e-01 | 0.980 |
| SIN3A | 7.7e-01 | 0.980 |
| DNAJC25 | 7.7e-01 | 0.980 |
| BAX | 7.7e-01 | 0.980 |
| SRSF11 | 7.7e-01 | 0.980 |
| SOX11 | 7.7e-01 | 0.980 |
| ZNF451 | 7.7e-01 | 0.980 |
| MED10 | 7.7e-01 | 0.980 |
| VPS18 | 7.7e-01 | 0.980 |
| FAM107B | 7.7e-01 | 0.980 |
| GATAD1 | 7.7e-01 | 0.980 |
| PKIA | 7.7e-01 | 0.980 |
| POLR2J3 | 7.7e-01 | 0.980 |
| RBM5 | 7.7e-01 | 0.980 |
| TTLL4 | 7.7e-01 | 0.980 |
| CNBD2 | 7.7e-01 | 0.980 |
| AP5M1 | 7.7e-01 | 0.980 |
| ZCCHC9 | 7.7e-01 | 0.980 |
| ATG101 | 7.7e-01 | 0.980 |
| HPS4 | 7.7e-01 | 0.980 |
| ZNF446 | 7.7e-01 | 0.980 |
| AR | 7.7e-01 | 0.980 |
| FXR1 | 7.7e-01 | 0.980 |
| DIAPH1 | 7.7e-01 | 0.980 |
| PAFAH1B1 | 7.7e-01 | 0.980 |
| MRPL41 | 7.7e-01 | 0.980 |
| CPXM1 | 7.7e-01 | 0.980 |
| GTF3C1 | 7.7e-01 | 0.980 |
| ZBTB38 | 7.7e-01 | 0.980 |
| BABAM1 | 7.7e-01 | 0.980 |
| PBK | 7.7e-01 | 0.980 |
| RPP38 | 7.7e-01 | 0.980 |
| SPATA5L1 | 7.7e-01 | 0.980 |
| CEP72 | 7.7e-01 | 0.980 |
| CCDC28B | 7.7e-01 | 0.980 |
| PXDN | 7.7e-01 | 0.980 |
| ATXN7L3B | 7.7e-01 | 0.980 |
| TM9SF3 | 7.7e-01 | 0.980 |
| SPATA6 | 7.7e-01 | 0.980 |
| DBT | 7.7e-01 | 0.980 |
| JMJD1C | 7.7e-01 | 0.980 |
| LARP4 | 7.7e-01 | 0.980 |
| ZBTB10 | 7.7e-01 | 0.980 |
| GPALPP1 | 7.7e-01 | 0.980 |
| TOM1L1 | 7.7e-01 | 0.990 |
| PRRC1 | 7.7e-01 | 0.990 |
| NOL7 | 7.7e-01 | 0.990 |
| ZW10 | 7.7e-01 | 0.990 |
| CCNY | 7.7e-01 | 0.990 |
| YJEFN3 | 7.7e-01 | 0.990 |
| HSP90B1 | 7.7e-01 | 0.990 |
| NLGN2 | 7.7e-01 | 0.990 |
| NIT1 | 7.7e-01 | 0.990 |
| YWHAZ | 7.7e-01 | 0.990 |
| TSPAN13 | 7.7e-01 | 0.990 |
| CHCHD6 | 7.7e-01 | 0.990 |
| NUS1 | 7.7e-01 | 0.990 |
| TOX4 | 7.7e-01 | 0.990 |
| ZNF138 | 7.7e-01 | 0.990 |
| MOCS2 | 7.7e-01 | 0.990 |
| LINC00461 | 7.7e-01 | 0.990 |
| PPP3CC | 7.7e-01 | 0.990 |
| DNAJC5 | 7.8e-01 | 0.990 |
| C9orf114 | 7.8e-01 | 0.990 |
| TPBG | 7.8e-01 | 0.990 |
| ZNF33B | 7.8e-01 | 0.990 |
| NDUFA11 | 7.8e-01 | 0.990 |
| SMIM3 | 7.8e-01 | 0.990 |
| CCNK | 7.8e-01 | 0.990 |
| C11orf68 | 7.8e-01 | 0.990 |
| ARMC6 | 7.8e-01 | 0.990 |
| NEK6 | 7.8e-01 | 0.990 |
| PHF19 | 7.8e-01 | 0.990 |
| KLHL28 | 7.8e-01 | 0.990 |
| SDHAF2 | 7.8e-01 | 0.990 |
| GTPBP6 | 7.8e-01 | 0.990 |
| DAD1 | 7.8e-01 | 0.990 |
| BCL2L13 | 7.8e-01 | 0.990 |
| SLC1A3 | 7.8e-01 | 0.990 |
| XPOT | 7.8e-01 | 0.990 |
| ASPSCR1 | 7.8e-01 | 0.990 |
| ZNF573 | 7.8e-01 | 0.990 |
| PRKRIP1 | 7.8e-01 | 0.990 |
| VPS13A | 7.8e-01 | 0.990 |
| PRPS1 | 7.8e-01 | 0.990 |
| ZNF821 | 7.8e-01 | 0.990 |
| NSDHL | 7.8e-01 | 0.990 |
| SNF8 | 7.8e-01 | 0.990 |
| COX6A1 | 7.8e-01 | 0.990 |
| AC005562.1 | 7.8e-01 | 0.990 |
| SETDB1 | 7.8e-01 | 0.990 |
| MTRF1 | 7.8e-01 | 0.990 |
| CCM2 | 7.8e-01 | 0.990 |
| MRPL51 | 7.8e-01 | 0.990 |
| SLC25A1 | 7.8e-01 | 0.990 |
| MRPS14 | 7.8e-01 | 0.990 |
| DNAJC7 | 7.8e-01 | 0.990 |
| SNAPC1 | 7.8e-01 | 0.990 |
| SNX11 | 7.8e-01 | 0.990 |
| AC068491.1 | 7.8e-01 | 0.990 |
| CHM | 7.8e-01 | 0.990 |
| EFCC1 | 7.8e-01 | 0.990 |
| MEIS3 | 7.8e-01 | 0.990 |
| MAP1A | 7.8e-01 | 0.990 |
| ITGAE | 7.8e-01 | 0.990 |
| HAUS3 | 7.8e-01 | 0.990 |
| HNRNPR | 7.8e-01 | 0.990 |
| YAE1D1 | 7.8e-01 | 0.990 |
| NMNAT1 | 7.8e-01 | 0.990 |
| MLF2 | 7.8e-01 | 0.990 |
| TCEAL1 | 7.8e-01 | 0.990 |
| MKS1 | 7.8e-01 | 0.990 |
| C8orf76 | 7.8e-01 | 0.990 |
| BCAS4 | 7.8e-01 | 0.990 |
| SLC35A3 | 7.8e-01 | 0.990 |
| SLC35B4 | 7.8e-01 | 0.990 |
| ALAS1 | 7.8e-01 | 0.990 |
| ATF6 | 7.8e-01 | 0.990 |
| SEPN1 | 7.8e-01 | 0.990 |
| MORF4L1 | 7.8e-01 | 0.990 |
| STAG1 | 7.8e-01 | 0.990 |
| FAM64A | 7.8e-01 | 0.990 |
| HSF2 | 7.8e-01 | 0.990 |
| ENOPH1 | 7.8e-01 | 0.990 |
| CDKN2AIP | 7.8e-01 | 0.990 |
| TTC37 | 7.8e-01 | 0.990 |
| SND1 | 7.9e-01 | 0.990 |
| GNG2 | 7.9e-01 | 0.990 |
| ZDHHC24 | 7.9e-01 | 0.990 |
| PTCH1 | 7.9e-01 | 0.990 |
| RP11-293G6.1 | 7.9e-01 | 0.990 |
| RPS16 | 7.9e-01 | 0.990 |
| DPYSL3 | 7.9e-01 | 0.990 |
| ABCC5 | 7.9e-01 | 0.990 |
| C2orf47 | 7.9e-01 | 0.990 |
| FBL | 7.9e-01 | 0.990 |
| ARL2 | 7.9e-01 | 0.990 |
| HOMER1 | 7.9e-01 | 0.990 |
| RALB | 7.9e-01 | 0.990 |
| CCDC88C | 7.9e-01 | 0.990 |
| TLK1 | 7.9e-01 | 0.990 |
| SLC25A14 | 7.9e-01 | 0.990 |
| PIP5K1A | 7.9e-01 | 0.990 |
| UBXN2B | 7.9e-01 | 0.990 |
| ITFG2 | 7.9e-01 | 0.990 |
| FSTL1 | 7.9e-01 | 0.990 |
| BRD4 | 7.9e-01 | 0.990 |
| PGAM5 | 7.9e-01 | 0.990 |
| MPG | 7.9e-01 | 0.990 |
| MRPL22 | 7.9e-01 | 0.990 |
| SLC7A6 | 7.9e-01 | 0.990 |
| SASH1 | 7.9e-01 | 0.990 |
| MT-CO3 | 7.9e-01 | 0.990 |
| SEP15 | 7.9e-01 | 0.990 |
| SAV1 | 7.9e-01 | 0.990 |
| FAM60A | 7.9e-01 | 0.990 |
| ASB8 | 7.9e-01 | 0.990 |
| NT5C2 | 7.9e-01 | 0.990 |
| MFSD2A | 7.9e-01 | 0.990 |
| IKBIP | 7.9e-01 | 0.990 |
| PDSS1 | 7.9e-01 | 0.990 |
| RPL30 | 7.9e-01 | 0.990 |
| CABIN1 | 7.9e-01 | 0.990 |
| PRR14 | 7.9e-01 | 0.990 |
| DAPK1 | 7.9e-01 | 0.990 |
| SPATS2 | 7.9e-01 | 0.990 |
| XRCC4 | 7.9e-01 | 0.990 |
| C12orf76 | 7.9e-01 | 0.990 |
| GALNT2 | 7.9e-01 | 0.990 |
| UPF2 | 7.9e-01 | 0.990 |
| NABP2 | 7.9e-01 | 0.990 |
| RAD21 | 7.9e-01 | 0.990 |
| MYO9A | 7.9e-01 | 0.990 |
| PSMD14 | 7.9e-01 | 0.990 |
| FAM234A | 7.9e-01 | 0.990 |
| WDR74 | 7.9e-01 | 0.990 |
| GALM | 7.9e-01 | 0.990 |
| RPL41 | 7.9e-01 | 0.990 |
| KDM5A | 7.9e-01 | 0.990 |
| NKAIN4 | 7.9e-01 | 0.990 |
| ZMAT2 | 7.9e-01 | 0.990 |
| SPC24 | 7.9e-01 | 0.990 |
| SAP30L | 7.9e-01 | 0.990 |
| CCNJ | 7.9e-01 | 0.990 |
| MT-CO2 | 7.9e-01 | 0.990 |
| UQCRHL | 7.9e-01 | 0.990 |
| NHSL2 | 7.9e-01 | 0.990 |
| PTPN9 | 7.9e-01 | 0.990 |
| TBRG4 | 7.9e-01 | 0.990 |
| BRD3 | 7.9e-01 | 0.990 |
| ANKRD13B | 7.9e-01 | 0.990 |
| NUDT14 | 7.9e-01 | 0.990 |
| LHPP | 7.9e-01 | 0.990 |
| DGCR8 | 7.9e-01 | 0.990 |
| BRF1 | 7.9e-01 | 0.990 |
| POLR2M | 7.9e-01 | 0.990 |
| PPP3CA | 7.9e-01 | 0.990 |
| IKZF4 | 7.9e-01 | 0.990 |
| LSS | 7.9e-01 | 0.990 |
| RNF150 | 7.9e-01 | 0.990 |
| HOXB4 | 7.9e-01 | 0.990 |
| C19orf53 | 7.9e-01 | 0.990 |
| ARID1A | 8.0e-01 | 0.990 |
| C16orf45 | 8.0e-01 | 0.990 |
| PSMA2 | 8.0e-01 | 0.990 |
| BOK | 8.0e-01 | 0.990 |
| MT-CYB | 8.0e-01 | 0.990 |
| MLLT1 | 8.0e-01 | 0.990 |
| IMPA1 | 8.0e-01 | 0.990 |
| RASSF2 | 8.0e-01 | 0.990 |
| LLGL1 | 8.0e-01 | 0.990 |
| AHSA1 | 8.0e-01 | 0.990 |
| DHX8 | 8.0e-01 | 0.990 |
| PPP4C | 8.0e-01 | 0.990 |
| TNKS1BP1 | 8.0e-01 | 0.990 |
| LRWD1 | 8.0e-01 | 0.990 |
| LRIG1 | 8.0e-01 | 0.990 |
| EID1 | 8.0e-01 | 0.990 |
| STMN2 | 8.0e-01 | 0.990 |
| CALD1 | 8.0e-01 | 0.990 |
| EVL | 8.0e-01 | 0.990 |
| PARD6G | 8.0e-01 | 0.990 |
| PTPRF | 8.0e-01 | 0.990 |
| MRPL35 | 8.0e-01 | 0.990 |
| RP13-942N8.1 | 8.0e-01 | 0.990 |
| RNF114 | 8.0e-01 | 0.990 |
| PTPN1 | 8.0e-01 | 0.990 |
| BBX | 8.0e-01 | 0.990 |
| APEX1 | 8.0e-01 | 0.990 |
| MAPRE3 | 8.0e-01 | 0.990 |
| RBM6 | 8.0e-01 | 0.990 |
| ZBTB40 | 8.0e-01 | 0.990 |
| PHYH | 8.0e-01 | 0.990 |
| RASL10B | 8.0e-01 | 0.990 |
| C5orf51 | 8.0e-01 | 0.990 |
| HS3ST3B1 | 8.0e-01 | 0.990 |
| MSRB2 | 8.0e-01 | 0.990 |
| PRDX1 | 8.0e-01 | 0.990 |
| CCDC102A | 8.0e-01 | 0.990 |
| RHOB | 8.0e-01 | 0.990 |
| GRB10 | 8.0e-01 | 0.990 |
| DHTKD1 | 8.0e-01 | 0.990 |
| GLMN | 8.0e-01 | 0.990 |
| ZNF574 | 8.0e-01 | 0.990 |
| GALNS | 8.0e-01 | 0.990 |
| RCN2 | 8.0e-01 | 0.990 |
| BRWD1 | 8.0e-01 | 0.990 |
| TCERG1 | 8.0e-01 | 0.990 |
| ZCCHC3 | 8.0e-01 | 0.990 |
| PPOX | 8.0e-01 | 0.990 |
| KIF9 | 8.0e-01 | 0.990 |
| CHCHD2 | 8.0e-01 | 0.990 |
| STX8 | 8.0e-01 | 0.990 |
| G6PD | 8.0e-01 | 0.990 |
| DTWD1 | 8.0e-01 | 0.990 |
| ELL | 8.0e-01 | 0.990 |
| KARS | 8.0e-01 | 0.990 |
| GNAZ | 8.0e-01 | 0.990 |
| SF3B2 | 8.0e-01 | 0.990 |
| ARMC8 | 8.0e-01 | 0.990 |
