ashr on sctransformed CROP-seq data
Yifan Zhou
6/16/2019
library(ashr)
library(ggplot2)
library(knitr)
library(kableExtra)
options(stringsAsFactors = F)
wkdir <- '~/Downloads/ASoC/singlecell/'
load(paste0(wkdir,'data/cropseq_expression.Rd'))Data property
nlocus <- colSums(exp.per.enhancer>0)
gene.percent <- rowMeans(gene.exp>0)
hist(gene.percent,xlab = '% of cells a gene is in',ylab = 'Count')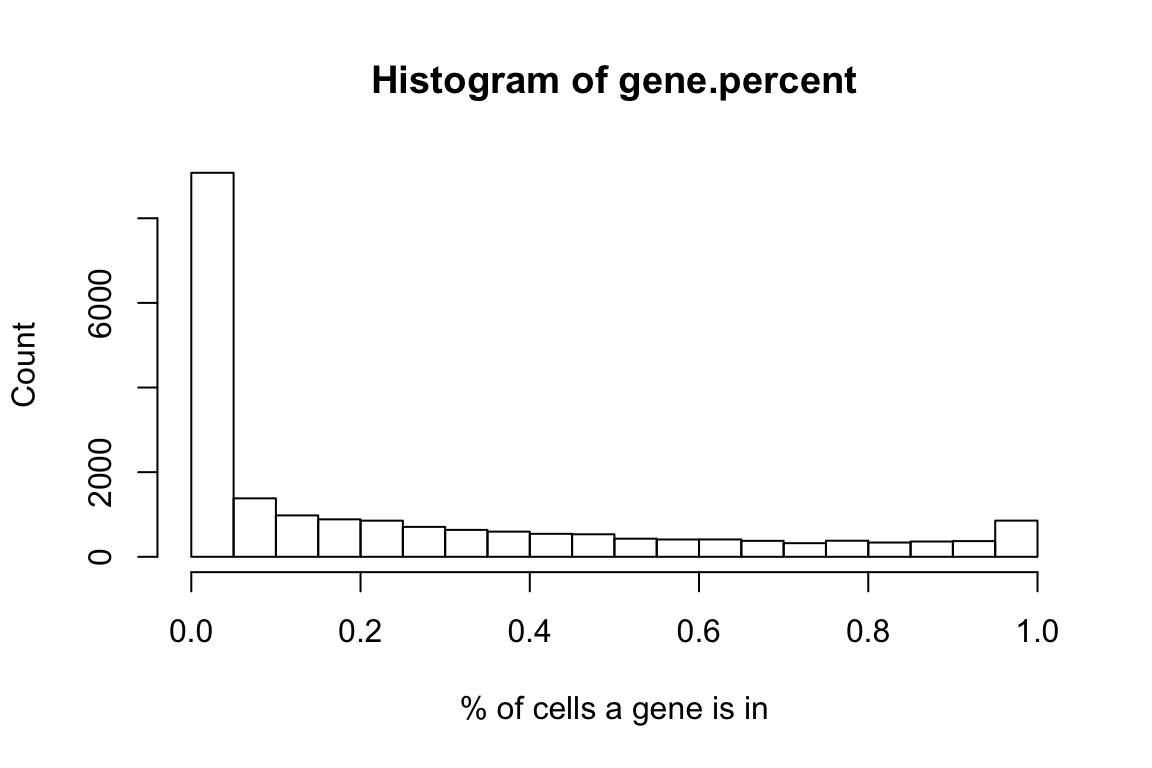
ncell_uniq <- rep(NA,nrow(exp.per.gRNA))
for (i in 1:nrow(exp.per.gRNA)){
glocus <- row.names(exp.per.gRNA)[i]
ncell_uniq[i]=sum(exp.per.gRNA[glocus,]>0 & nlocus==1)
}
tmp <- sapply(strsplit(row.names(exp.per.gRNA),split = '_'),
function(x){paste(x[1],x[2],sep = '_')})
names(ncell_uniq) <- tmp
cat('Number of cells uniquely targeted by each gRNA:')Number of cells uniquely targeted by each gRNA:print(ncell_uniq) BAG5_1 BAG5_2 BAG5_3 BCL11B_1 BCL11B_2
36 33 31 13 33
BCL11B_3 CHRNA3_1 CHRNA3_2 CHRNA3_3 GALNT10_1
13 3 23 13 38
GALNT10_2 GALNT10_3 KCTD13_1 KCTD13_2 KCTD13_3
37 50 30 32 15
KMT2E_1 KMT2E_2 KMT2E_3 LOC105376975_1 LOC105376975_2
11 19 18 35 50
LOC105376975_3 miR137_1 miR137_2 miR137_3 NAB2_1
8 44 30 54 36
NAB2_2 NAB2_3 PCDHA123_1 PCDHA123_2 PCDHA123_3
24 25 53 64 62
RERE_1 RERE_2 RERE_3 SETD1A_1st SETD1A_1st
48 46 25 23 28
SETD1A_1st STAT6_1 STAT6_2 STAT6_3 TRANK1_1
4 24 38 53 3
TRANK1_2 TRANK1_3 UBE2Q2P1_1 UBE2Q2P1_2 UBE2Q2P1_3
19 65 11 8 11
VPS45_1 VPS45_2 VPS45_3 NGEF_1 NGEF_2
14 79 9 11 2
NGEF_3 DPYD_1 DPYD_2 DPYD_3 LOC100507431_1
0 37 68 28 15
LOC100507431_2 LOC100507431_3 LINC00637_1 LINC00637_2 LINC00637_3
29 49 45 34 53
PBRM1_1 PBRM1_2 PBRM1_3 PPP1R16B_1 PPP1R16B_2
4 12 4 44 31
PPP1R16B_3 TCF4-ITF2_1 TCF4-ITF2_2 TCF4-ITF2_3 pos_SNAP91
35 50 45 12 3
pos_SNAP91 neg_EGFP neg_EGFP neg_EGFP neg_CTRL
26 5 29 35 17
neg_CTRL
26 Categorical regression on sctransformed data
We transformed the single-cell raw count data using sctransform to Pearson residuals that supposedly follow \(N(0,1)\) distribution per gene.
We filtered the genes to be those present in at least 20% of all cells (8117 in total).