| LMO4 | 8.0e-01 | 0.990 |
| HEMK1 | 8.0e-01 | 0.990 |
| LARP1B | 8.0e-01 | 0.990 |
| KIF1C | 8.0e-01 | 0.990 |
| MSH3 | 8.0e-01 | 0.990 |
| CELSR2 | 8.0e-01 | 0.990 |
| TSHZ2 | 8.0e-01 | 0.990 |
| WDR77 | 8.0e-01 | 0.990 |
| MESDC2 | 8.0e-01 | 0.990 |
| PPP6R3 | 8.0e-01 | 0.990 |
| LIN28B | 8.0e-01 | 0.990 |
| CIAO1 | 8.0e-01 | 0.990 |
| CDH4 | 8.0e-01 | 0.990 |
| C6orf120 | 8.0e-01 | 0.990 |
| INTS14 | 8.0e-01 | 0.990 |
| PRIM2 | 8.0e-01 | 0.990 |
| CSRP1 | 8.0e-01 | 0.990 |
| SEC14L1 | 8.0e-01 | 0.990 |
| RNF34 | 8.0e-01 | 0.990 |
| CH17-340M24.3 | 8.0e-01 | 0.990 |
| NELFCD | 8.0e-01 | 0.990 |
| TSTD2 | 8.0e-01 | 0.990 |
| TMEM63B | 8.0e-01 | 0.990 |
| WFIKKN1 | 8.0e-01 | 0.990 |
| NHP2 | 8.0e-01 | 0.990 |
| CDK5RAP3 | 8.0e-01 | 0.990 |
| MRPL15 | 8.0e-01 | 0.990 |
| TXNIP | 8.0e-01 | 0.990 |
| WBP2 | 8.0e-01 | 0.990 |
| ICT1 | 8.0e-01 | 0.990 |
| RIOK3 | 8.0e-01 | 0.990 |
| FANCG | 8.0e-01 | 0.990 |
| COL4A2 | 8.0e-01 | 0.990 |
| ASAH1 | 8.0e-01 | 0.990 |
| ZNF280D | 8.0e-01 | 0.990 |
| ZNF121 | 8.0e-01 | 0.990 |
| CDK12 | 8.0e-01 | 0.990 |
| SORD | 8.0e-01 | 0.990 |
| KRT10 | 8.0e-01 | 0.990 |
| PSTK | 8.0e-01 | 0.990 |
| RAB14 | 8.0e-01 | 0.990 |
| KIF22 | 8.0e-01 | 0.990 |
| RPL17 | 8.1e-01 | 0.990 |
| DLST | 8.1e-01 | 0.990 |
| B4GALT4 | 8.1e-01 | 0.990 |
| PSMF1 | 8.1e-01 | 0.990 |
| FAM199X | 8.1e-01 | 0.990 |
| IARS | 8.1e-01 | 0.990 |
| FAM110A | 8.1e-01 | 0.990 |
| ATP5J2 | 8.1e-01 | 0.990 |
| ZEB2 | 8.1e-01 | 0.990 |
| PFDN4 | 8.1e-01 | 0.990 |
| GTPBP8 | 8.1e-01 | 0.990 |
| JAM3 | 8.1e-01 | 0.990 |
| TTLL7 | 8.1e-01 | 0.990 |
| FAM172A | 8.1e-01 | 0.990 |
| TSC2 | 8.1e-01 | 0.990 |
| WDR4 | 8.1e-01 | 0.990 |
| MELK | 8.1e-01 | 0.990 |
| BORA | 8.1e-01 | 0.990 |
| TMEM131 | 8.1e-01 | 0.990 |
| SSU72 | 8.1e-01 | 0.990 |
| NAT9 | 8.1e-01 | 0.990 |
| TUBB2B | 8.1e-01 | 0.990 |
| NDN | 8.1e-01 | 0.990 |
| AK3 | 8.1e-01 | 0.990 |
| CNDP2 | 8.1e-01 | 0.990 |
| MSI1 | 8.1e-01 | 0.990 |
| MAPK8 | 8.1e-01 | 0.990 |
| TSTA3 | 8.1e-01 | 0.990 |
| TMEM209 | 8.1e-01 | 0.990 |
| AARSD1 | 8.1e-01 | 0.990 |
| ZNF621 | 8.1e-01 | 0.990 |
| MTMR9 | 8.1e-01 | 0.990 |
| VDAC3 | 8.1e-01 | 0.990 |
| NUP205 | 8.1e-01 | 0.990 |
| MDM2 | 8.1e-01 | 0.990 |
| NCALD | 8.1e-01 | 0.990 |
| ATP6V1H | 8.1e-01 | 0.990 |
| CCDC140 | 8.1e-01 | 0.990 |
| TPT1 | 8.1e-01 | 0.990 |
| SAT1 | 8.1e-01 | 0.990 |
| SCRN3 | 8.1e-01 | 0.990 |
| SH3BP4 | 8.1e-01 | 0.990 |
| PANK1 | 8.1e-01 | 0.990 |
| PARP11 | 8.1e-01 | 0.990 |
| MKKS | 8.1e-01 | 0.990 |
| CLASRP | 8.1e-01 | 0.990 |
| WDHD1 | 8.1e-01 | 0.990 |
| KPNA3 | 8.1e-01 | 0.990 |
| ASB13 | 8.1e-01 | 0.990 |
| PPP4R3B | 8.1e-01 | 0.990 |
| FEM1A | 8.1e-01 | 0.990 |
| ELOVL5 | 8.1e-01 | 0.990 |
| GOLGA8B | 8.1e-01 | 0.990 |
| ATXN1L | 8.1e-01 | 0.990 |
| FASTK | 8.1e-01 | 0.990 |
| FKBP4 | 8.1e-01 | 0.990 |
| C12orf57 | 8.1e-01 | 0.990 |
| TRMT61B | 8.1e-01 | 0.990 |
| KCTD20 | 8.1e-01 | 0.990 |
| ASCC1 | 8.1e-01 | 0.990 |
| SPHK2 | 8.1e-01 | 0.990 |
| NTN1 | 8.1e-01 | 0.990 |
| ZNF770 | 8.1e-01 | 0.990 |
| MFSD10 | 8.1e-01 | 0.990 |
| KIAA2013 | 8.1e-01 | 0.990 |
| ALG6 | 8.1e-01 | 0.990 |
| SEC61A2 | 8.1e-01 | 0.990 |
| UCHL5 | 8.1e-01 | 0.990 |
| MKNK1 | 8.1e-01 | 0.990 |
| ACER3 | 8.1e-01 | 0.990 |
| AIF1L | 8.1e-01 | 0.990 |
| EIF5 | 8.1e-01 | 0.990 |
| CAAP1 | 8.1e-01 | 0.990 |
| YWHAB | 8.1e-01 | 0.990 |
| RERE | 8.1e-01 | 0.990 |
| MTA3 | 8.1e-01 | 0.990 |
| NME6 | 8.1e-01 | 0.990 |
| NTPCR | 8.1e-01 | 0.990 |
| SGCB | 8.1e-01 | 0.990 |
| HOXB3 | 8.1e-01 | 0.990 |
| HIGD1A | 8.1e-01 | 0.990 |
| SMARCE1 | 8.1e-01 | 0.990 |
| COQ5 | 8.1e-01 | 0.990 |
| SUCLA2 | 8.1e-01 | 0.990 |
| JAKMIP2 | 8.1e-01 | 0.990 |
| SFSWAP | 8.2e-01 | 0.990 |
| DNAJC8 | 8.2e-01 | 0.990 |
| UBR5 | 8.2e-01 | 0.990 |
| EIF4E2 | 8.2e-01 | 0.990 |
| ATG7 | 8.2e-01 | 0.990 |
| POLR3A | 8.2e-01 | 0.990 |
| YWHAG | 8.2e-01 | 0.990 |
| RAB22A | 8.2e-01 | 0.990 |
| RABEP1 | 8.2e-01 | 0.990 |
| ZNF430 | 8.2e-01 | 0.990 |
| TERF2 | 8.2e-01 | 0.990 |
| PHB2 | 8.2e-01 | 0.990 |
| MTUS1 | 8.2e-01 | 0.990 |
| CANX | 8.2e-01 | 0.990 |
| ATP6V0A2 | 8.2e-01 | 0.990 |
| SLX4IP | 8.2e-01 | 0.990 |
| SNX18 | 8.2e-01 | 0.990 |
| PAM | 8.2e-01 | 0.990 |
| MLLT10 | 8.2e-01 | 0.990 |
| ZNF587 | 8.2e-01 | 0.990 |
| DNPH1 | 8.2e-01 | 0.990 |
| VPS52 | 8.2e-01 | 0.990 |
| ETFB | 8.2e-01 | 0.990 |
| MKRN2 | 8.2e-01 | 0.990 |
| CHPT1 | 8.2e-01 | 0.990 |
| MDM4 | 8.2e-01 | 0.990 |
| GAS2L1 | 8.2e-01 | 0.990 |
| ARHGEF17 | 8.2e-01 | 0.990 |
| ECI2 | 8.2e-01 | 0.990 |
| NVL | 8.2e-01 | 0.990 |
| STMN1 | 8.2e-01 | 0.990 |
| WWP2 | 8.2e-01 | 0.990 |
| TOR1AIP1 | 8.2e-01 | 0.990 |
| NDUFS4 | 8.2e-01 | 0.990 |
| H1FX | 8.2e-01 | 0.990 |
| OBSL1 | 8.2e-01 | 0.990 |
| PSME1 | 8.2e-01 | 0.990 |
| PMAIP1 | 8.2e-01 | 0.990 |
| BBIP1 | 8.2e-01 | 0.990 |
| CEP95 | 8.2e-01 | 0.990 |
| VPS72 | 8.2e-01 | 0.990 |
| SCD5 | 8.2e-01 | 0.990 |
| FAM173B | 8.2e-01 | 0.990 |
| MRPS2 | 8.2e-01 | 0.990 |
| MED19 | 8.2e-01 | 0.990 |
| ACSF3 | 8.2e-01 | 0.990 |
| LRTOMT | 8.2e-01 | 0.990 |
| INTS8 | 8.2e-01 | 0.990 |
| EPRS | 8.2e-01 | 0.990 |
| SLC3A2 | 8.2e-01 | 0.990 |
| SOBP | 8.2e-01 | 0.990 |
| FAM162A | 8.2e-01 | 0.990 |
| CMIP | 8.2e-01 | 0.990 |
| AGFG1 | 8.2e-01 | 0.990 |
| UBFD1 | 8.2e-01 | 0.990 |
| LATS1 | 8.2e-01 | 0.990 |
| SYDE1 | 8.2e-01 | 0.990 |
| CCDC88A | 8.2e-01 | 0.990 |
| NDUFAB1 | 8.2e-01 | 0.990 |
| LAMTOR5 | 8.2e-01 | 0.990 |
| CDIP1 | 8.2e-01 | 0.990 |
| COX7B | 8.2e-01 | 0.990 |
| ZNF577 | 8.2e-01 | 0.990 |
| ATP9A | 8.2e-01 | 0.990 |
| AASDHPPT | 8.2e-01 | 0.990 |
| MRPS15 | 8.2e-01 | 0.990 |
| SLC2A1 | 8.3e-01 | 0.990 |
| KCNIP4 | 8.3e-01 | 0.990 |
| EIF2AK2 | 8.3e-01 | 0.990 |
| FANCD2 | 8.3e-01 | 0.990 |
| ISYNA1 | 8.3e-01 | 0.990 |
| COX5B | 8.3e-01 | 0.990 |
| UBE2Z | 8.3e-01 | 0.990 |
| TMEM30A | 8.3e-01 | 0.990 |
| CTBP1 | 8.3e-01 | 0.990 |
| ATP6V1A | 8.3e-01 | 0.990 |
| CTD-2037K23.2 | 8.3e-01 | 0.990 |
| PPP2R5C | 8.3e-01 | 0.990 |
| PPP1R9A | 8.3e-01 | 0.990 |
| VTA1 | 8.3e-01 | 0.990 |
| CALR | 8.3e-01 | 0.990 |
| APPL1 | 8.3e-01 | 0.990 |
| SLC35B3 | 8.3e-01 | 0.990 |
| GTF2B | 8.3e-01 | 0.990 |
| SLC25A11 | 8.3e-01 | 0.990 |
| NDUFV3 | 8.3e-01 | 0.990 |
| PPIC | 8.3e-01 | 0.990 |
| FADS1 | 8.3e-01 | 0.990 |
| TWNK | 8.3e-01 | 0.990 |
| CPSF6 | 8.3e-01 | 0.990 |
| DBNL | 8.3e-01 | 0.990 |
| RPS15A | 8.3e-01 | 0.990 |
| IRF2BP1 | 8.3e-01 | 0.990 |
| TMEM203 | 8.3e-01 | 0.990 |
| LRRC27 | 8.3e-01 | 0.990 |
| ANAPC4 | 8.3e-01 | 0.990 |
| RIPK2 | 8.3e-01 | 0.990 |
| TMEM55B | 8.3e-01 | 0.990 |
| PARN | 8.3e-01 | 0.990 |
| CLIC4 | 8.3e-01 | 0.990 |
| RAB11A | 8.3e-01 | 0.990 |
| PPIP5K2 | 8.3e-01 | 0.990 |
| SERTAD3 | 8.3e-01 | 0.990 |
| PRPF6 | 8.3e-01 | 0.990 |
| EXOC6 | 8.3e-01 | 0.990 |
| TOP1 | 8.3e-01 | 0.990 |
| MAPK6 | 8.3e-01 | 0.990 |
| EPB41L5 | 8.3e-01 | 0.990 |
| RAB3C | 8.3e-01 | 0.990 |
| CAV2 | 8.3e-01 | 0.990 |
| POLR3GL | 8.3e-01 | 0.990 |
| CERS1 | 8.3e-01 | 0.990 |
| SLC39A9 | 8.3e-01 | 0.990 |
| UNK | 8.3e-01 | 0.990 |
| DAZAP1 | 8.3e-01 | 0.990 |
| RAP1B | 8.3e-01 | 0.990 |
| VAPA | 8.3e-01 | 0.990 |
| RAB9A | 8.3e-01 | 0.990 |
| C21orf33 | 8.3e-01 | 0.990 |
| POLR3H | 8.3e-01 | 0.990 |
| BEX3 | 8.3e-01 | 0.990 |
| DEXI | 8.3e-01 | 0.990 |
| ATP6V1B2 | 8.3e-01 | 0.990 |
| STX3 | 8.3e-01 | 0.990 |
| PSMA3 | 8.3e-01 | 0.990 |
| ZFAND1 | 8.3e-01 | 0.990 |
| CARM1 | 8.3e-01 | 0.990 |
| WDR89 | 8.3e-01 | 0.990 |
| EML4 | 8.3e-01 | 0.990 |
| NIPBL | 8.3e-01 | 0.990 |
| NFU1 | 8.3e-01 | 0.990 |
| CEP250 | 8.3e-01 | 0.990 |
| PHF12 | 8.3e-01 | 0.990 |