Then, for each target gRNA, we selected 2 groups of cells: cells that only contain the target gRNAs, and cells that only contain the negative control gRNAs. We conducted categorical regression on the Pearson residuals (~ 2 conditions) to obtain the summary statistics reflecting how differentially expressed each gene is under the 2 conditions. 70 (gRNA vs neg ctrl) pairs were tested.
categoric <- function(mtx,tg.cells,neg.cells){
pval <- rep(NA,nrow(mtx))
beta <- rep(NA,nrow(mtx))
se <- rep(NA,nrow(mtx))
subset <- mtx[,c(tg.cells,neg.cells)]
for (i in 1:nrow(subset)){
df <- data.frame(row.names = c(tg.cells,neg.cells),
residual = as.numeric(subset[i,]),
condition=c(rep(1,length(tg.cells)),
rep(0,length(neg.cells))))
cat.lm <- summary(lm(residual~factor(condition), data = df))
beta[i] <- cat.lm$coefficients[2,1]
se[i] <- cat.lm$coefficients[2,2]
pval[i] <- cat.lm$coefficients[2,4]
}
return(list(beta=beta,se=se,pval=pval))
}
vst_out <- readRDS(paste0(wkdir,'data/sctransformed_object.rds'))
sctransform_data <- vst_out$y
## Filter the genes to be present in > 20% cells:
subset.genes <- names(gene.percent)[gene.percent>0.2]
subset.sctransform_data <- sctransform_data[subset.genes,]
dim(subset.sctransform_data)
## Categorical regression on sctransformed data
summary_per_gene <- list()
neg.cells <- colnames(exp.per.enhancer)[exp.per.enhancer["neg",]>0 & nlocus==1]
# btw each gRNA condition and neg ctrl
for (glocus in row.names(exp.per.gRNA)[-c(51,72:76)]){
print(paste('gRNA target:',glocus))
tg.cells <- colnames(exp.per.gRNA)[exp.per.gRNA[glocus,]>0 & nlocus==1]
print(paste('# of targeted cells:',length(tg.cells)))
summary_per_gene[[glocus]] <- categoric(subset.sctransform_data,tg.cells,neg.cells)
}
saveRDS(summary_per_gene,file = 'data/categoric_summary.sctrans_gene0.2.rds')ashr meta-analysis on regression summary statistics
For each gene, we combined the 70 DE effect sizes and standard errors estimated in the previous step, and used an adaptive shrinkage method, ashr, to discern which effect sizes are truly non-zero (significant). Here, we used a cutoff of 0.1 for lfsr (the local false sign rate).
summary_per_gene <- readRDS(paste0(wkdir,'data/categoric_summary.sctrans_gene0.2.rds'))
beta.mtx <- sapply(summary_per_gene, function(x){x$beta})
se.mtx <- sapply(summary_per_gene, function(x){x$se})
pval.mtx <- sapply(summary_per_gene, function(x){x$pval})
subset.genes <- names(gene.percent)[gene.percent>0.2]
rownames(beta.mtx) <- subset.genes
rownames(se.mtx) <- subset.genes
rownames(pval.mtx) <- subset.geneslfsr.mtx <- data.frame(matrix(nrow = nrow(beta.mtx),ncol = ncol(beta.mtx)),
row.names = rownames(beta.mtx))
names(lfsr.mtx) <- colnames(beta.mtx)
betaest.mtx <- lfsr.mtx
for (i in 1:nrow(beta.mtx)){
ash.tmp <- ash(beta.mtx[i,],se.mtx[i,])
lfsr.mtx[i,] <- get_lfsr(ash.tmp)
betaest.mtx[i,] <- get_pm(ash.tmp)
}
save(betaest.mtx,lfsr.mtx,file = 'data/ashr_estimation.Rdata')load(paste0(wkdir,'data/ashr_estimation.Rdata'))
tmp.gRNA <- colSums(lfsr.mtx<0.1)
tmp.gene <- rowSums(lfsr.mtx<0.1)
signif_genes <- row.names(betaest.mtx)[tmp.gene>0]
cat('There are',length(signif_genes),'significant genes in total.')There are 462 significant genes in total.gRNA targets per significant gene
Here are all the significant genes (\(lfsr<0.1\)) and the gRNA groups where they are differentially expressed:
signif_genes.gRNA.lst <- list()
for(i in signif_genes){
gRNA.group <- colnames(betaest.mtx)[lfsr.mtx[i,]<0.1]
signif_genes.gRNA.lst[[i]] <- sapply(strsplit(gRNA.group,split = '_'),
function(x){paste(x[1],x[2],sep = '_')})
}
# Make table:
compact.genes.lst <- sapply(signif_genes.gRNA.lst,FUN = function(x){paste(x,collapse = ', ')})
compact.genes.df <- as.data.frame(compact.genes.lst)
tmp.indx <- order(rownames(compact.genes.df))
compact.genes.df <- data.frame(signif_gene=rownames(compact.genes.df)[tmp.indx],
gRNA_targets=compact.genes.df[tmp.indx,])
# Present table:
kable(compact.genes.df) %>%
kable_styling() %>%
scroll_box(width = "100%", height = "400px")| signif_gene | gRNA_targets |
|---|---|
| A2M | VPS45_1 |
| ABHD17A | CHRNA3_1 |
| AC084018.1 | CHRNA3_1 |
| ACBD3 | LOC105376975_3 |
| ACER3 | UBE2Q2P1_3 |
| ADGRL2 | KCTD13_1, pos_SNAP91 |
| AFF4 | UBE2Q2P1_2 |
| AHCTF1 | PBRM1_1 |
| AIFM1 | LOC105376975_2 |
| AKAP11 | pos_SNAP91 |
| ALDH1B1 | TRANK1_2 |
| AMN1 | UBE2Q2P1_3, PPP1R16B_2 |
| AMPH | pos_SNAP91 |
| ANKMY2 | PBRM1_3 |
| ANLN | SETD1A_1st |
| APC | KMT2E_2, DPYD_1 |
| APMAP | miR137_1 |
| ARAF | BAG5_1, BAG5_2, CHRNA3_3, GALNT10_1, GALNT10_3, KMT2E_1, KMT2E_3, miR137_1, NAB2_1, NAB2_2, RERE_1, UBE2Q2P1_3, VPS45_2, NGEF_1, DPYD_2, LOC100507431_1, LOC100507431_2, LINC00637_1, LINC00637_3, TCF4-ITF2_1 |
| ARFGAP1 | BAG5_2, NAB2_2, VPS45_2, LOC100507431_2, TCF4-ITF2_1 |
| ARHGAP11A | NGEF_2 |
| ARHGEF9 | PBRM1_3 |
| ARMCX1 | GALNT10_3 |
| ARMCX2 | VPS45_3 |
| ARMCX6 | BCL11B_1, miR137_1, miR137_3, NAB2_2, PCDHA123_1, DPYD_1, LOC100507431_1, LOC100507431_2, TCF4-ITF2_2 |
| ARNT | LINC00637_1 |
| ASPM | PBRM1_1 |
| ASRGL1 | BCL11B_2, GALNT10_1, miR137_2, PCDHA123_1, SETD1A_1st |
| ATAD1 | LOC105376975_2 |
| ATF2 | PPP1R16B_2 |
| ATG4C | TRANK1_1 |
| ATP5B | UBE2Q2P1_2 |
| ATPIF1 | LOC105376975_2, TCF4-ITF2_2 |
| AURKB | NGEF_2 |
| BAG1 | BAG5_2 |
| BBIP1 | NGEF_2 |
| BCL10 | STAT6_1, DPYD_2 |
| BIRC6 | SETD1A_1st |
| BORA | NGEF_2 |
| BRAF | SETD1A_1st |
| BTBD1 | CHRNA3_3 |
| BTBD7 | CHRNA3_1 |
| BUD13 | GALNT10_1, PCDHA123_1 |
| C16orf72 | TRANK1_1 |
| C17orf80 | VPS45_2 |
| C19orf47 | PBRM1_2, PPP1R16B_3 |
| C1orf131 | NGEF_2 |
| C1orf56 | KMT2E_3, PCDHA123_1 |
| C2orf49 | CHRNA3_3, PCDHA123_1, DPYD_3, LINC00637_3 |
| C2orf69 | NGEF_2 |
| C7orf73 | STAT6_3, LINC00637_1, LINC00637_2 |
| CAMSAP1 | TCF4-ITF2_1 |
| CAPN15 | BCL11B_2 |
| CASC4 | PBRM1_3 |
| CASD1 | DPYD_2 |
| CBWD1 | CHRNA3_3 |
| CBX4 | CHRNA3_2 |