| ZNF480 | 8.3e-01 | 0.990 |
| PACSIN3 | 8.4e-01 | 0.990 |
| NFXL1 | 8.4e-01 | 0.990 |
| THAP3 | 8.4e-01 | 0.990 |
| RWDD4 | 8.4e-01 | 0.990 |
| SLC35C2 | 8.4e-01 | 0.990 |
| ZNHIT6 | 8.4e-01 | 0.990 |
| MED8 | 8.4e-01 | 0.990 |
| FBXO45 | 8.4e-01 | 0.990 |
| STK33 | 8.4e-01 | 0.990 |
| DYNC2LI1 | 8.4e-01 | 0.990 |
| SFRP1 | 8.4e-01 | 0.990 |
| RPS4X | 8.4e-01 | 0.990 |
| VAMP7 | 8.4e-01 | 0.990 |
| UBXN6 | 8.4e-01 | 0.990 |
| IER3IP1 | 8.4e-01 | 0.990 |
| KLHL20 | 8.4e-01 | 0.990 |
| UBTF | 8.4e-01 | 0.990 |
| PAX6 | 8.4e-01 | 0.990 |
| PAPOLA | 8.4e-01 | 0.990 |
| NT5C | 8.4e-01 | 0.990 |
| PTX3 | 8.4e-01 | 0.990 |
| PCGF6 | 8.4e-01 | 0.990 |
| RPS12 | 8.4e-01 | 0.990 |
| KIAA0114 | 8.4e-01 | 0.990 |
| BPTF | 8.4e-01 | 0.990 |
| SSRP1 | 8.4e-01 | 0.990 |
| CNP | 8.4e-01 | 0.990 |
| NFKBIE | 8.4e-01 | 0.990 |
| GTF3C4 | 8.4e-01 | 0.990 |
| CTC-228N24.3 | 8.4e-01 | 0.990 |
| VEPH1 | 8.4e-01 | 0.990 |
| RGS2 | 8.4e-01 | 0.990 |
| UBAP2L | 8.4e-01 | 0.990 |
| POU3F3 | 8.4e-01 | 0.990 |
| MRPS18A | 8.4e-01 | 0.990 |
| ZNF140 | 8.4e-01 | 0.990 |
| AC018730.1 | 8.4e-01 | 0.990 |
| NCBP3 | 8.4e-01 | 0.990 |
| XPR1 | 8.4e-01 | 0.990 |
| PEX13 | 8.4e-01 | 0.990 |
| B4GALT7 | 8.4e-01 | 0.990 |
| PCF11 | 8.4e-01 | 0.990 |
| CCDC94 | 8.4e-01 | 0.990 |
| ST3GAL3 | 8.4e-01 | 0.990 |
| LYPLA1 | 8.4e-01 | 0.990 |
| SRSF3 | 8.4e-01 | 0.990 |
| TRIAP1 | 8.4e-01 | 0.990 |
| CDC26 | 8.4e-01 | 0.990 |
| OFD1 | 8.4e-01 | 0.990 |
| LSM8 | 8.4e-01 | 0.990 |
| PCGF2 | 8.4e-01 | 0.990 |
| YARS2 | 8.4e-01 | 0.990 |
| VEZT | 8.4e-01 | 0.990 |
| NDUFA1 | 8.4e-01 | 0.990 |
| PHF21B | 8.4e-01 | 0.990 |
| MSRB3 | 8.4e-01 | 0.990 |
| TAX1BP3 | 8.4e-01 | 0.990 |
| ERP44 | 8.4e-01 | 0.990 |
| SACS | 8.4e-01 | 0.990 |
| YIPF2 | 8.4e-01 | 0.990 |
| DIRC2 | 8.4e-01 | 0.990 |
| RPL9 | 8.4e-01 | 0.990 |
| CSNK1G3 | 8.4e-01 | 0.990 |
| HNRNPA1 | 8.4e-01 | 0.990 |
| IMMP1L | 8.4e-01 | 0.990 |
| UQCRB | 8.4e-01 | 0.990 |
| CDPF1 | 8.4e-01 | 0.990 |
| SKIV2L2 | 8.4e-01 | 0.990 |
| ISCU | 8.4e-01 | 0.990 |
| DDX49 | 8.4e-01 | 0.990 |
| GSTA4 | 8.4e-01 | 0.990 |
| MXD3 | 8.4e-01 | 0.990 |
| RASSF4 | 8.4e-01 | 0.990 |
| SEC23IP | 8.4e-01 | 0.990 |
| KIF3B | 8.4e-01 | 0.990 |
| RAP1A | 8.4e-01 | 0.990 |
| GPATCH3 | 8.4e-01 | 0.990 |
| C3orf17 | 8.4e-01 | 0.990 |
| SPCS2 | 8.4e-01 | 0.990 |
| CHMP1A | 8.4e-01 | 0.990 |
| PSMD1 | 8.4e-01 | 0.990 |
| ASGR1 | 8.4e-01 | 0.990 |
| CTNNA1 | 8.4e-01 | 0.990 |
| SIN3B | 8.4e-01 | 0.990 |
| FXR2 | 8.4e-01 | 0.990 |
| FZD9 | 8.4e-01 | 0.990 |
| MTX2 | 8.4e-01 | 0.990 |
| GPR137 | 8.4e-01 | 0.990 |
| RARS | 8.5e-01 | 0.990 |
| ZEB1 | 8.5e-01 | 0.990 |
| MRS2 | 8.5e-01 | 0.990 |
| GSPT1 | 8.5e-01 | 0.990 |
| SLC25A13 | 8.5e-01 | 0.990 |
| PIGF | 8.5e-01 | 0.990 |
| BAG6 | 8.5e-01 | 0.990 |
| RPL8 | 8.5e-01 | 0.990 |
| ARHGAP12 | 8.5e-01 | 0.990 |
| ZNF768 | 8.5e-01 | 0.990 |
| TTC5 | 8.5e-01 | 0.990 |
| ARIH1 | 8.5e-01 | 0.990 |
| CHMP5 | 8.5e-01 | 0.990 |
| COPRS | 8.5e-01 | 0.990 |
| SMIM7 | 8.5e-01 | 0.990 |
| DIO3OS | 8.5e-01 | 0.990 |
| SNX12 | 8.5e-01 | 0.990 |
| DHX16 | 8.5e-01 | 0.990 |
| RAI14 | 8.5e-01 | 0.990 |
| KIAA0586 | 8.5e-01 | 0.990 |
| RP11-6N17.4 | 8.5e-01 | 0.990 |
| CXorf40B | 8.5e-01 | 0.990 |
| AGO1 | 8.5e-01 | 0.990 |
| EIF4H | 8.5e-01 | 0.990 |
| UBR4 | 8.5e-01 | 0.990 |
| DENND5B | 8.5e-01 | 0.990 |
| PKN1 | 8.5e-01 | 0.990 |
| EXOC6B | 8.5e-01 | 0.990 |
| GSPT2 | 8.5e-01 | 0.990 |
| C16orf70 | 8.5e-01 | 0.990 |
| HMGXB3 | 8.5e-01 | 0.990 |
| FZD10 | 8.5e-01 | 0.990 |
| PPP2R5D | 8.5e-01 | 0.990 |
| ARHGEF1 | 8.5e-01 | 0.990 |
| SRGAP3 | 8.5e-01 | 0.990 |
| ZNF688 | 8.5e-01 | 0.990 |
| TARS2 | 8.5e-01 | 0.990 |
| MRPL3 | 8.5e-01 | 0.990 |
| EMC1 | 8.5e-01 | 0.990 |
| UBA2 | 8.5e-01 | 0.990 |
| MBD3 | 8.5e-01 | 0.990 |
| ECE1 | 8.5e-01 | 0.990 |
| XPO6 | 8.5e-01 | 0.990 |
| FRYL | 8.5e-01 | 0.990 |
| NDE1 | 8.5e-01 | 0.990 |
| PATZ1 | 8.5e-01 | 0.990 |
| SLC25A37 | 8.5e-01 | 0.990 |
| STARD3NL | 8.5e-01 | 0.990 |
| ZNF714 | 8.5e-01 | 0.990 |
| IL6ST | 8.5e-01 | 0.990 |
| DPYSL4 | 8.5e-01 | 0.990 |
| ZNF513 | 8.5e-01 | 0.990 |
| NDUFA9 | 8.5e-01 | 0.990 |
| ODF2 | 8.5e-01 | 0.990 |
| GTF2A1 | 8.5e-01 | 0.990 |
| BIRC5 | 8.5e-01 | 0.990 |
| HECTD2 | 8.5e-01 | 0.990 |
| B4GALT5 | 8.5e-01 | 0.990 |
| PRMT3 | 8.5e-01 | 0.990 |
| HARS2 | 8.5e-01 | 0.990 |
| IKBKG | 8.5e-01 | 0.990 |
| TACO1 | 8.5e-01 | 0.990 |
| ABT1 | 8.5e-01 | 0.990 |
| NARFL | 8.5e-01 | 0.990 |
| IFT20 | 8.5e-01 | 0.990 |
| HADH | 8.5e-01 | 0.990 |
| TUBB6 | 8.5e-01 | 0.990 |
| ZNF664 | 8.5e-01 | 0.990 |
| TACC3 | 8.5e-01 | 0.990 |
| USP42 | 8.5e-01 | 0.990 |
| PAX7 | 8.5e-01 | 0.990 |
| GSS | 8.5e-01 | 0.990 |
| ERCC2 | 8.5e-01 | 0.990 |
| COQ10B | 8.6e-01 | 0.990 |
| DUS4L | 8.6e-01 | 0.990 |
| WDR20 | 8.6e-01 | 0.990 |
| SEMA4C | 8.6e-01 | 0.990 |
| PFDN2 | 8.6e-01 | 0.990 |
| PAFAH1B2 | 8.6e-01 | 0.990 |
| INO80E | 8.6e-01 | 0.990 |
| CSNK1A1 | 8.6e-01 | 0.990 |
| VPS45 | 8.6e-01 | 0.990 |
| EIF3K | 8.6e-01 | 0.990 |
| SMEK1 | 8.6e-01 | 0.990 |
| PDLIM5 | 8.6e-01 | 0.990 |
| RNF4 | 8.6e-01 | 0.990 |
| TXNL4B | 8.6e-01 | 0.990 |
| KLHL18 | 8.6e-01 | 0.990 |
| ZNF652 | 8.6e-01 | 0.990 |
| PAK4 | 8.6e-01 | 0.990 |
| SFXN4 | 8.6e-01 | 0.990 |
| ZNF326 | 8.6e-01 | 0.990 |
| ZBTB43 | 8.6e-01 | 0.990 |
| UBP1 | 8.6e-01 | 0.990 |
| RBM10 | 8.6e-01 | 0.990 |
| REXO2 | 8.6e-01 | 0.990 |
| FAM3C | 8.6e-01 | 0.990 |
| EIF4G3 | 8.6e-01 | 0.990 |
| DDX10 | 8.6e-01 | 0.990 |
| RAB40B | 8.6e-01 | 0.990 |
| ZDHHC9 | 8.6e-01 | 0.990 |
| MOB3A | 8.6e-01 | 0.990 |
| CDKAL1 | 8.6e-01 | 0.990 |
| PSD3 | 8.6e-01 | 0.990 |
| EPN2 | 8.6e-01 | 0.990 |
| IVD | 8.6e-01 | 0.990 |
| RDX | 8.6e-01 | 0.990 |
| SNHG3 | 8.6e-01 | 0.990 |
| SALL4 | 8.6e-01 | 0.990 |
| NAB2 | 8.6e-01 | 0.990 |
| ARHGAP35 | 8.6e-01 | 0.990 |
| SEC22C | 8.6e-01 | 0.990 |
| MAP7D3 | 8.6e-01 | 0.990 |
| DIP2C | 8.6e-01 | 0.990 |
| C6orf1 | 8.6e-01 | 0.990 |
| FBXO11 | 8.6e-01 | 0.990 |
| SLC39A6 | 8.6e-01 | 0.990 |
| C17orf63 | 8.6e-01 | 0.990 |
| METTL5 | 8.6e-01 | 0.990 |
| LUC7L3 | 8.6e-01 | 0.990 |
| SLC25A46 | 8.6e-01 | 0.990 |
| KANSL3 | 8.6e-01 | 0.990 |
| TAOK2 | 8.6e-01 | 0.990 |
| CCDC138 | 8.6e-01 | 0.990 |
| SLC39A1 | 8.6e-01 | 0.990 |
| KAT5 | 8.6e-01 | 0.990 |
| KIAA0100 | 8.6e-01 | 0.990 |
| EXOSC4 | 8.6e-01 | 0.990 |
| CTNND2 | 8.6e-01 | 0.990 |
| SLC12A9 | 8.6e-01 | 0.990 |
| MAP3K7 | 8.6e-01 | 0.990 |
| HDAC2 | 8.6e-01 | 0.990 |
| COG5 | 8.6e-01 | 0.990 |
| PGS1 | 8.6e-01 | 0.990 |
| DTNA | 8.6e-01 | 0.990 |
| MRFAP1 | 8.6e-01 | 0.990 |
| YME1L1 | 8.6e-01 | 0.990 |
| TDRP | 8.6e-01 | 0.990 |
| PTAR1 | 8.6e-01 | 0.990 |
| RAB18 | 8.6e-01 | 0.990 |
| ADIPOR1 | 8.6e-01 | 0.990 |
| HGSNAT | 8.6e-01 | 0.990 |
| VGLL4 | 8.6e-01 | 0.990 |
| FAM57A | 8.6e-01 | 0.990 |
| PTRHD1 | 8.6e-01 | 0.990 |
| LMNB2 | 8.6e-01 | 0.990 |
| RFX4 | 8.6e-01 | 0.990 |
| SPATA2L | 8.6e-01 | 0.990 |
| MRPL21 | 8.6e-01 | 0.990 |
| SLAIN1 | 8.6e-01 | 0.990 |
| PRR7 | 8.6e-01 | 0.990 |
| DDX17 | 8.6e-01 | 0.990 |
| MAT2B | 8.6e-01 | 0.990 |
| ZBTB2 | 8.6e-01 | 0.990 |
| PRELID3B | 8.7e-01 | 0.990 |
| ZNF346 | 8.7e-01 | 0.990 |
| RSBN1 | 8.7e-01 | 0.990 |
| NEK9 | 8.7e-01 | 0.990 |
| SYF2 | 8.7e-01 | 0.990 |
| AKT1 | 8.7e-01 | 0.990 |
| BUB3 | 8.7e-01 | 0.990 |
| EIF2B1 | 8.7e-01 | 0.990 |
| PPP2R3A | 8.7e-01 | 0.990 |
| SORBS3 | 8.7e-01 | 0.990 |
| SENP2 | 8.7e-01 | 0.990 |
| MT-ND1 | 8.7e-01 | 0.990 |
| COX7A2L | 8.7e-01 | 0.990 |
| FKBP14 | 8.7e-01 | 0.990 |
| TMEM11 | 8.7e-01 | 0.990 |
| TERF1 | 8.7e-01 | 0.990 |
| PCMTD2 | 8.7e-01 | 0.990 |