| CCDC126 | NGEF_2 |
| CCDC136 | CHRNA3_2, GALNT10_1, STAT6_2, PBRM1_2, PPP1R16B_1 |
| CCDC167 | PBRM1_1 |
| CCDC50 | GALNT10_1 |
| CCDC84 | SETD1A_1st, STAT6_3, TCF4-ITF2_3 |
| CCNF | NGEF_2 |
| CCNH | STAT6_2, UBE2Q2P1_3 |
| CDC25B | PBRM1_1 |
| CDC6 | BCL11B_2, miR137_1 |
| CDCA2 | NGEF_2 |
| CDCA3 | NGEF_2 |
| CDCA7 | PCDHA123_2, TRANK1_3, DPYD_2 |
| CDKN3 | BAG5_3 |
| CECR5 | LOC105376975_2 |
| CENPA | NGEF_2 |
| CEP170 | TRANK1_1 |
| CEP250 | BAG5_1, BAG5_3, KCTD13_1, PPP1R16B_2 |
| CHAMP1 | LOC105376975_1 |
| CHEK2 | PBRM1_1 |
| CHTF18 | KCTD13_3, TCF4-ITF2_3 |
| CKAP2L | TRANK1_1 |
| CLCN6 | TRANK1_1 |
| CLCN7 | miR137_1 |
| CLK3 | TRANK1_3, TCF4-ITF2_1 |
| CLN8 | DPYD_2 |
| CLPTM1L | PCDHA123_2, DPYD_2 |
| CMBL | PBRM1_3 |
| CNPY4 | KMT2E_1, PCDHA123_3 |
| CNRIP1 | KMT2E_1 |
| CNTROB | NGEF_1 |
| COMMD6 | miR137_2 |
| COQ7 | KMT2E_2 |
| CPNE1 | KCTD13_1 |
| CPPED1 | BAG5_1, BAG5_3, CHRNA3_3, miR137_2, NAB2_3, VPS45_3, NGEF_1, PBRM1_2, pos_SNAP91 |
| CRYBB2 | LOC105376975_3 |
| CWC25 | KCTD13_1 |
| CWF19L2 | BAG5_1, BAG5_3, BCL11B_1, BCL11B_2, miR137_1, miR137_3, NAB2_3, PCDHA123_3, STAT6_3, LOC100507431_2, TCF4-ITF2_2 |
| CYFIP2 | STAT6_2 |
| DBI | PPP1R16B_3 |
| DBT | KCTD13_3 |
| DDX3Y | TCF4-ITF2_3 |
| DEPDC1 | NGEF_2 |
| DERA | DPYD_2 |
| DHFR | STAT6_2, DPYD_1 |
| DHRS7 | CHRNA3_3, KCTD13_1, VPS45_3, LOC100507431_2 |
| DLGAP1 | SETD1A_1st |
| DNAH14 | TCF4-ITF2_2 |
| DNAJC16 | VPS45_2, LOC100507431_3 |
| DNAJC9 | BCL11B_2, RERE_1 |
| DOHH | DPYD_2 |
| DST | miR137_3 |
| DVL2 | PCDHA123_2 |
| E2F5 | LOC100507431_2, TCF4-ITF2_2, TCF4-ITF2_3 |
| ECT2 | PBRM1_1 |
| EDC3 | pos_SNAP91 |
| EEF2KMT | TRANK1_1 |
| EFCAB11 | PBRM1_2 |
| EIF2AK2 | NGEF_1 |
| EIF2S3 | DPYD_3 |
| EIF5AL1 | KMT2E_3, LOC105376975_2, TCF4-ITF2_1, TCF4-ITF2_3 |
| EMG1 | NGEF_2 |
| EML1 | GALNT10_3 |
| ERMARD | BCL11B_1 |
| ETV1 | NAB2_2 |
| EVA1B | NAB2_3 |
| EXOC6 | KMT2E_1 |
| EZR | KMT2E_2 |
| FAM105A | TRANK1_3 |
| FAM134C | UBE2Q2P1_2 |
| FAM13B | TRANK1_1 |
| FAM193B | PBRM1_3 |
| FAM228B | BCL11B_1, GALNT10_3, KCTD13_3, KMT2E_3, miR137_1, miR137_3, RERE_3, SETD1A_1st, VPS45_1, VPS45_2, VPS45_3, DPYD_1, LOC100507431_1, LOC100507431_2, LINC00637_3, TCF4-ITF2_1 |
| FAM69B | LINC00637_1, pos_SNAP91 |
| FAM83D | NGEF_2 |
| FANCD2 | NGEF_2 |
| FBXO9 | LOC105376975_3 |
| FBXW11 | CHRNA3_1 |
| FLNB | pos_SNAP91 |
| FMR1 | CHRNA3_2, VPS45_3, pos_SNAP91 |
| FNDC4 | BCL11B_1 |
| FOXM1 | BAG5_3 |
| FRS2 | UBE2Q2P1_2 |
| FXYD6 | DPYD_2 |
| FZD3 | GALNT10_2, STAT6_3, TRANK1_2 |
| FZD7 | NAB2_2, TRANK1_2 |
| GAP43 | NAB2_2 |
| GCAT | GALNT10_1, DPYD_2, PPP1R16B_1 |
| GEMIN2 | KCTD13_3 |
| GEMIN4 | RERE_1 |
| GGA3 | BCL11B_3 |
| GINS2 | DPYD_1 |
| GLOD4 | NGEF_2 |
| GLT25D1 | BCL11B_2, GALNT10_2, LINC00637_3 |
| GNL3 | PCDHA123_3 |
| GOLGB1 | KCTD13_3, UBE2Q2P1_2 |
| GOSR1 | PCDHA123_1 |
| GPATCH2L | RERE_2, SETD1A_1st |
| GPM6B | PCDHA123_3 |
| GPN1 | miR137_3 |
| GRHPR | GALNT10_1 |
| GRIA1 | GALNT10_3, miR137_3, PCDHA123_1 |
| GRSF1 | PCDHA123_2, SETD1A_1st |
| GTF2F2 | PPP1R16B_3 |
| GTF3A | miR137_3 |
| GTF3C2 | NAB2_1, NAB2_2, PCDHA123_2, SETD1A_1st, LOC100507431_3, PPP1R16B_1 |
| GUF1 | pos_SNAP91 |
| HADHB | CHRNA3_2 |
| HCCS | NAB2_3 |
| HEBP1 | PBRM1_3 |
| HECTD4 | TCF4-ITF2_2 |
| HEXIM1 | BAG5_1, NGEF_2, TCF4-ITF2_1 |
| HMBS | TCF4-ITF2_1 |
| HPS4 | CHRNA3_3 |
| HSD17B4 | miR137_1, miR137_2, RERE_3, DPYD_1 |
| HSPG2 | VPS45_2 |
| HYOU1 | pos_SNAP91 |
| IDH3A | UBE2Q2P1_2 |
| IFI27L1 | NAB2_2 |
| IFITM3 | VPS45_1 |
| IGF2BP1 | PCDHA123_3 |
| IGF2BP3 | TRANK1_1 |
| IKBKAP | miR137_2 |
| IMPA2 | PCDHA123_2, STAT6_2 |
| INIP | TRANK1_1 |
| INPP5F | BCL11B_3 |
| INTU | PBRM1_2 |
| IPO11 | NGEF_2 |
| IPO9 | KCTD13_3 |
| IRX5 | PPP1R16B_3 |
| ISCA2 | CHRNA3_3 |
| ISY1 | LOC105376975_1, NAB2_1, VPS45_1 |
| ITGA6 | DPYD_3, LOC100507431_1, PBRM1_2 |
| JOSD1 | VPS45_1 |
| JUN | PBRM1_1 |
| KAT6B | BCL11B_1 |
| KBTBD2 | NGEF_2 |
| KCTD10 | GALNT10_1, GALNT10_3, LOC105376975_2, miR137_2, PCDHA123_2, PCDHA123_3, RERE_2, SETD1A_1st, STAT6_2, TRANK1_3, VPS45_2, DPYD_2, PPP1R16B_1, TCF4-ITF2_2 |
| KCTD2 | STAT6_1, UBE2Q2P1_2 |
| KDELR2 | RERE_2 |
| KDM4B | UBE2Q2P1_2 |
| KIF3B | VPS45_3 |
| KLHL5 | CHRNA3_2 |
| KMT5A | NGEF_1 |
| LAPTM4A | LOC100507431_2 |
| LATS2 | KCTD13_3 |
| LDOC1 | DPYD_1 |
| LIN28B | KMT2E_3, pos_SNAP91 |
| LMF2 | TCF4-ITF2_3 |
| LPIN2 | KMT2E_2, SETD1A_1st, STAT6_3, LINC00637_2, pos_SNAP91 |
| LRP3 | pos_SNAP91 |
| LRRC45 | DPYD_2 |
| LSM10 | LINC00637_3 |
| LTN1 | UBE2Q2P1_2 |
| MAD1L1 | KMT2E_2 |
| MAF1 | SETD1A_1st |
| MALAT1 | UBE2Q2P1_2, LINC00637_1 |
| MANEAL | TRANK1_2 |
| MAP3K11 | VPS45_2 |
| MAP3K4 | KCTD13_3, LOC105376975_2, miR137_2, SETD1A_1st, UBE2Q2P1_3, TCF4-ITF2_2 |
| MAPK8IP3 | KCTD13_1, LOC105376975_2, LOC105376975_3, PCDHA123_1, PPP1R16B_2, PPP1R16B_3 |
| MARCH7 | LOC105376975_2, NAB2_1, PCDHA123_2, DPYD_1 |
| MARK4 | CHRNA3_3 |
| MAX | DPYD_3, TCF4-ITF2_1 |
| MCM5 | LOC105376975_2, PCDHA123_2, LINC00637_3, TCF4-ITF2_2 |
| MED12 | BAG5_1, miR137_1, PCDHA123_3, SETD1A_1st, TRANK1_1, LOC100507431_2 |
| MED13 | BAG5_2, PBRM1_2 |
| METAP2 | GALNT10_1 |
| METTL18 | TRANK1_1 |
| MGME1 | pos_SNAP91 |
| MGRN1 | SETD1A_1st |
| MIS12 | GALNT10_2 |
| MIS18BP1 | NGEF_2 |
| MKI67 | NGEF_2 |
| MMP2 | SETD1A_1st |
| MMS22L | SETD1A_1st, TCF4-ITF2_3 |
| MOK | BCL11B_1 |
| MPHOSPH6 | LOC100507431_3 |
| MPRIP | SETD1A_1st |
| MPST | DPYD_1 |
| MPZL1 | BAG5_1, BCL11B_2, CHRNA3_2, PCDHA123_2, SETD1A_1st, STAT6_2, STAT6_3, PBRM1_3 |
| MRPL45 | PPP1R16B_3 |
| MSANTD2 | NGEF_2 |
| MSRA | GALNT10_3, PCDHA123_2, PCDHA123_3, VPS45_2 |
| MSTO1 | SETD1A_1st |
| MT-CO2 | pos_SNAP91 |
| MT-CYB | pos_SNAP91 |
| MT-ND4 | CHRNA3_1 |
| MYCL | PBRM1_1 |
| MYO9A | NGEF_2 |
| MZT1 | PBRM1_1 |
| NAT9 | LOC105376975_2 |
| NAV1 | UBE2Q2P1_3 |
| NCAPD3 | PBRM1_1 |
| NCAPG | PCDHA123_3, RERE_1 |
| NCK2 | BAG5_1, KCTD13_2, PCDHA123_1 |