| PRCP | 8.7e-01 | 0.990 |
| SMC5 | 8.7e-01 | 0.990 |
| CLIP1 | 8.7e-01 | 0.990 |
| ARL10 | 8.7e-01 | 0.990 |
| PTP4A1 | 8.7e-01 | 0.990 |
| TCTN1 | 8.7e-01 | 0.990 |
| CCNB1IP1 | 8.7e-01 | 0.990 |
| GRB2 | 8.7e-01 | 0.990 |
| SREK1IP1 | 8.7e-01 | 0.990 |
| PRMT5 | 8.7e-01 | 0.990 |
| TFPI2 | 8.7e-01 | 0.990 |
| TP53INP1 | 8.7e-01 | 0.990 |
| ATXN10 | 8.7e-01 | 0.990 |
| MAPRE1 | 8.7e-01 | 0.990 |
| PSMD13 | 8.7e-01 | 0.990 |
| LUZP1 | 8.7e-01 | 0.990 |
| DNAJB12 | 8.7e-01 | 0.990 |
| NUCB2 | 8.7e-01 | 0.990 |
| DCLRE1C | 8.7e-01 | 0.990 |
| KXD1 | 8.7e-01 | 0.990 |
| UGGT2 | 8.7e-01 | 0.990 |
| UHRF2 | 8.7e-01 | 0.990 |
| SVBP | 8.7e-01 | 0.990 |
| DNAAF2 | 8.7e-01 | 0.990 |
| GDAP2 | 8.7e-01 | 0.990 |
| ANKFY1 | 8.7e-01 | 0.990 |
| DTD1 | 8.7e-01 | 0.990 |
| DHX38 | 8.7e-01 | 0.990 |
| DLG1 | 8.7e-01 | 0.990 |
| MYL6 | 8.7e-01 | 0.990 |
| GSTO1 | 8.7e-01 | 0.990 |
| HYI | 8.7e-01 | 0.990 |
| HLA-E | 8.7e-01 | 0.990 |
| USP22 | 8.7e-01 | 0.990 |
| MRPS9 | 8.7e-01 | 0.990 |
| EBP | 8.7e-01 | 0.990 |
| MRPS31 | 8.7e-01 | 0.990 |
| FGFR1OP | 8.7e-01 | 0.990 |
| RCAN1 | 8.7e-01 | 0.990 |
| ARRDC1 | 8.7e-01 | 0.990 |
| EPC1 | 8.7e-01 | 0.990 |
| PTTG1 | 8.7e-01 | 0.990 |
| IK | 8.7e-01 | 0.990 |
| WDR12 | 8.7e-01 | 0.990 |
| ERCC1 | 8.7e-01 | 0.990 |
| NCBP2 | 8.7e-01 | 0.990 |
| CAPN5 | 8.7e-01 | 0.990 |
| CDC42EP1 | 8.7e-01 | 0.990 |
| RP11-477B16.5 | 8.7e-01 | 0.990 |
| SLC25A17 | 8.7e-01 | 0.990 |
| C11orf83 | 8.7e-01 | 0.990 |
| MRPS11 | 8.7e-01 | 0.990 |
| KAT2A | 8.7e-01 | 0.990 |
| WDR70 | 8.7e-01 | 0.990 |
| EPHX1 | 8.7e-01 | 0.990 |
| TULP4 | 8.7e-01 | 0.990 |
| MTA1 | 8.7e-01 | 0.990 |
| AGAP3 | 8.7e-01 | 0.990 |
| TMEM138 | 8.7e-01 | 0.990 |
| RASSF7 | 8.8e-01 | 0.990 |
| RNF145 | 8.8e-01 | 0.990 |
| SOX4 | 8.8e-01 | 0.990 |
| TARS | 8.8e-01 | 0.990 |
| RPS5 | 8.8e-01 | 0.990 |
| UBA52 | 8.8e-01 | 0.990 |
| NUDT4 | 8.8e-01 | 0.990 |
| ZYX | 8.8e-01 | 0.990 |
| GLCE | 8.8e-01 | 0.990 |
| FBXL12 | 8.8e-01 | 0.990 |
| RNF5 | 8.8e-01 | 0.990 |
| SAP18 | 8.8e-01 | 0.990 |
| C14orf132 | 8.8e-01 | 0.990 |
| RMI1 | 8.8e-01 | 0.990 |
| RP11-490M8.1 | 8.8e-01 | 0.990 |
| SFRP2 | 8.8e-01 | 0.990 |
| ZNF426 | 8.8e-01 | 0.990 |
| ITSN2 | 8.8e-01 | 0.990 |
| KIAA1143 | 8.8e-01 | 0.990 |
| FAM83D | 8.8e-01 | 0.990 |
| KHK | 8.8e-01 | 0.990 |
| RNF138 | 8.8e-01 | 0.990 |
| TOGARAM1 | 8.8e-01 | 0.990 |
| UFM1 | 8.8e-01 | 0.990 |
| DNAJC21 | 8.8e-01 | 0.990 |
| HDGF | 8.8e-01 | 0.990 |
| DICER1 | 8.8e-01 | 0.990 |
| RMND5B | 8.8e-01 | 0.990 |
| LMAN2L | 8.8e-01 | 0.990 |
| SLC25A6 | 8.8e-01 | 0.990 |
| FBXO7 | 8.8e-01 | 0.990 |
| HOXA2 | 8.8e-01 | 0.990 |
| BTG2 | 8.8e-01 | 0.990 |
| AVEN | 8.8e-01 | 0.990 |
| ARPC1B | 8.8e-01 | 0.990 |
| SNTB2 | 8.8e-01 | 0.990 |
| STRN3 | 8.8e-01 | 0.990 |
| GLYR1 | 8.8e-01 | 0.990 |
| PMF1 | 8.8e-01 | 0.990 |
| CARS2 | 8.8e-01 | 0.990 |
| FBRS | 8.8e-01 | 0.990 |
| MICAL1 | 8.8e-01 | 0.990 |
| RP11-343C2.12 | 8.8e-01 | 0.990 |
| LIN52 | 8.8e-01 | 0.990 |
| TPMT | 8.8e-01 | 0.990 |
| PABPN1 | 8.8e-01 | 0.990 |
| SDCCAG8 | 8.8e-01 | 0.990 |
| NKAIN3 | 8.8e-01 | 0.990 |
| SGO1 | 8.8e-01 | 0.990 |
| ZC3H10 | 8.8e-01 | 0.990 |
| RILPL2 | 8.8e-01 | 0.990 |
| THBS1 | 8.8e-01 | 0.990 |
| DLG3 | 8.8e-01 | 0.990 |
| GPC1 | 8.8e-01 | 0.990 |
| MTR | 8.8e-01 | 0.990 |
| AP4S1 | 8.8e-01 | 0.990 |
| TTC13 | 8.8e-01 | 0.990 |
| NDFIP1 | 8.8e-01 | 0.990 |
| AGO3 | 8.8e-01 | 0.990 |
| SDF4 | 8.8e-01 | 0.990 |
| SFXN2 | 8.8e-01 | 0.990 |
| SLAIN2 | 8.8e-01 | 0.990 |
| CDK10 | 8.8e-01 | 0.990 |
| RP11-446H18.3 | 8.8e-01 | 0.990 |
| BCAP31 | 8.8e-01 | 0.990 |
| THAP12 | 8.8e-01 | 0.990 |
| NNAT | 8.8e-01 | 0.990 |
| ALS2 | 8.8e-01 | 0.990 |
| VPS26B | 8.8e-01 | 0.990 |
| C12orf29 | 8.8e-01 | 0.990 |
| CHMP4A | 8.8e-01 | 0.990 |
| C9orf16 | 8.8e-01 | 0.990 |
| RTN4IP1 | 8.8e-01 | 0.990 |
| ARG2 | 8.8e-01 | 0.990 |
| NDUFA7 | 8.8e-01 | 0.990 |
| BRMS1L | 8.8e-01 | 0.990 |
| QTRT2 | 8.8e-01 | 0.990 |
| KIAA1147 | 8.8e-01 | 0.990 |
| PAPSS1 | 8.8e-01 | 0.990 |
| SRM | 8.8e-01 | 0.990 |
| RALY | 8.8e-01 | 0.990 |
| WRB | 8.9e-01 | 0.990 |
| RPL36A | 8.9e-01 | 0.990 |
| MUL1 | 8.9e-01 | 0.990 |
| PSME4 | 8.9e-01 | 0.990 |
| SENP6 | 8.9e-01 | 0.990 |
| WDR1 | 8.9e-01 | 0.990 |
| STK25 | 8.9e-01 | 0.990 |
| SCAMP4 | 8.9e-01 | 0.990 |
| RALGAPA1 | 8.9e-01 | 0.990 |
| TBCA | 8.9e-01 | 0.990 |
| TMEM80 | 8.9e-01 | 0.990 |
| VHL | 8.9e-01 | 0.990 |
| TCTA | 8.9e-01 | 0.990 |
| ERI2 | 8.9e-01 | 0.990 |
| SLC25A44 | 8.9e-01 | 0.990 |
| TTLL12 | 8.9e-01 | 0.990 |
| ZDHHC7 | 8.9e-01 | 0.990 |
| GOT1 | 8.9e-01 | 0.990 |
| CNOT2 | 8.9e-01 | 0.990 |
| SIRT1 | 8.9e-01 | 0.990 |
| GUCD1 | 8.9e-01 | 0.990 |
| ZRSR2 | 8.9e-01 | 0.990 |
| NAA40 | 8.9e-01 | 0.990 |
| SLC4A7 | 8.9e-01 | 0.990 |
| EXT2 | 8.9e-01 | 0.990 |
| MRPL32 | 8.9e-01 | 0.990 |
| SNHG11 | 8.9e-01 | 0.990 |
| LRRC41 | 8.9e-01 | 0.990 |
| MYPOP | 8.9e-01 | 0.990 |
| ZNF16 | 8.9e-01 | 0.990 |
| RRP36 | 8.9e-01 | 0.990 |
| MPP6 | 8.9e-01 | 0.990 |
| RHEB | 8.9e-01 | 0.990 |
| HDAC6 | 8.9e-01 | 0.990 |
| UBC | 8.9e-01 | 0.990 |
| ZNF670 | 8.9e-01 | 0.990 |
| DECR1 | 8.9e-01 | 0.990 |
| ST3GAL2 | 8.9e-01 | 0.990 |
| SSB | 8.9e-01 | 0.990 |
| SQSTM1 | 8.9e-01 | 0.990 |
| CENPA | 8.9e-01 | 0.990 |
| FUZ | 8.9e-01 | 0.990 |
| APLP2 | 8.9e-01 | 0.990 |
| PKM | 8.9e-01 | 0.990 |
| CNOT7 | 8.9e-01 | 0.990 |
| MKRN1 | 8.9e-01 | 0.990 |
| RBM14 | 8.9e-01 | 0.990 |
| PRTG | 8.9e-01 | 0.990 |
| ATP6V1F | 8.9e-01 | 0.990 |
| MYLK | 8.9e-01 | 0.990 |
| TMEM179B | 8.9e-01 | 0.990 |
| PRRC2C | 8.9e-01 | 0.990 |
| ASTN2 | 8.9e-01 | 0.990 |
| BRD9 | 8.9e-01 | 0.990 |
| NDUFAF3 | 8.9e-01 | 0.990 |
| HEATR3 | 8.9e-01 | 0.990 |
| UFSP2 | 8.9e-01 | 0.990 |
| SLC35F5 | 8.9e-01 | 0.990 |
| CLN6 | 8.9e-01 | 0.990 |
| NENF | 8.9e-01 | 0.990 |
| APOOL | 8.9e-01 | 0.990 |
| HIRA | 8.9e-01 | 0.990 |
| RNF130 | 8.9e-01 | 0.990 |
| APOM | 8.9e-01 | 0.990 |
| PRKAA1 | 8.9e-01 | 0.990 |
| MAP1LC3B | 8.9e-01 | 0.990 |
| GRINA | 8.9e-01 | 0.990 |
| ITM2B | 8.9e-01 | 0.990 |
| SH3BP5L | 8.9e-01 | 0.990 |
| SRP68 | 8.9e-01 | 0.990 |
| PSMD8 | 8.9e-01 | 0.990 |
| EIF3G | 8.9e-01 | 0.990 |
| FAM175A | 8.9e-01 | 0.990 |
| SRGAP1 | 8.9e-01 | 0.990 |
| GLIPR2 | 8.9e-01 | 0.990 |
| SUPT4H1 | 8.9e-01 | 0.990 |
| GNAI3 | 8.9e-01 | 0.990 |
| ATF1 | 8.9e-01 | 0.990 |
| TRIP10 | 8.9e-01 | 0.990 |
| EIF3M | 8.9e-01 | 0.990 |
| C6orf89 | 8.9e-01 | 0.990 |
| RINT1 | 8.9e-01 | 0.990 |
| SLC35E3 | 8.9e-01 | 0.990 |
| ANKRD46 | 8.9e-01 | 0.990 |
| MPRIP | 8.9e-01 | 0.990 |
| MRPL4 | 8.9e-01 | 0.990 |
| ERCC8 | 8.9e-01 | 0.990 |
| IGFBP2 | 8.9e-01 | 0.990 |
| MRPL33 | 8.9e-01 | 0.990 |
| UBE2E1 | 8.9e-01 | 0.990 |
| BTF3 | 8.9e-01 | 0.990 |
| RPS10 | 8.9e-01 | 0.990 |
| SLC38A7 | 8.9e-01 | 0.990 |
| ZNF599 | 8.9e-01 | 0.990 |
| NRM | 9.0e-01 | 0.990 |
| COX14 | 9.0e-01 | 0.990 |
| MAPK1 | 9.0e-01 | 0.990 |
| VKORC1L1 | 9.0e-01 | 0.990 |
| REV1 | 9.0e-01 | 0.990 |
| FAM208A | 9.0e-01 | 0.990 |
| PUF60 | 9.0e-01 | 0.990 |
| CC2D2A | 9.0e-01 | 0.990 |
| FAM216A | 9.0e-01 | 0.990 |
| DHDDS | 9.0e-01 | 0.990 |
| GTPBP1 | 9.0e-01 | 0.990 |
| NDUFA10 | 9.0e-01 | 0.990 |
| ALDOA | 9.0e-01 | 0.990 |
| BSDC1 | 9.0e-01 | 0.990 |
| FGFR1 | 9.0e-01 | 0.990 |
| U2AF2 | 9.0e-01 | 0.990 |
| AP3S2 | 9.0e-01 | 0.990 |
| MAP2K4 | 9.0e-01 | 0.990 |
| GDAP1 | 9.0e-01 | 0.990 |
| RAI1 | 9.0e-01 | 0.990 |
| AC016747.3 | 9.0e-01 | 0.990 |
| SLC35E2 | 9.0e-01 | 0.990 |
| STK17A | 9.0e-01 | 0.990 |
| MT-ND3 | 9.0e-01 | 0.990 |
| MEIS2 | 9.0e-01 | 0.990 |
| NDEL1 | 9.0e-01 | 0.990 |