| NCOA4 | LOC105376975_2, TCF4-ITF2_3, pos_SNAP91 |
| NDC80 | RERE_1, NGEF_2, PBRM1_1 |
| NDFIP2 | SETD1A_1st, LOC100507431_2 |
| NDN | PBRM1_3 |
| NDUFAF4 | KCTD13_1, LINC00637_3 |
| NDUFAF5 | BAG5_2, KCTD13_3, RERE_2 |
| NEDD9 | VPS45_1 |
| NEK2 | PBRM1_1 |
| NHLRC3 | pos_SNAP91 |
| NHSL2 | NGEF_2 |
| NINJ1 | BCL11B_2, KCTD13_2, miR137_1, PCDHA123_2, RERE_1, SETD1A_1st, VPS45_2 |
| NME2 | pos_SNAP91 |
| NOLC1 | DPYD_2 |
| NOTCH3 | NGEF_2 |
| NPRL3 | NGEF_2 |
| NR2F2 | GALNT10_2, LOC105376975_2, miR137_1, PPP1R16B_3 |
| NR6A1 | KMT2E_1 |
| NRDC | PCDHA123_3 |
| NRIP1 | BAG5_3, TRANK1_1 |
| NSMAF | LOC100507431_2 |
| NSUN2 | BAG5_1, KMT2E_2 |
| NUB1 | BAG5_1 |
| NUDCD2 | PBRM1_1 |
| NUF2 | PBRM1_1 |
| NUP155 | TRANK1_3 |
| NUP58 | PCDHA123_1 |
| NUP98 | CHRNA3_3, STAT6_1, UBE2Q2P1_2, pos_SNAP91 |
| OMA1 | RERE_3 |
| PAK3 | NAB2_3 |
| PAN3 | UBE2Q2P1_3 |
| PAPD5 | RERE_1, PBRM1_1, PPP1R16B_2 |
| PAQR4 | VPS45_2 |
| PARD6G | SETD1A_1st |
| PAXBP1 | BCL11B_1, KCTD13_3, RERE_3, DPYD_3 |
| PDE6D | PCDHA123_2 |
| PDLIM2 | PCDHA123_1 |
| PGGT1B | DPYD_2, LOC100507431_2 |
| PHLDA1 | BCL11B_2 |
| PIF1 | NGEF_2 |
| PIGM | BAG5_2 |
| PITRM1 | DPYD_3 |
| PKMYT1 | miR137_1, TRANK1_3, LINC00637_1 |
| PKN2 | NAB2_3, RERE_1 |
| PLK1 | PBRM1_1 |
| PLPP1 | CHRNA3_2, VPS45_1 |
| PLPP3 | KCTD13_1, LOC105376975_1, LOC105376975_2, miR137_1, RERE_1, RERE_2, SETD1A_1st, STAT6_1, STAT6_2, STAT6_3, NGEF_1, DPYD_2, DPYD_3, LOC100507431_3 |
| PLPP5 | BAG5_2 |
| PLXNB1 | KCTD13_1, miR137_3, RERE_1, RERE_2, SETD1A_1st, STAT6_3, PPP1R16B_1, PPP1R16B_3 |
| POFUT1 | SETD1A_1st |
| POLR2E | SETD1A_1st |
| PPIF | miR137_1, LINC00637_1, LINC00637_3 |
| PPP1R2 | UBE2Q2P1_2 |
| PPP2R3A | PBRM1_1 |
| PPP2R5E | NAB2_2, NAB2_3 |
| PRC1 | PBRM1_2 |
| PRCC | miR137_2, NAB2_1, NAB2_2, PCDHA123_2, SETD1A_1st, LINC00637_1, LINC00637_2, LINC00637_3, PPP1R16B_1 |
| PREPL | CHRNA3_1, VPS45_3 |
| PRKAB2 | BCL11B_3 |
| PRKAR1A | CHRNA3_2, GALNT10_1, PCDHA123_2 |
| PRR11 | BAG5_3, BCL11B_1, CHRNA3_3, miR137_1, miR137_3, NAB2_3, NGEF_1, DPYD_3, TCF4-ITF2_3 |
| PSMA5 | miR137_1, NAB2_1, RERE_1, TRANK1_3, LINC00637_1 |
| PSMA6 | GALNT10_3, TRANK1_3, VPS45_2 |
| PSRC1 | NGEF_2, PBRM1_1 |
| PTPN11 | PCDHA123_3 |
| PTTG1 | PBRM1_1 |
| PVT1 | BAG5_1, BCL11B_1, BCL11B_2, CHRNA3_3, GALNT10_3, miR137_2, NAB2_2, STAT6_1, LINC00637_2, pos_SNAP91 |
| PYCR1 | TRANK1_1 |
| QRICH1 | BAG5_3, BCL11B_1, GALNT10_3, KCTD13_1, KCTD13_3, KMT2E_1, LOC105376975_1, LOC105376975_2, PCDHA123_1, PCDHA123_2, PCDHA123_3, RERE_2, STAT6_2, STAT6_3, VPS45_2, DPYD_1, LOC100507431_3, LINC00637_2, PBRM1_2, TCF4-ITF2_1 |
| RAB28 | STAT6_1 |
| RAB6B | SETD1A_1st |
| RACGAP1 | BAG5_3, GALNT10_2, LINC00637_3 |
| RANBP1 | BCL11B_2 |
| RBAK-RBAKDN | miR137_2, miR137_3, NAB2_3, PBRM1_1 |
| RBM41 | GALNT10_3, DPYD_1 |
| RCOR3 | miR137_2, miR137_3, LINC00637_1 |
| REEP4 | NGEF_2 |