| TCEAL7 | 9.0e-01 | 0.990 |
| ATP2B1 | 9.0e-01 | 0.990 |
| SCYL2 | 9.0e-01 | 0.990 |
| UBL7 | 9.0e-01 | 0.990 |
| NUSAP1 | 9.0e-01 | 0.990 |
| ATG12 | 9.0e-01 | 0.990 |
| ZFAND2B | 9.0e-01 | 0.990 |
| PDIA3 | 9.0e-01 | 0.990 |
| BCS1L | 9.0e-01 | 0.990 |
| RPL10A | 9.0e-01 | 0.990 |
| SNRNP40 | 9.0e-01 | 0.990 |
| TIMM17A | 9.0e-01 | 0.990 |
| ERV3-1 | 9.0e-01 | 0.990 |
| LRRC20 | 9.0e-01 | 0.990 |
| IPO5 | 9.0e-01 | 0.990 |
| GTF2H1 | 9.0e-01 | 0.990 |
| TTPAL | 9.0e-01 | 0.990 |
| TIAL1 | 9.0e-01 | 0.990 |
| UPRT | 9.0e-01 | 0.990 |
| AIG1 | 9.0e-01 | 0.990 |
| YTHDC2 | 9.0e-01 | 0.990 |
| HUS1 | 9.0e-01 | 0.990 |
| PLPP1 | 9.0e-01 | 0.990 |
| CECR5 | 9.0e-01 | 0.990 |
| COX7A2 | 9.0e-01 | 0.990 |
| SCAMP3 | 9.0e-01 | 0.990 |
| STYXL1 | 9.0e-01 | 0.990 |
| SPAG9 | 9.0e-01 | 0.990 |
| ERAL1 | 9.0e-01 | 0.990 |
| CTU2 | 9.0e-01 | 0.990 |
| PNPLA8 | 9.0e-01 | 0.990 |
| CHAC1 | 9.0e-01 | 0.990 |
| RND2 | 9.0e-01 | 0.990 |
| CHORDC1 | 9.0e-01 | 0.990 |
| TRMT13 | 9.0e-01 | 0.990 |
| RSRC1 | 9.0e-01 | 0.990 |
| TRIM41 | 9.0e-01 | 0.990 |
| CRYBA1 | 9.0e-01 | 0.990 |
| WASF3 | 9.0e-01 | 0.990 |
| GCA | 9.0e-01 | 0.990 |
| MAU2 | 9.0e-01 | 0.990 |
| RHOT2 | 9.0e-01 | 0.990 |
| PRR36 | 9.0e-01 | 0.990 |
| KLHDC10 | 9.0e-01 | 0.990 |
| PDE6D | 9.0e-01 | 0.990 |
| CHCHD10 | 9.0e-01 | 0.990 |
| ZBTB33 | 9.0e-01 | 0.990 |
| CCND1 | 9.1e-01 | 0.990 |
| ARID2 | 9.1e-01 | 0.990 |
| FBXO46 | 9.1e-01 | 0.990 |
| ETF1 | 9.1e-01 | 0.990 |
| CDS2 | 9.1e-01 | 0.990 |
| DBR1 | 9.1e-01 | 0.990 |
| CASP2 | 9.1e-01 | 0.990 |
| ZNF692 | 9.1e-01 | 0.990 |
| KIF1BP | 9.1e-01 | 0.990 |
| SLC39A13 | 9.1e-01 | 0.990 |
| C22orf29 | 9.1e-01 | 0.990 |
| RNFT2 | 9.1e-01 | 0.990 |
| DDX1 | 9.1e-01 | 0.990 |
| GYPC | 9.1e-01 | 0.990 |
| DFNA5 | 9.1e-01 | 0.990 |
| DNASE2 | 9.1e-01 | 0.990 |
| PCSK1N | 9.1e-01 | 0.990 |
| NDUFAF1 | 9.1e-01 | 0.990 |
| CCT8 | 9.1e-01 | 0.990 |
| CDCA5 | 9.1e-01 | 0.990 |
| LPGAT1 | 9.1e-01 | 0.990 |
| GATAD2B | 9.1e-01 | 0.990 |
| WAC | 9.1e-01 | 0.990 |
| SNHG8 | 9.1e-01 | 0.990 |
| MYCN | 9.1e-01 | 0.990 |
| POGLUT1 | 9.1e-01 | 0.990 |
| UTP14A | 9.1e-01 | 0.990 |
| FLOT1 | 9.1e-01 | 0.990 |
| UBE2I | 9.1e-01 | 0.990 |
| DPH2 | 9.1e-01 | 0.990 |
| TXNRD2 | 9.1e-01 | 0.990 |
| OSGEPL1 | 9.1e-01 | 0.990 |
| ORC1 | 9.1e-01 | 0.990 |
| HEBP1 | 9.1e-01 | 0.990 |
| PSMC4 | 9.1e-01 | 0.990 |
| UBE2W | 9.1e-01 | 0.990 |
| RNF185 | 9.1e-01 | 0.990 |
| PREP | 9.1e-01 | 0.990 |
| VPS53 | 9.1e-01 | 0.990 |
| NACA2 | 9.1e-01 | 0.990 |
| PPDPF | 9.1e-01 | 0.990 |
| PSMC5 | 9.1e-01 | 0.990 |
| UBE2R2 | 9.1e-01 | 0.990 |
| UBXN7 | 9.1e-01 | 0.990 |
| PHF14 | 9.1e-01 | 0.990 |
| CCDC181 | 9.1e-01 | 0.990 |
| BCAT1 | 9.1e-01 | 0.990 |
| SLC22A17 | 9.1e-01 | 0.990 |
| PCMT1 | 9.1e-01 | 0.990 |
| RPS3 | 9.1e-01 | 0.990 |
| SPAG16 | 9.1e-01 | 0.990 |
| RPL37A | 9.1e-01 | 0.990 |
| AXIN1 | 9.1e-01 | 0.990 |
| TXLNG | 9.1e-01 | 0.990 |
| DGUOK | 9.1e-01 | 0.990 |
| FLNB | 9.1e-01 | 0.990 |
| MTHFD2L | 9.1e-01 | 0.990 |
| DDX28 | 9.1e-01 | 0.990 |
| ASNA1 | 9.1e-01 | 0.990 |
| PIP4K2A | 9.1e-01 | 0.990 |
| WDR3 | 9.1e-01 | 0.990 |
| GSK3B | 9.1e-01 | 0.990 |
| RP11-452F19.3 | 9.1e-01 | 0.990 |
| CEPT1 | 9.1e-01 | 0.990 |
| PGAP2 | 9.1e-01 | 0.990 |
| AQR | 9.1e-01 | 0.990 |
| KIF20B | 9.1e-01 | 0.990 |
| ELMO2 | 9.1e-01 | 0.990 |
| NOL11 | 9.1e-01 | 0.990 |
| GORASP2 | 9.1e-01 | 0.990 |
| C14orf156 | 9.1e-01 | 0.990 |
| SZRD1 | 9.1e-01 | 0.990 |
| MTX3 | 9.1e-01 | 0.990 |
| UBE2A | 9.1e-01 | 0.990 |
| MARK3 | 9.1e-01 | 0.990 |
| ORMDL2 | 9.1e-01 | 0.990 |
| USF2 | 9.1e-01 | 0.990 |
| TIMMDC1 | 9.1e-01 | 0.990 |
| EID2 | 9.1e-01 | 0.990 |
| IFITM2 | 9.1e-01 | 0.990 |
| EXOC7 | 9.2e-01 | 0.990 |
| AP3M1 | 9.2e-01 | 0.990 |
| ATL2 | 9.2e-01 | 0.990 |
| CRELD2 | 9.2e-01 | 0.990 |
| TXN | 9.2e-01 | 0.990 |
| EXTL2 | 9.2e-01 | 0.990 |
| GSK3A | 9.2e-01 | 0.990 |
| LSM3 | 9.2e-01 | 0.990 |
| ZNF791 | 9.2e-01 | 0.990 |
| PTRF | 9.2e-01 | 0.990 |
| INTS10 | 9.2e-01 | 0.990 |
| PGP | 9.2e-01 | 0.990 |
| RPL34 | 9.2e-01 | 0.990 |
| HMGB2 | 9.2e-01 | 0.990 |
| ZNF136 | 9.2e-01 | 0.990 |
| GPATCH4 | 9.2e-01 | 0.990 |
| PIGS | 9.2e-01 | 0.990 |
| CCSER2 | 9.2e-01 | 0.990 |
| PARP16 | 9.2e-01 | 0.990 |
| FOXK2 | 9.2e-01 | 0.990 |
| ATE1 | 9.2e-01 | 0.990 |
| TMEM184B | 9.2e-01 | 0.990 |
| PAX3 | 9.2e-01 | 0.990 |
| CEP104 | 9.2e-01 | 0.990 |
| ESYT1 | 9.2e-01 | 0.990 |
| DCP2 | 9.2e-01 | 0.990 |
| KIAA0930 | 9.2e-01 | 0.990 |
| EXOC3 | 9.2e-01 | 0.990 |
| DNAL1 | 9.2e-01 | 0.990 |
| ZBTB1 | 9.2e-01 | 0.990 |
| SLC25A23 | 9.2e-01 | 0.990 |
| RPL35A | 9.2e-01 | 0.990 |
| PYCR2 | 9.2e-01 | 0.990 |
| FAM53C | 9.2e-01 | 0.990 |
| FKBP15 | 9.2e-01 | 0.990 |
| SENP5 | 9.2e-01 | 0.990 |
| CHP1 | 9.2e-01 | 0.990 |
| RP11-1094M14.11 | 9.2e-01 | 0.990 |
| SERF2 | 9.2e-01 | 0.990 |
| SHOC2 | 9.2e-01 | 0.990 |
| EIF4G2 | 9.2e-01 | 0.990 |
| DGAT1 | 9.2e-01 | 0.990 |
| TUBGCP4 | 9.2e-01 | 0.990 |
| PEPD | 9.2e-01 | 0.990 |
| MED26 | 9.2e-01 | 0.990 |
| DDT | 9.2e-01 | 0.990 |
| RBMXL1 | 9.2e-01 | 0.990 |
| LATS2 | 9.2e-01 | 0.990 |
| CHST14 | 9.2e-01 | 0.990 |
| MAPK1IP1L | 9.2e-01 | 0.990 |
| KIF1A | 9.2e-01 | 0.990 |
| PEX10 | 9.2e-01 | 0.990 |
| MYC | 9.2e-01 | 0.990 |
| SMYD4 | 9.2e-01 | 0.990 |
| ADAM15 | 9.2e-01 | 0.990 |
| CPNE2 | 9.2e-01 | 0.990 |
| TAX1BP1 | 9.2e-01 | 0.990 |
| R3HDM2 | 9.2e-01 | 0.990 |
| PPIG | 9.2e-01 | 0.990 |
| GFM1 | 9.2e-01 | 0.990 |
| Z83851.1 | 9.2e-01 | 0.990 |
| RAP2C | 9.2e-01 | 0.990 |
| TIGAR | 9.2e-01 | 0.990 |
| LAP3 | 9.2e-01 | 0.990 |
| MOB4 | 9.3e-01 | 0.990 |
| THEM4 | 9.3e-01 | 0.990 |
| MRPS22 | 9.3e-01 | 0.990 |
| CDIPT | 9.3e-01 | 0.990 |
| NEAT1 | 9.3e-01 | 0.990 |
| LACTB | 9.3e-01 | 0.990 |
| CEP135 | 9.3e-01 | 0.990 |
| SOCS6 | 9.3e-01 | 0.990 |
| BCCIP | 9.3e-01 | 0.990 |
| ZNF330 | 9.3e-01 | 0.990 |
| SPTSSA | 9.3e-01 | 0.990 |
| GNPAT | 9.3e-01 | 0.990 |
| C16orf58 | 9.3e-01 | 0.990 |
| SMARCC2 | 9.3e-01 | 0.990 |
| BIRC2 | 9.3e-01 | 0.990 |
| IRF7 | 9.3e-01 | 0.990 |
| PHRF1 | 9.3e-01 | 0.990 |
| NR2F6 | 9.3e-01 | 0.990 |
| C16orf52 | 9.3e-01 | 0.990 |
| CACFD1 | 9.3e-01 | 0.990 |
| MED15 | 9.3e-01 | 1.000 |
| POLB | 9.3e-01 | 1.000 |
| NSMCE1 | 9.3e-01 | 1.000 |
| POLR2A | 9.3e-01 | 1.000 |
| CHML | 9.3e-01 | 1.000 |
| UBLCP1 | 9.3e-01 | 1.000 |
| METTL18 | 9.3e-01 | 1.000 |
| ZNF219 | 9.3e-01 | 1.000 |
| CCDC77 | 9.3e-01 | 1.000 |
| RAB3GAP1 | 9.3e-01 | 1.000 |
| SLC26A6 | 9.3e-01 | 1.000 |
| DHX34 | 9.3e-01 | 1.000 |
| DNAAF5 | 9.3e-01 | 1.000 |
| IFNGR1 | 9.3e-01 | 1.000 |
| RP11-395A13.2 | 9.3e-01 | 1.000 |
| NF1 | 9.3e-01 | 1.000 |
| KPNA1 | 9.3e-01 | 1.000 |
| GPATCH2 | 9.3e-01 | 1.000 |
| ROCK1 | 9.3e-01 | 1.000 |
| MED13L | 9.3e-01 | 1.000 |
| SOX12 | 9.3e-01 | 1.000 |
| ATP1B3 | 9.3e-01 | 1.000 |
| DDX51 | 9.3e-01 | 1.000 |
| BAZ2A | 9.3e-01 | 1.000 |
| POLI | 9.3e-01 | 1.000 |
| JUNB | 9.3e-01 | 1.000 |
| TKT | 9.3e-01 | 1.000 |
| CMTR2 | 9.3e-01 | 1.000 |
| PSMA1 | 9.3e-01 | 1.000 |
| C16orf62 | 9.3e-01 | 1.000 |
| RABAC1 | 9.3e-01 | 1.000 |
| SET | 9.3e-01 | 1.000 |
| EIF4EBP2 | 9.3e-01 | 1.000 |
| EPC2 | 9.3e-01 | 1.000 |
| RFLNB | 9.3e-01 | 1.000 |
| TIMP1 | 9.3e-01 | 1.000 |
| NUTF2 | 9.3e-01 | 1.000 |
| PPIE | 9.3e-01 | 1.000 |
| DNMT3A | 9.3e-01 | 1.000 |
| RP11-339B21.12 | 9.3e-01 | 1.000 |
| METTL21A | 9.4e-01 | 1.000 |
| DDX50 | 9.4e-01 | 1.000 |
| INIP | 9.4e-01 | 1.000 |
| ZNF521 | 9.4e-01 | 1.000 |
| QSOX2 | 9.4e-01 | 1.000 |
| PHF20L1 | 9.4e-01 | 1.000 |
| FIS1 | 9.4e-01 | 1.000 |
| TMEM206 | 9.4e-01 | 1.000 |
| RP11-480D4.3 | 9.4e-01 | 1.000 |
| GUSB | 9.4e-01 | 1.000 |
| BRAF | 9.4e-01 | 1.000 |
| CENPQ | 9.4e-01 | 1.000 |
| FAM98B | 9.4e-01 | 1.000 |
| BOD1 | 9.4e-01 | 1.000 |
| ESD | 9.4e-01 | 1.000 |
| KRCC1 | 9.4e-01 | 1.000 |
| MAGI1 | 9.4e-01 | 1.000 |
| TRAM1 | 9.4e-01 | 1.000 |
| FAM189B | 9.4e-01 | 1.000 |
| WDFY1 | 9.4e-01 | 1.000 |
| EIF5B | 9.4e-01 | 1.000 |
| TPP1 | 9.4e-01 | 1.000 |
| NADSYN1 | 9.4e-01 | 1.000 |
| RMST | 9.4e-01 | 1.000 |
| AP5Z1 | 9.4e-01 | 1.000 |
| CDC42BPB | 9.4e-01 | 1.000 |
| FLRT3 | 9.4e-01 | 1.000 |
| POC1B | 9.4e-01 | 1.000 |
| MAPK12 | 9.4e-01 | 1.000 |
| C22orf39 | 9.4e-01 | 1.000 |
| RAB3GAP2 | 9.4e-01 | 1.000 |
| RAB21 | 9.4e-01 | 1.000 |
| TSPAN12 | 9.4e-01 | 1.000 |
| FGFRL1 | 9.4e-01 | 1.000 |
| TCEAL4 | 9.4e-01 | 1.000 |
| CYCS | 9.4e-01 | 1.000 |
| SCAF4 | 9.4e-01 | 1.000 |
| HDAC7 | 9.4e-01 | 1.000 |
| LRIF1 | 9.4e-01 | 1.000 |
| PQLC2 | 9.4e-01 | 1.000 |
| SMARCAL1 | 9.4e-01 | 1.000 |
| RPL21 | 9.4e-01 | 1.000 |
| RPS6KA2 | 9.4e-01 | 1.000 |
| PELI1 | 9.4e-01 | 1.000 |
| TTC39C | 9.4e-01 | 1.000 |
| NLN | 9.4e-01 | 1.000 |
| RAB3IP | 9.4e-01 | 1.000 |
| VAC14 | 9.4e-01 | 1.000 |
| EFS | 9.4e-01 | 1.000 |
| KIDINS220 | 9.4e-01 | 1.000 |
| PREX1 | 9.4e-01 | 1.000 |
| TMEM136 | 9.4e-01 | 1.000 |
| COMMD3 | 9.4e-01 | 1.000 |
| DCPS | 9.4e-01 | 1.000 |
| PRAF2 | 9.4e-01 | 1.000 |
| RBCK1 | 9.4e-01 | 1.000 |
| TMED9 | 9.4e-01 | 1.000 |
| FNBP4 | 9.4e-01 | 1.000 |
| AKAP13 | 9.4e-01 | 1.000 |
| CDK11B | 9.4e-01 | 1.000 |
| CGGBP1 | 9.4e-01 | 1.000 |
| ING5 | 9.4e-01 | 1.000 |
| PTK2 | 9.4e-01 | 1.000 |
| FBXO22 | 9.4e-01 | 1.000 |
| WWTR1 | 9.4e-01 | 1.000 |
| ZNF75A | 9.4e-01 | 1.000 |
| TRAF7 | 9.4e-01 | 1.000 |
| SMARCAD1 | 9.4e-01 | 1.000 |
| STK19 | 9.4e-01 | 1.000 |
| GUK1 | 9.4e-01 | 1.000 |
| FXN | 9.4e-01 | 1.000 |
| DDAH2 | 9.4e-01 | 1.000 |
| HMGCL | 9.4e-01 | 1.000 |
| MCTS1 | 9.4e-01 | 1.000 |
| ZSCAN5A | 9.4e-01 | 1.000 |
| MMS22L | 9.4e-01 | 1.000 |
| MTRF1L | 9.5e-01 | 1.000 |
| FAM98C | 9.5e-01 | 1.000 |
| TTK | 9.5e-01 | 1.000 |
| C1orf174 | 9.5e-01 | 1.000 |
| MVD | 9.5e-01 | 1.000 |
| CNN3 | 9.5e-01 | 1.000 |
| NUDT11 | 9.5e-01 | 1.000 |
| GADD45B | 9.5e-01 | 1.000 |
| GPR161 | 9.5e-01 | 1.000 |
| FBXO18 | 9.5e-01 | 1.000 |
| PHF1 | 9.5e-01 | 1.000 |
| ADSL | 9.5e-01 | 1.000 |
| MED20 | 9.5e-01 | 1.000 |
| NOP16 | 9.5e-01 | 1.000 |
| PBRM1 | 9.5e-01 | 1.000 |
| UBE2Q2 | 9.5e-01 | 1.000 |
| PSMD4 | 9.5e-01 | 1.000 |
| CPT2 | 9.5e-01 | 1.000 |
| TNPO3 | 9.5e-01 | 1.000 |
| ATP13A3 | 9.5e-01 | 1.000 |
| SLC26A11 | 9.5e-01 | 1.000 |
| KPNA5 | 9.5e-01 | 1.000 |
| IRX3 | 9.5e-01 | 1.000 |
| ZNF292 | 9.5e-01 | 1.000 |
| PTTG1IP | 9.5e-01 | 1.000 |
| RNF41 | 9.5e-01 | 1.000 |
| SDE2 | 9.5e-01 | 1.000 |
| USO1 | 9.5e-01 | 1.000 |
| WSCD1 | 9.5e-01 | 1.000 |
| FAM173A | 9.5e-01 | 1.000 |
| LRR1 | 9.5e-01 | 1.000 |
| COCH | 9.5e-01 | 1.000 |
| FKBP5 | 9.5e-01 | 1.000 |
| GON4L | 9.5e-01 | 1.000 |
| ORMDL3 | 9.5e-01 | 1.000 |
| HAX1 | 9.5e-01 | 1.000 |
| STAU1 | 9.5e-01 | 1.000 |
| ANTXR1 | 9.5e-01 | 1.000 |
| HEXB | 9.5e-01 | 1.000 |
| TAGLN2 | 9.5e-01 | 1.000 |
| BLOC1S2 | 9.5e-01 | 1.000 |
| SF3A1 | 9.5e-01 | 1.000 |
| RNF167 | 9.5e-01 | 1.000 |
| RBM26 | 9.5e-01 | 1.000 |
| PPP1R10 | 9.5e-01 | 1.000 |
| NEDD1 | 9.5e-01 | 1.000 |
| DNAL4 | 9.5e-01 | 1.000 |
| PLIN3 | 9.5e-01 | 1.000 |
| TBC1D22A | 9.5e-01 | 1.000 |
| RPGRIP1L | 9.5e-01 | 1.000 |
| ALKBH6 | 9.5e-01 | 1.000 |
| NEMF | 9.5e-01 | 1.000 |
| RPIA | 9.5e-01 | 1.000 |
| TFRC | 9.5e-01 | 1.000 |
| ELAC1 | 9.5e-01 | 1.000 |
| RPS23 | 9.5e-01 | 1.000 |
| PCGF5 | 9.5e-01 | 1.000 |
| TMEM261 | 9.5e-01 | 1.000 |
| NPPC | 9.5e-01 | 1.000 |
| ERGIC1 | 9.5e-01 | 1.000 |
| FBXO44 | 9.5e-01 | 1.000 |
| EHD4 | 9.5e-01 | 1.000 |
| HTATSF1 | 9.5e-01 | 1.000 |
| PGAP1 | 9.5e-01 | 1.000 |
| TMX4 | 9.5e-01 | 1.000 |
| NEK2 | 9.5e-01 | 1.000 |
| PLTP | 9.5e-01 | 1.000 |
| LDOC1 | 9.5e-01 | 1.000 |
| CWC15 | 9.5e-01 | 1.000 |
| DPCD | 9.5e-01 | 1.000 |
| VPS41 | 9.5e-01 | 1.000 |
| EFHC1 | 9.5e-01 | 1.000 |
| APCDD1 | 9.5e-01 | 1.000 |
| LENG1 | 9.5e-01 | 1.000 |
| HTRA2 | 9.5e-01 | 1.000 |
| AKR7A2 | 9.5e-01 | 1.000 |
| TIMM44 | 9.5e-01 | 1.000 |
| ACSL3 | 9.5e-01 | 1.000 |
| SPG11 | 9.5e-01 | 1.000 |
| ANKRD17 | 9.5e-01 | 1.000 |
| PIGT | 9.5e-01 | 1.000 |
| RMDN1 | 9.5e-01 | 1.000 |
| GTF2A2 | 9.5e-01 | 1.000 |
| CD81 | 9.5e-01 | 1.000 |
| PPP1R14B | 9.5e-01 | 1.000 |
| SLC27A4 | 9.6e-01 | 1.000 |
| WDR48 | 9.6e-01 | 1.000 |
| MARCH5 | 9.6e-01 | 1.000 |
| RPS14 | 9.6e-01 | 1.000 |
| NRG1 | 9.6e-01 | 1.000 |
| ERBB2 | 9.6e-01 | 1.000 |
| USB1 | 9.6e-01 | 1.000 |
| MRPL28 | 9.6e-01 | 1.000 |
| NECAP1 | 9.6e-01 | 1.000 |
| RHOBTB3 | 9.6e-01 | 1.000 |
| CHEK2 | 9.6e-01 | 1.000 |
| REEP5 | 9.6e-01 | 1.000 |
| FADS3 | 9.6e-01 | 1.000 |
| EXOG | 9.6e-01 | 1.000 |
| RPAP2 | 9.6e-01 | 1.000 |
| INPPL1 | 9.6e-01 | 1.000 |
| LRRIQ1 | 9.6e-01 | 1.000 |
| ADA | 9.6e-01 | 1.000 |
| RPS29 | 9.6e-01 | 1.000 |
| OIP5 | 9.6e-01 | 1.000 |
| ATP6V0E2 | 9.6e-01 | 1.000 |
| LMAN2 | 9.6e-01 | 1.000 |
| ZNF93 | 9.6e-01 | 1.000 |
| PLXDC2 | 9.6e-01 | 1.000 |
| FASTKD3 | 9.6e-01 | 1.000 |
| GIT2 | 9.6e-01 | 1.000 |
| PLBD2 | 9.6e-01 | 1.000 |
| GSR | 9.6e-01 | 1.000 |
| CCDC47 | 9.6e-01 | 1.000 |
| NSMCE3 | 9.6e-01 | 1.000 |
| GIN1 | 9.6e-01 | 1.000 |
| MANBAL | 9.6e-01 | 1.000 |
| METTL1 | 9.6e-01 | 1.000 |
| WARS | 9.6e-01 | 1.000 |
| TMEM53 | 9.6e-01 | 1.000 |
| AIFM1 | 9.6e-01 | 1.000 |
| PUM3 | 9.6e-01 | 1.000 |
| ACTR2 | 9.6e-01 | 1.000 |
| DUSP16 | 9.6e-01 | 1.000 |
| CTD-2001E22.2 | 9.6e-01 | 1.000 |
| ANKRD54 | 9.6e-01 | 1.000 |
| FAM114A2 | 9.6e-01 | 1.000 |
| ZUFSP | 9.6e-01 | 1.000 |
| IPO8 | 9.6e-01 | 1.000 |
| SPG7 | 9.6e-01 | 1.000 |
| APBB1 | 9.6e-01 | 1.000 |
| ASCC2 | 9.6e-01 | 1.000 |
| RPS17 | 9.6e-01 | 1.000 |
| TSR1 | 9.6e-01 | 1.000 |
| RPS26 | 9.6e-01 | 1.000 |
| FGGY | 9.6e-01 | 1.000 |
| PDK2 | 9.6e-01 | 1.000 |
| KIAA1967 | 9.6e-01 | 1.000 |
| C17orf101 | 9.6e-01 | 1.000 |
| TRIM36 | 9.6e-01 | 1.000 |
| EPHB2 | 9.6e-01 | 1.000 |
| BLM | 9.6e-01 | 1.000 |
| MITD1 | 9.6e-01 | 1.000 |
| RP11-884K10.5 | 9.6e-01 | 1.000 |
| PSMD6 | 9.6e-01 | 1.000 |
| SNHG5 | 9.6e-01 | 1.000 |
| CEBPB | 9.6e-01 | 1.000 |
| POLR1A | 9.6e-01 | 1.000 |
| RGS3 | 9.6e-01 | 1.000 |
| SIKE1 | 9.6e-01 | 1.000 |
| PLXNB1 | 9.6e-01 | 1.000 |
| FAM195B | 9.6e-01 | 1.000 |
| EAF1 | 9.6e-01 | 1.000 |
| CD276 | 9.6e-01 | 1.000 |
| COA6 | 9.6e-01 | 1.000 |
| NUP160 | 9.6e-01 | 1.000 |
| VPS4B | 9.6e-01 | 1.000 |
| APH1A | 9.6e-01 | 1.000 |
| CYSTM1 | 9.6e-01 | 1.000 |
| SLC2A11 | 9.6e-01 | 1.000 |
| YLPM1 | 9.6e-01 | 1.000 |
| PRNP | 9.6e-01 | 1.000 |
| MYEOV2 | 9.6e-01 | 1.000 |
| IGSF8 | 9.6e-01 | 1.000 |
| RHNO1 | 9.6e-01 | 1.000 |
| C18orf8 | 9.6e-01 | 1.000 |
| UTP15 | 9.6e-01 | 1.000 |
| VWA1 | 9.6e-01 | 1.000 |
| CNPY3 | 9.6e-01 | 1.000 |
| PRRC2A | 9.6e-01 | 1.000 |
| MAPK8IP2 | 9.6e-01 | 1.000 |
| HDHD2 | 9.7e-01 | 1.000 |
| U2AF1L4 | 9.7e-01 | 1.000 |
| MAPKAP1 | 9.7e-01 | 1.000 |
| PACS1 | 9.7e-01 | 1.000 |
| NPDC1 | 9.7e-01 | 1.000 |
| HMGA1 | 9.7e-01 | 1.000 |
| NCKAP1 | 9.7e-01 | 1.000 |
| TOR2A | 9.7e-01 | 1.000 |
| TAF3 | 9.7e-01 | 1.000 |
| BROX | 9.7e-01 | 1.000 |
| PSMD9 | 9.7e-01 | 1.000 |
| OARD1 | 9.7e-01 | 1.000 |
| NFKBIL1 | 9.7e-01 | 1.000 |
| VARS | 9.7e-01 | 1.000 |
| PIR | 9.7e-01 | 1.000 |
| SMURF2 | 9.7e-01 | 1.000 |
| RNF181 | 9.7e-01 | 1.000 |
| DONSON | 9.7e-01 | 1.000 |
| EYA3 | 9.7e-01 | 1.000 |
| KIFC1 | 9.7e-01 | 1.000 |
| CREB3L2 | 9.7e-01 | 1.000 |
| DNAJA2 | 9.7e-01 | 1.000 |