| RGS5 | BCL11B_2, CHRNA3_1, GALNT10_2, miR137_3, PCDHA123_1, SETD1A_1st, UBE2Q2P1_1, DPYD_2, PBRM1_2, PBRM1_3, pos_SNAP91 |
| RHBDL3 | UBE2Q2P1_3 |
| RHNO1 | DPYD_1 |
| RND3 | miR137_3 |
| RNF111 | CHRNA3_1 |
| RNF115 | BAG5_2, LINC00637_3, TCF4-ITF2_2, pos_SNAP91 |
| ROMO1 | NAB2_1 |
| RP11-231E4.4 | BCL11B_1 |
| RP11-339B21.12 | PBRM1_1 |
| RP11-343C2.12 | NGEF_2 |
| RP11-357C3.3 | pos_SNAP91 |
| RP11-46C20.1 | LOC105376975_3 |
| RP11-75C9.1 | UBE2Q2P1_1 |
| RPE | RERE_1 |
| RPS6KA1 | UBE2Q2P1_2, PPP1R16B_3 |
| RPUSD1 | TRANK1_3 |
| RRP9 | CHRNA3_3 |
| RSRP1 | pos_SNAP91 |
| RTN4IP1 | PBRM1_1 |
| RUFY2 | UBE2Q2P1_2 |
| SAMD4A | GALNT10_1, GALNT10_2, KMT2E_3, miR137_3, PCDHA123_2, VPS45_1, LINC00637_3 |
| SCD | PBRM1_3 |
| SCD5 | TCF4-ITF2_1 |
| SCML1 | DPYD_2 |
| SCPEP1 | SETD1A_1st |
| SDC2 | LOC105376975_3, LOC100507431_2 |
| SEC11A | LOC105376975_1 |
| SEC61A1 | STAT6_2 |
| SEMA3A | BAG5_2, BCL11B_2, GALNT10_1, KMT2E_2, miR137_2, miR137_3, NAB2_1, SETD1A_1st, STAT6_3, LOC100507431_2, LINC00637_3 |
| SEPN1 | CHRNA3_1 |
| SEPT11 | LOC100507431_2 |
| SEPT2 | PCDHA123_2 |
| SERPINH1 | UBE2Q2P1_1 |
| SESN3 | PBRM1_3 |
| SGO2 | NGEF_2 |
| SH2B1 | LINC00637_1 |
| SHOC2 | SETD1A_1st |
| SHPRH | KCTD13_1, DPYD_2 |
| SKA2 | PBRM1_1 |
| SKA3 | NGEF_2 |
| SKP2 | BCL11B_2, PPP1R16B_1 |
| SLC25A46 | KMT2E_2 |
| SLC2A3 | BCL11B_2, miR137_3 |
| SLC37A3 | LINC00637_3 |
| SLC38A1 | LOC100507431_3 |
| SLC44A2 | LOC105376975_2, PPP1R16B_2 |
| SLF2 | VPS45_2 |
| SMAD1 | LINC00637_1, pos_SNAP91 |
| SMG1 | KCTD13_3, miR137_3, PCDHA123_3, PPP1R16B_1, PPP1R16B_3 |
| SNAPC2 | PPP1R16B_3 |
| SNAPC3 | BCL11B_2, SETD1A_1st, VPS45_2 |
| SNRNP27 | BCL11B_1, KCTD13_3 |
| SNU13 | BCL11B_2 |
| SNX27 | KCTD13_1, KCTD13_2, NAB2_2, RERE_1, VPS45_2, TCF4-ITF2_1 |
| SOX21 | miR137_1, NAB2_1, STAT6_1, TRANK1_3, LINC00637_2 |
| SPARC | PBRM1_3 |
| SPATA5L1 | VPS45_3 |
| SPECC1L | BAG5_1 |
| SPOCK1 | LOC105376975_3, UBE2Q2P1_2, LOC100507431_2 |
| SPTLC1 | CHRNA3_3 |
| SRD5A1 | LINC00637_2 |
| SRD5A3 | NGEF_2 |
| SSR3 | PCDHA123_1 |
| ST7L | GALNT10_2 |
| STAMBP | PPP1R16B_1 |
| STK26 | miR137_1, STAT6_2, LINC00637_3 |
| STT3B | miR137_3, RERE_2, LOC100507431_3, PPP1R16B_1 |
| STX2 | SETD1A_1st |
| STXBP3 | LINC00637_1 |
| SUMO2 | PCDHA123_1 |
| SUMO3 | VPS45_2 |
| TAMM41 | TRANK1_3 |
| TCF19 | pos_SNAP91 |
| TCF7L2 | BAG5_1, BCL11B_2, CHRNA3_2, KCTD13_1, SETD1A_1st |
| TDP2 | TCF4-ITF2_2 |
| TEAD1 | KMT2E_2 |
| TERF2IP | UBE2Q2P1_2 |
| TEX10 | NGEF_1 |
| TFDP1 | BCL11B_2, STAT6_2, PPP1R16B_1 |
| THG1L | BAG5_1, BAG5_2, BAG5_3, GALNT10_2, KMT2E_3, LOC105376975_2, LOC105376975_3, miR137_1, NAB2_2, PCDHA123_1, RERE_3, SETD1A_1st, STAT6_3, TRANK1_2, TRANK1_3, UBE2Q2P1_3, VPS45_2, DPYD_2, LOC100507431_3, LINC00637_1, LINC00637_3, PBRM1_2, PPP1R16B_3, TCF4-ITF2_2, pos_SNAP91 |
| TIMP2 | pos_SNAP91 |
| TLE4 | KCTD13_3, PBRM1_2, PPP1R16B_2 |
| TMEM101 | GALNT10_1, NAB2_2, PCDHA123_1, PCDHA123_3, PBRM1_2, PPP1R16B_3 |
| TMEM159 | BCL11B_1 |
| TMEM165 | miR137_1 |