| OS9 | 9.7e-01 | 1.000 |
| TMCO1 | 9.7e-01 | 1.000 |
| ZNF721 | 9.7e-01 | 1.000 |
| RPL29 | 9.7e-01 | 1.000 |
| SNAPIN | 9.7e-01 | 1.000 |
| CHTOP | 9.7e-01 | 1.000 |
| NUCKS1 | 9.7e-01 | 1.000 |
| GLT8D1 | 9.7e-01 | 1.000 |
| PHF3 | 9.7e-01 | 1.000 |
| POP5 | 9.7e-01 | 1.000 |
| TRAPPC13 | 9.7e-01 | 1.000 |
| ARNT2 | 9.7e-01 | 1.000 |
| VTI1A | 9.7e-01 | 1.000 |
| PCSK7 | 9.7e-01 | 1.000 |
| MOB3B | 9.7e-01 | 1.000 |
| SEPHS2 | 9.7e-01 | 1.000 |
| MAGED1 | 9.7e-01 | 1.000 |
| C19orf24 | 9.7e-01 | 1.000 |
| TBC1D1 | 9.7e-01 | 1.000 |
| FKRP | 9.7e-01 | 1.000 |
| STX10 | 9.7e-01 | 1.000 |
| IFI27L2 | 9.7e-01 | 1.000 |
| CHST11 | 9.7e-01 | 1.000 |
| PPP2R3C | 9.7e-01 | 1.000 |
| BRPF3 | 9.7e-01 | 1.000 |
| CLGN | 9.7e-01 | 1.000 |
| ATP5G2 | 9.7e-01 | 1.000 |
| C3orf38 | 9.7e-01 | 1.000 |
| STOML2 | 9.7e-01 | 1.000 |
| NAA16 | 9.7e-01 | 1.000 |
| KPNA4 | 9.7e-01 | 1.000 |
| PRPSAP2 | 9.7e-01 | 1.000 |
| GNB1L | 9.7e-01 | 1.000 |
| ZMYM2 | 9.7e-01 | 1.000 |
| EXO1 | 9.7e-01 | 1.000 |
| ACVR2A | 9.7e-01 | 1.000 |
| ARFIP1 | 9.7e-01 | 1.000 |
| CLDND1 | 9.7e-01 | 1.000 |
| CAND1 | 9.7e-01 | 1.000 |
| SMIM8 | 9.7e-01 | 1.000 |
| KRTCAP2 | 9.7e-01 | 1.000 |
| ATP13A2 | 9.7e-01 | 1.000 |
| THRAP3 | 9.7e-01 | 1.000 |
| EIF4A1 | 9.7e-01 | 1.000 |
| C7orf26 | 9.7e-01 | 1.000 |
| ANO10 | 9.7e-01 | 1.000 |
| CDC20 | 9.7e-01 | 1.000 |
| SHARPIN | 9.7e-01 | 1.000 |
| ANGEL2 | 9.7e-01 | 1.000 |
| SMOX | 9.7e-01 | 1.000 |
| NFKBIB | 9.7e-01 | 1.000 |
| PYM1 | 9.7e-01 | 1.000 |
| RPL26L1 | 9.7e-01 | 1.000 |
| RPS7 | 9.8e-01 | 1.000 |
| NAA20 | 9.8e-01 | 1.000 |
| NCKAP5L | 9.8e-01 | 1.000 |
| CDC42EP4 | 9.8e-01 | 1.000 |
| RGMA | 9.8e-01 | 1.000 |
| RBM12B | 9.8e-01 | 1.000 |
| SEC61A1 | 9.8e-01 | 1.000 |
| RNF220 | 9.8e-01 | 1.000 |
| RPP21 | 9.8e-01 | 1.000 |
| CHST12 | 9.8e-01 | 1.000 |
| DRAM1 | 9.8e-01 | 1.000 |
| RP11-676J15.1 | 9.8e-01 | 1.000 |
| UBXN2A | 9.8e-01 | 1.000 |
| BBS7 | 9.8e-01 | 1.000 |
| ACOT8 | 9.8e-01 | 1.000 |
| RALYL | 9.8e-01 | 1.000 |
| ZC3H14 | 9.8e-01 | 1.000 |
| MT-ND6 | 9.8e-01 | 1.000 |
| RGS19 | 9.8e-01 | 1.000 |
| JUND | 9.8e-01 | 1.000 |
| AC073346.2 | 9.8e-01 | 1.000 |
| AXIN2 | 9.8e-01 | 1.000 |
| MYO10 | 9.8e-01 | 1.000 |
| URM1 | 9.8e-01 | 1.000 |
| ABHD11 | 9.8e-01 | 1.000 |
| ECHS1 | 9.8e-01 | 1.000 |
| STIM2 | 9.8e-01 | 1.000 |
| HIF1A | 9.8e-01 | 1.000 |
| RFXAP | 9.8e-01 | 1.000 |
| MVB12B | 9.8e-01 | 1.000 |
| ALG5 | 9.8e-01 | 1.000 |
| EXOSC8 | 9.8e-01 | 1.000 |
| METTL6 | 9.8e-01 | 1.000 |
| FAM76B | 9.8e-01 | 1.000 |
| FRS2 | 9.8e-01 | 1.000 |
| GLOD4 | 9.8e-01 | 1.000 |
| MIER2 | 9.8e-01 | 1.000 |
| AKAP8 | 9.8e-01 | 1.000 |
| JMJD6 | 9.8e-01 | 1.000 |
| ZNF493 | 9.8e-01 | 1.000 |
| AP1S2 | 9.8e-01 | 1.000 |
| NSUN4 | 9.8e-01 | 1.000 |
| HS1BP3 | 9.8e-01 | 1.000 |
| VPS51 | 9.8e-01 | 1.000 |
| POC1A | 9.8e-01 | 1.000 |
| ASF1A | 9.8e-01 | 1.000 |
| KMT2D | 9.8e-01 | 1.000 |
| TKFC | 9.8e-01 | 1.000 |
| ILVBL | 9.8e-01 | 1.000 |
| SH2B1 | 9.8e-01 | 1.000 |
| SETX | 9.8e-01 | 1.000 |
| SUGP1 | 9.8e-01 | 1.000 |
| LZIC | 9.8e-01 | 1.000 |
| RP11-220I1.1 | 9.8e-01 | 1.000 |
| ADNP | 9.8e-01 | 1.000 |
| SETMAR | 9.8e-01 | 1.000 |
| TCHP | 9.8e-01 | 1.000 |
| LIN28A | 9.8e-01 | 1.000 |
| BAG5 | 9.8e-01 | 1.000 |
| MBD4 | 9.8e-01 | 1.000 |
| KPTN | 9.8e-01 | 1.000 |
| METTL23 | 9.8e-01 | 1.000 |
| C19orf47 | 9.8e-01 | 1.000 |
| ZNF579 | 9.8e-01 | 1.000 |
| HSPBP1 | 9.8e-01 | 1.000 |
| UBE2S | 9.8e-01 | 1.000 |
| ASAP3 | 9.8e-01 | 1.000 |
| PIAS3 | 9.8e-01 | 1.000 |
| KIF3A | 9.8e-01 | 1.000 |
| C10orf88 | 9.8e-01 | 1.000 |
| SKP1 | 9.8e-01 | 1.000 |
| IFRD2 | 9.8e-01 | 1.000 |
| MT-ND4 | 9.8e-01 | 1.000 |
| FASN | 9.8e-01 | 1.000 |
| RNF26 | 9.8e-01 | 1.000 |
| PITPNA | 9.8e-01 | 1.000 |
| SLC29A2 | 9.9e-01 | 1.000 |
| TMED8 | 9.9e-01 | 1.000 |
| DAGLB | 9.9e-01 | 1.000 |
| OAF | 9.9e-01 | 1.000 |
| BNIP3L | 9.9e-01 | 1.000 |
| GBA | 9.9e-01 | 1.000 |
| PRPF3 | 9.9e-01 | 1.000 |
| CYB5R4 | 9.9e-01 | 1.000 |
| RNF24 | 9.9e-01 | 1.000 |
| CTSB | 9.9e-01 | 1.000 |
| FAM213B | 9.9e-01 | 1.000 |
| ABL2 | 9.9e-01 | 1.000 |
| EIF2B5 | 9.9e-01 | 1.000 |
| FAM213A | 9.9e-01 | 1.000 |
| STK40 | 9.9e-01 | 1.000 |
| MOSPD3 | 9.9e-01 | 1.000 |
| TINF2 | 9.9e-01 | 1.000 |
| PEA15 | 9.9e-01 | 1.000 |
| PPP2R1A | 9.9e-01 | 1.000 |
| RAB40C | 9.9e-01 | 1.000 |
| ATP5A1 | 9.9e-01 | 1.000 |
| USP11 | 9.9e-01 | 1.000 |
| ATAD3B | 9.9e-01 | 1.000 |
| SUPT6H | 9.9e-01 | 1.000 |
| SQLE | 9.9e-01 | 1.000 |
| IDH1 | 9.9e-01 | 1.000 |
| YARS | 9.9e-01 | 1.000 |
| ADGRL3 | 9.9e-01 | 1.000 |
| ALG13 | 9.9e-01 | 1.000 |
| DCP1B | 9.9e-01 | 1.000 |
| SARS | 9.9e-01 | 1.000 |
| PUM1 | 9.9e-01 | 1.000 |
| GPAA1 | 9.9e-01 | 1.000 |
| TBC1D13 | 9.9e-01 | 1.000 |
| PRPF40A | 9.9e-01 | 1.000 |
| ANKRD12 | 9.9e-01 | 1.000 |
| FABP7 | 9.9e-01 | 1.000 |
| CCDC189 | 9.9e-01 | 1.000 |
| SSX2IP | 9.9e-01 | 1.000 |
| SUPT3H | 9.9e-01 | 1.000 |
| MRPL50 | 9.9e-01 | 1.000 |
| B3GAT1 | 9.9e-01 | 1.000 |
| AMPD2 | 9.9e-01 | 1.000 |
| INSIG1 | 9.9e-01 | 1.000 |
| BMP7 | 9.9e-01 | 1.000 |
| SPRED1 | 9.9e-01 | 1.000 |
| AP1G1 | 9.9e-01 | 1.000 |
| CNIH1 | 9.9e-01 | 1.000 |
| KLHDC8A | 9.9e-01 | 1.000 |
| UTP23 | 9.9e-01 | 1.000 |
| ATP5F1 | 9.9e-01 | 1.000 |
| SETD5 | 9.9e-01 | 1.000 |
| RPSA | 9.9e-01 | 1.000 |
| PSMG3 | 9.9e-01 | 1.000 |
| SIL1 | 9.9e-01 | 1.000 |
| TCF3 | 9.9e-01 | 1.000 |
| ALG12 | 9.9e-01 | 1.000 |
| CUL1 | 9.9e-01 | 1.000 |
| DIS3 | 9.9e-01 | 1.000 |
| RP4-564F22.2 | 9.9e-01 | 1.000 |
| MLX | 9.9e-01 | 1.000 |
| CEP41 | 9.9e-01 | 1.000 |
| COX8A | 9.9e-01 | 1.000 |
| ZNF35 | 9.9e-01 | 1.000 |
| XRCC6 | 9.9e-01 | 1.000 |
| LAMTOR3 | 9.9e-01 | 1.000 |
| EIF3J | 9.9e-01 | 1.000 |
| MFSD1 | 9.9e-01 | 1.000 |
| ACTR6 | 9.9e-01 | 1.000 |
| TRIM44 | 9.9e-01 | 1.000 |
| GATS | 9.9e-01 | 1.000 |
| TANK | 9.9e-01 | 1.000 |
| CLPTM1 | 1.0e+00 | 1.000 |
| APPL2 | 1.0e+00 | 1.000 |
| SEC13 | 1.0e+00 | 1.000 |
| RIDA | 1.0e+00 | 1.000 |
| SH3PXD2B | 1.0e+00 | 1.000 |
| SOX3 | 1.0e+00 | 1.000 |
| CH507-254M2.3 | 1.0e+00 | 1.000 |
| PIN4 | 1.0e+00 | 1.000 |
| C8orf33 | 1.0e+00 | 1.000 |
| ACTN4 | 1.0e+00 | 1.000 |
| SMG6 | 1.0e+00 | 1.000 |
| SERAC1 | 1.0e+00 | 1.000 |
| CAMKK2 | 1.0e+00 | 1.000 |
| TRAK1 | 1.0e+00 | 1.000 |
| NIN | 1.0e+00 | 1.000 |
| SHC3 | 1.0e+00 | 1.000 |
| TSPAN4 | 1.0e+00 | 1.000 |
| NDUFAF4 | 1.0e+00 | 1.000 |
| DDR1 | 1.0e+00 | 1.000 |
| RALGAPB | 1.0e+00 | 1.000 |
| CDK20 | 1.0e+00 | 1.000 |
| PTN | 1.0e+00 | 1.000 |
| NONO | 1.0e+00 | 1.000 |
| AC012146.7 | 1.0e+00 | 1.000 |
| USP3 | 1.0e+00 | 1.000 |
| RUVBL1 | 1.0e+00 | 1.000 |
| CWC25 | 1.0e+00 | 1.000 |
| ALDH2 | 1.0e+00 | 1.000 |
| CEP63 | 1.0e+00 | 1.000 |
| C3orf14 | 1.0e+00 | 1.000 |
| PEF1 | 1.0e+00 | 1.000 |
| KIF1B | 1.0e+00 | 1.000 |
| MSANTD4 | 1.0e+00 | 1.000 |
| C19orf40 | 1.0e+00 | 1.000 |
| ACTR1B | 1.0e+00 | 1.000 |
| FBXL15 | 1.0e+00 | 1.000 |
Top DE genes detected
plot_gene <- function(glocus,testgene,tg.cells,neg.cells,expression,residual){
paired.df <- data.frame(row.names = c(tg.cells,neg.cells),
expression = expression,
residual = residual,
condition=c(rep(glocus,length(tg.cells)),
rep('neg',length(neg.cells))))
paired.df$condition <- relevel(as.factor(paired.df$condition),glocus)
plot1 <- ggplot(data = paired.df, aes(expression,fill=condition)) +
geom_histogram(position = "dodge",bins = 20) + theme_bw() +
labs(title=paste('test gene:',testgene), x = 'UMI count') +
theme(plot.title = element_text(size=14),
axis.text=element_text(size=12), axis.title=element_text(size=14),
legend.position = 'none')
plot2 <- ggplot(data = paired.df, aes(residual,fill=condition)) +
geom_histogram(position = "dodge",bins = 20) + theme_bw() +
labs(title=paste0('(',glocus,' locus)'), x = 'Pearson residual') +
theme(plot.title = element_text(size=14),
axis.text=element_text(size=12), axis.title=element_text(size=14),
legend.title = element_text(size=12),
legend.text = element_text(size=11),
legend.position = c(0.8,0.85))
grid.arrange(plot1,plot2, ncol=2)
}
for (testgene in BCL11B.pval$gene[order(BCL11B.pval$pval)][1:4]){
test.expression = as.numeric(count.gene.exp[testgene,])
test.residual = as.numeric(sctransform_data[testgene, c(tg.cells,neg.cells)])
plot_gene(glocus,testgene,tg.cells,neg.cells,test.expression,test.residual)
}
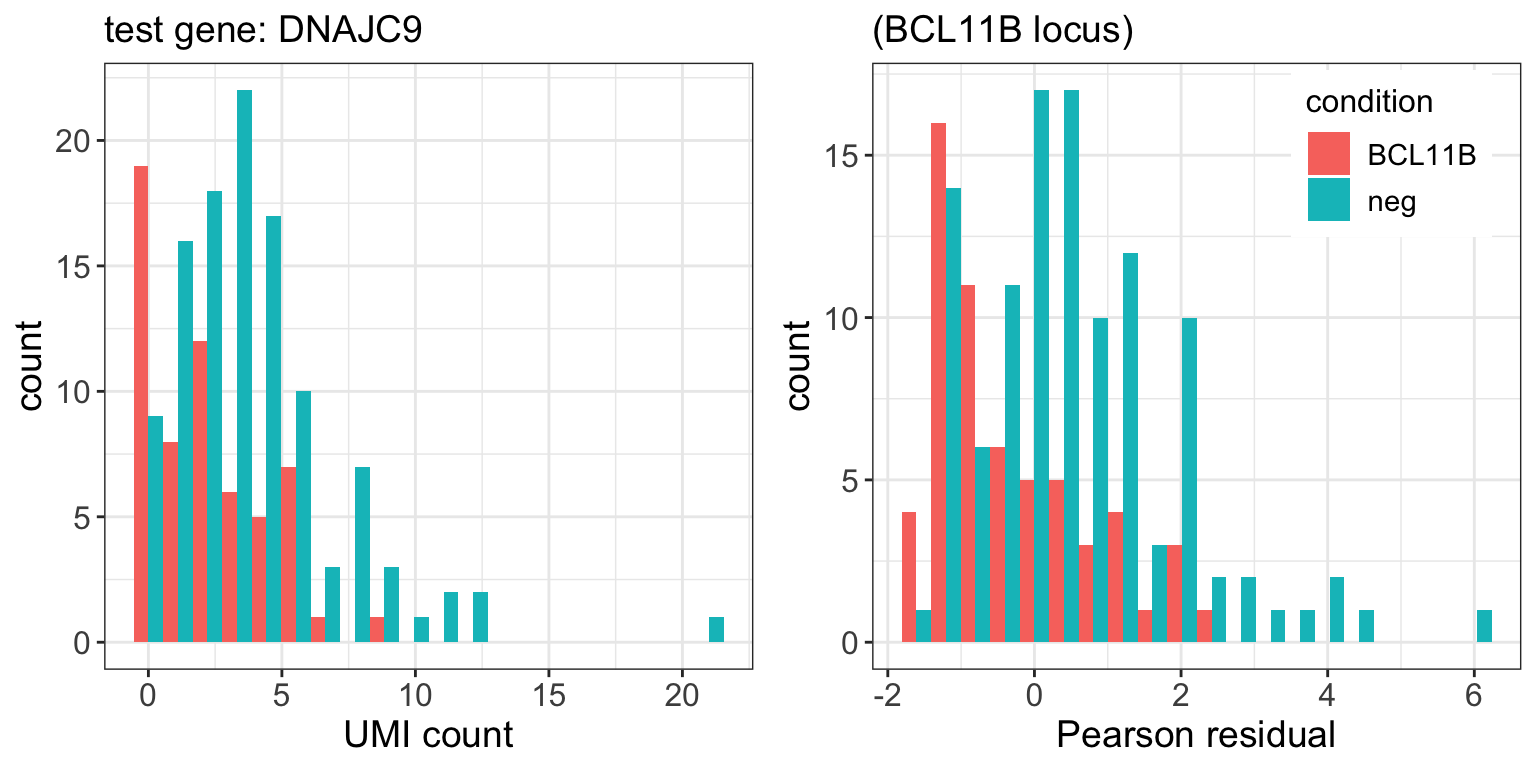
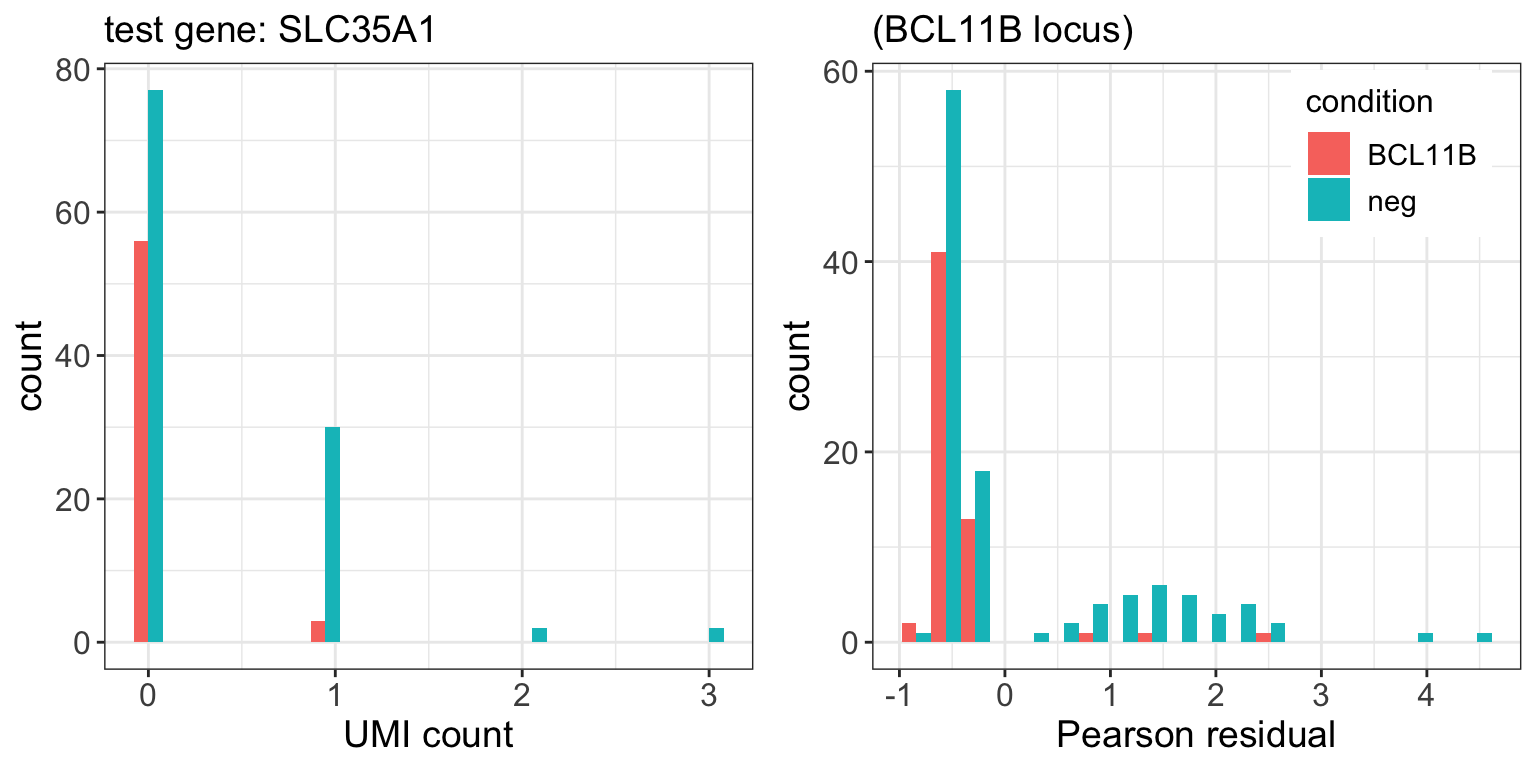
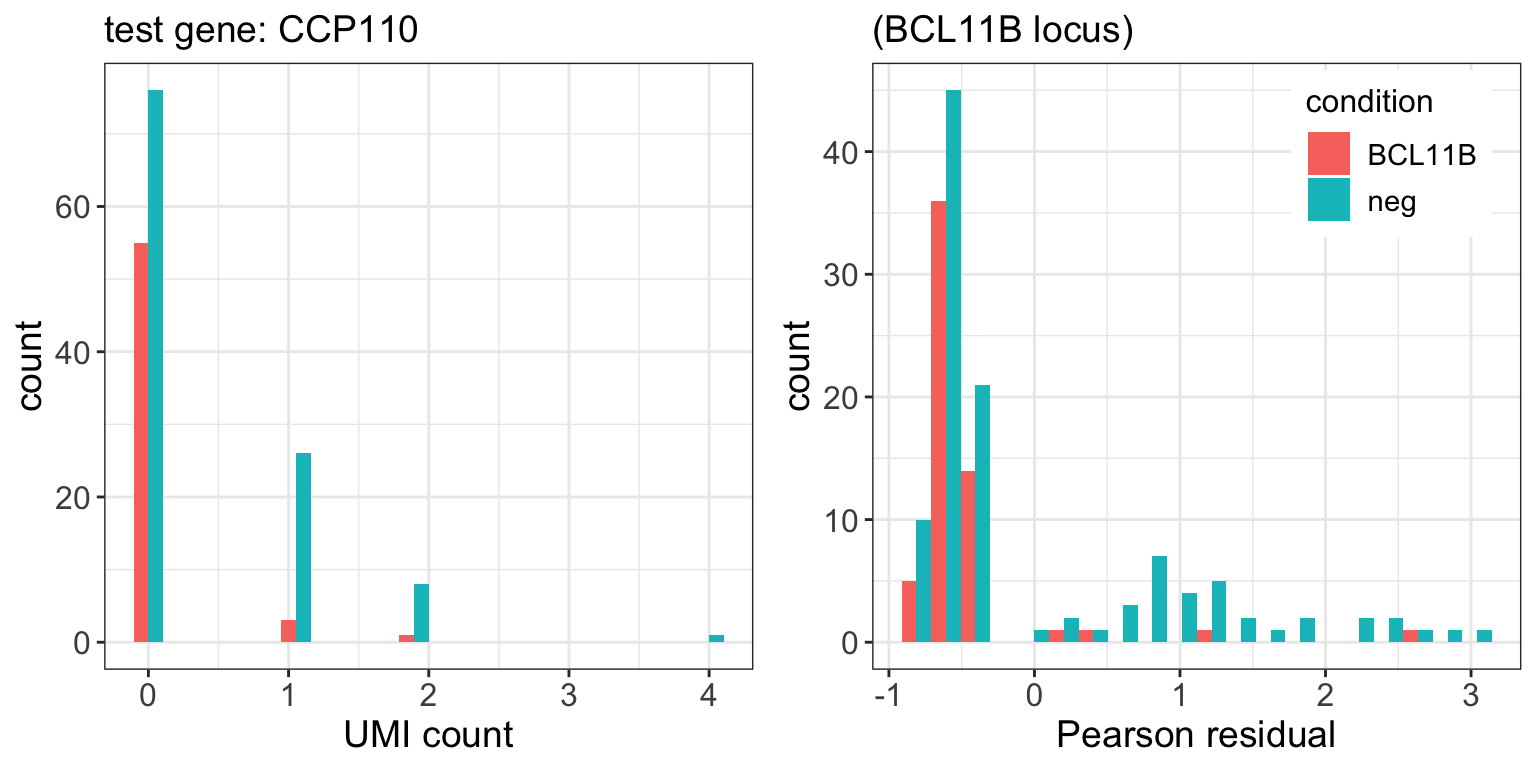
Permutation test
Comparison with using MASTcpm
Other perturbed target loci
enh_loci <- row.names(exp.per.enhancer)[-c(2,13,17,20,22)]
neg.cells <- colnames(exp.per.enhancer)[exp.per.enhancer["neg",]>0 & nlocus==1]
for (glocus in enh_loci){
cat('Target locus:',glocus)
cat(' \n')
nlocus <- colSums(exp.per.enhancer>0)
tg.cells <- colnames(exp.per.enhancer)[exp.per.enhancer[glocus,]>0 & nlocus==1]
cat('Number of targeted cells:',length(tg.cells))
cat(' \n')
ttest_loop(subset.sctransform_data,tg.cells,neg.cells,3)
cat('--------------------------------------')
cat('\n')
}Number of targeted cells: 97
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| CWF19L2 | 0.00010 | 0.81 |
| ASB3 | 0.00028 | 0.90 |
| THG1L | 0.00033 | 0.90 |
Target locus: CHRNA3
Number of targeted cells: 39
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| DPH7 | 5.2e-05 | 0.42 |
| ARFGAP3 | 2.2e-04 | 0.48 |
| RABL6 | 3.3e-04 | 0.48 |
Target locus: DPYD
Number of targeted cells: 133
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| PGGT1B | 9.1e-05 | 0.58 |
| ZNF48 | 2.2e-04 | 0.58 |
| DHODH | 4.9e-04 | 0.58 |
Target locus: GALNT10
Number of targeted cells: 124
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| TRAK1 | 8.6e-05 | 0.64 |
| ZNF512 | 2.3e-04 | 0.64 |
| KCTD10 | 2.3e-04 | 0.64 |
Target locus: KCTD13
Number of targeted cells: 76
1 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| SLC20A2 | 8.2e-06 | 0.066 |
| BLOC1S1 | 2.3e-04 | 0.790 |
| NINJ1 | 3.8e-04 | 0.790 |
Target locus: KMT2E
Number of targeted cells: 48
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| LIN7C | 4.2e-05 | 0.19 |
| SERTAD2 | 4.7e-05 | 0.19 |
| SOWAHC | 1.2e-04 | 0.27 |
Target locus: LINC00637
Number of targeted cells: 128
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| PRCC | 1.7e-05 | 0.14 |
| C7orf73 | 9.9e-05 | 0.40 |
| SYNC | 4.3e-04 | 0.68 |
Target locus: LOC100507431
Number of targeted cells: 92
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| CBR4 | 0.00024 | 0.88 |
| FAM228B | 0.00039 | 0.88 |
| C11orf58 | 0.00060 | 0.88 |
Target locus: LOC105376975
Number of targeted cells: 90
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| YIPF4 | 0.00016 | 0.93 |
| PARD3 | 0.00064 | 0.93 |
| KIAA0907 | 0.00068 | 0.93 |
Target locus: miR137
Number of targeted cells: 128
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| ARMCX6 | 3.8e-05 | 0.31 |
| CPPED1 | 2.1e-04 | 0.48 |
| FAM228B | 3.2e-04 | 0.48 |
Target locus: NAB2
Number of targeted cells: 85
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| PRCC | 2.7e-05 | 0.18 |
| GNL3 | 4.3e-05 | 0.18 |
| GNL1 | 1.7e-04 | 0.37 |
Target locus: NGEF
Number of targeted cells: 13
141 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| SPIN2B | 0 | 7.9e-06 |
| TPT1-AS1 | 0 | 7.9e-06 |
| CXorf40B | 0 | 1.2e-05 |
Target locus: PBRM1
Number of targeted cells: 20
54 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| NEK1 | 0e+00 | 5.6e-05 |
| ZNF544 | 0e+00 | 7.9e-05 |
| BRAT1 | 1e-07 | 2.2e-04 |
Target locus: PCDHA123
Number of targeted cells: 178
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| KCTD10 | 0.00012 | 0.67 |
| SMG1 | 0.00021 | 0.67 |
| PDLIM2 | 0.00056 | 0.67 |
Target locus: PPP1R16B
Number of targeted cells: 110
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| TMX3 | 5.8e-05 | 0.47 |
| GCAT | 2.1e-04 | 0.62 |
| SMG1 | 3.8e-04 | 0.62 |
Target locus: RERE
Number of targeted cells: 115
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| PLXNB1 | 0.00018 | 0.76 |
| USMG5 | 0.00025 | 0.76 |
| RPE | 0.00028 | 0.76 |
Target locus: STAT6
Number of targeted cells: 115
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| QRICH1 | 0.00015 | 1 |
| PRKCZ | 0.00038 | 1 |
| CEP70 | 0.00054 | 1 |
Target locus: TRANK1
Number of targeted cells: 86
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| RPUSD1 | 2.5e-05 | 0.21 |
| CLK3 | 1.8e-04 | 0.61 |
| KCTD10 | 2.6e-04 | 0.61 |
Target locus: UBE2Q2P1
Number of targeted cells: 30
13 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| FAM171A1 | 4.0e-07 | 0.003 |
| FAM199X | 1.2e-06 | 0.005 |
| PLCXD1 | 4.1e-06 | 0.011 |
Target locus: VPS45
Number of targeted cells: 101
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| SUMO3 | 0.00012 | 0.72 |
| ARAF | 0.00018 | 0.72 |
| FAM228B | 0.00027 | 0.72 |
Positive controls
pos_loci <- row.names(exp.per.enhancer)[c(17,20,22)]
# neg.cells <- colnames(exp.per.enhancer)[exp.per.enhancer["neg",]>0 & nlocus==1]
for (glocus in pos_loci){
if (glocus=='pos'){
cat('Positive locus:','SNAP91')
cat(' \n')
} else {
cat('Positive locus:',glocus)
cat(' \n')
}
nlocus <- colSums(exp.per.enhancer>0)
tg.cells <- colnames(exp.per.enhancer)[exp.per.enhancer[glocus,]>0 & nlocus==1]
cat('Number of targeted cells:',length(tg.cells))
cat(' \n')
ttest_loop(subset.sctransform_data,tg.cells,neg.cells,4)
cat('--------------------------------------')
cat('\n')
}Number of targeted cells: 29
4 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| SLC25A17 | 0.0e+00 | 0.00033 |
| SCARA3 | 3.0e-06 | 0.01200 |
| TMEM259 | 1.5e-05 | 0.04100 |
| SF3B4 | 4.5e-05 | 0.09200 |
Positive locus: SETD1A
Number of targeted cells: 54
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| PRCC | 3.1e-05 | 0.25 |
| TRIM26 | 4.1e-04 | 0.75 |
| ASRGL1 | 4.8e-04 | 0.75 |
| USP1 | 7.3e-04 | 0.75 |
Positive locus: TCF4-ITF2
Number of targeted cells: 105
0 genes passed FDR=0.1 cutoff
| gene | pval | adj |
|---|---|---|
| HMBS | 0.00020 | 0.99 |
| SCD5 | 0.00076 | 0.99 |
| AGPAT5 | 0.00082 | 0.99 |
| KCTD10 | 0.00100 | 0.99 |
Session Information
sessionInfo()R version 3.5.2 (2018-12-20)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] lattice_0.20-38 ggplot2_3.1.0 sctransform_0.2.0 Matrix_1.2-16
[5] gridExtra_2.3 kableExtra_1.0.1 knitr_1.22
loaded via a namespace (and not attached):
[1] tidyselect_0.2.5 xfun_0.5 purrr_0.3.1
[4] reshape2_1.4.3 listenv_0.7.0 colorspace_1.4-0
[7] htmltools_0.3.6 viridisLite_0.3.0 yaml_2.2.0
[10] rlang_0.3.1 pillar_1.3.1 glue_1.3.0
[13] withr_2.1.2 plyr_1.8.4 stringr_1.4.0
[16] munsell_0.5.0 gtable_0.2.0 rvest_0.3.2
[19] future_1.13.0 codetools_0.2-16 evaluate_0.13
[22] labeling_0.3 parallel_3.5.2 highr_0.7
[25] Rcpp_1.0.0 readr_1.3.1 scales_1.0.0
[28] webshot_0.5.1 hms_0.4.2 digest_0.6.18
[31] stringi_1.3.1 dplyr_0.8.0.1 tools_3.5.2
[34] magrittr_1.5 lazyeval_0.2.1 tibble_2.0.1
[37] crayon_1.3.4 future.apply_1.2.0 pkgconfig_2.0.2
[40] MASS_7.3-51.1 xml2_1.2.0 assertthat_0.2.0
[43] rmarkdown_1.15 httr_1.4.0 rstudioapi_0.9.0
[46] R6_2.4.0 globals_0.12.4 compiler_3.5.2