| TMEM50B | LOC105376975_1, RERE_2 |
| TMEM69 | PCDHA123_1 |
| TMSB10 | SETD1A_1st |
| TMX3 | PPP1R16B_1 |
| TPGS1 | BAG5_1 |
| TPM2 | PBRM1_3 |
| TPPP3 | PBRM1_3 |
| TPT1-AS1 | VPS45_3 |
| TPX2 | PBRM1_1 |
| TRIM24 | RERE_2, PPP1R16B_3 |
| TRIM9 | SETD1A_1st |
| TRIO | CHRNA3_2, VPS45_1, LINC00637_2, PBRM1_3, PPP1R16B_1, TCF4-ITF2_1 |
| TRIP12 | GALNT10_3, KCTD13_1, LOC105376975_2, VPS45_1 |
| TRIT1 | DPYD_3 |
| TRMT112 | VPS45_2 |
| TRMT2A | VPS45_2 |
| TRMT61A | GALNT10_1, TCF4-ITF2_1 |
| TROAP | NGEF_2 |
| TROVE2 | RERE_1 |
| TSC1 | VPS45_3 |
| TSPAN31 | BCL11B_1 |
| TTC17 | VPS45_1 |
| TTC31 | GALNT10_2 |
| TTC32 | UBE2Q2P1_2 |
| TTC39C | NGEF_1 |
| TTK | NGEF_2 |
| UBAP1 | BCL11B_2, PCDHA123_1, SETD1A_1st, LOC100507431_3 |
| UBE3D | RERE_3 |
| UBXN11 | TRANK1_1 |
| UNC119B | PBRM1_1 |
| UPRT | pos_SNAP91 |
| USMG5 | RERE_2 |
| USP12 | pos_SNAP91 |
| USP34 | SETD1A_1st |
| USP42 | UBE2Q2P1_2 |
| VOPP1 | BCL11B_2, PPP1R16B_1 |
| WDR11 | VPS45_1 |
| WDR26 | PCDHA123_2, VPS45_3 |
| WDR45 | PPP1R16B_1 |
| ZNF134 | LOC105376975_3, NAB2_2, TRANK1_2, VPS45_2, PBRM1_2, TCF4-ITF2_3 |
| ZNF205 | PPP1R16B_3 |
| ZNF24 | BCL11B_2, LOC105376975_2, RERE_1, TRANK1_2, DPYD_1, DPYD_3 |
| ZNF444 | UBE2Q2P1_2 |
| ZNF48 | BAG5_2, BCL11B_3, KCTD13_1, LOC105376975_2, miR137_1, RERE_2, TRANK1_3, VPS45_2, DPYD_2, DPYD_3, PBRM1_1, PBRM1_2, TCF4-ITF2_2 |
| ZNF496 | CHRNA3_1 |
| ZNF512 | GALNT10_3, PPP1R16B_2 |
| ZNF660 | miR137_3, PCDHA123_2 |
| ZNF687 | PBRM1_1 |
| ZNF738 | LOC105376975_2 |
| ZNF787 | GALNT10_2, miR137_3, STAT6_3, PPP1R16B_1 |
num_gRNA_pergene <- sapply(signif_genes.gRNA.lst,FUN = function(x){length(x)})
hist(num_gRNA_pergene,xlab = 'Number of gRNA targets per gene',ylab = 'Count',
main = 'Distribution of 462 significant genes')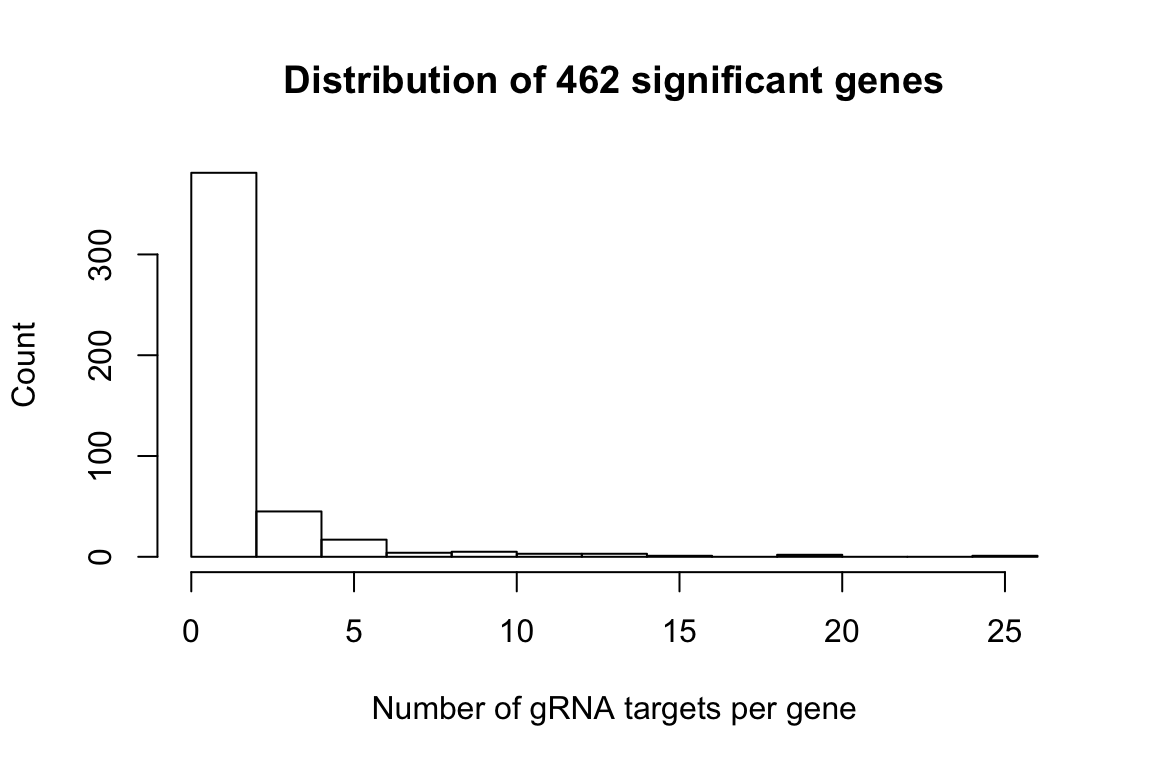
Examples of ashr shrinkage on estimated effect sizes of selected genes:
- Genes with high frequency of appearance
library(gridExtra)
plot_beta <- function(gene){
num <- num_gRNA_pergene[gene]
log10_lfsr <- log10(as.numeric(lfsr.mtx[gene,]))
beta.plot <- qplot(as.numeric(beta.mtx[gene,]),as.numeric(betaest.mtx[gene,]),
color=log10_lfsr, main = paste0(gene,' (',num,')'),
xlab = 'beta from regression',
ylab = 'beta after shrinkage') +
geom_abline(intercept = 0,slope = 1,linetype = "dotted")
return(beta.plot)
}
plot1 <- plot_beta('ARAF')
plot2 <- plot_beta('CWF19L2')
plot3 <- plot_beta('THG1L')
plot4 <- plot_beta('ZNF48')
grid.arrange(plot1,plot2,plot3,plot4,ncol=2)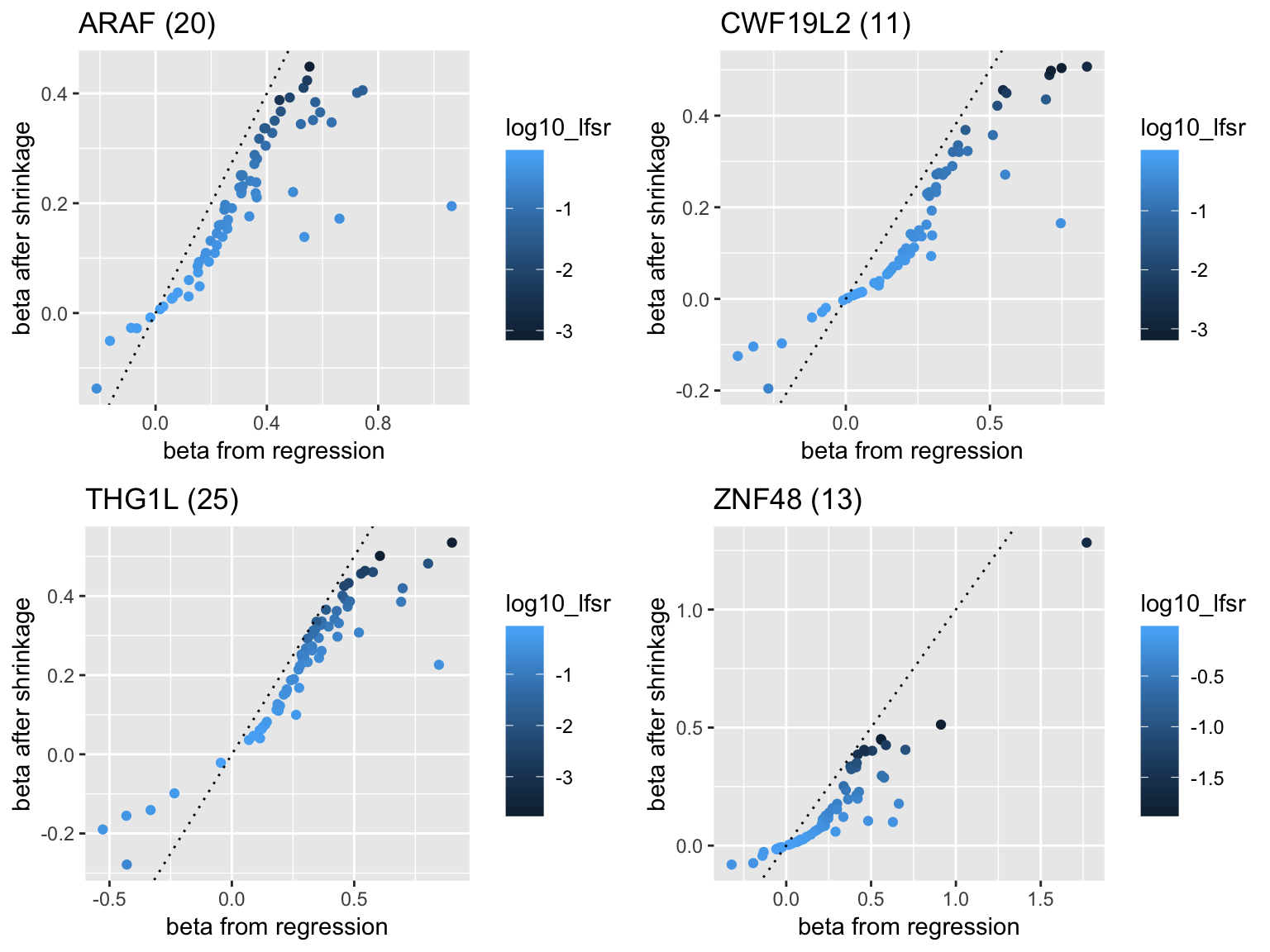
- Genes with low frequency of appearance
plot1 <- plot_beta('A2M')
plot2 <- plot_beta('SUMO3')
plot3 <- plot_beta('DNAJC9')
plot4 <- plot_beta('APC')
grid.arrange(plot1,plot2,plot3,plot4,ncol=2)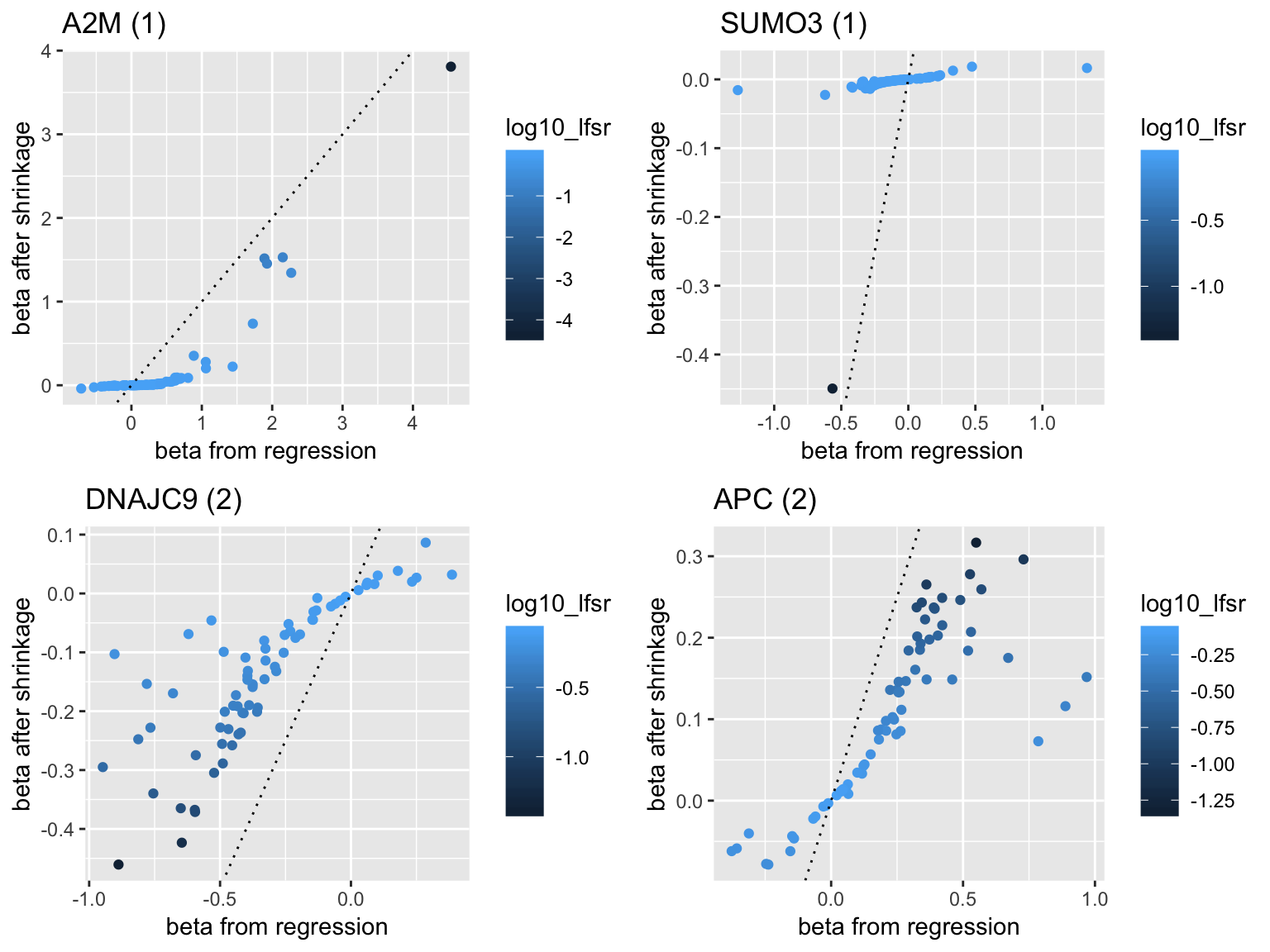
Examples of regression z score distributions for selected genes:
- Genes with high frequency of appearance
plot_zscore <- function(gene){
num <- num_gRNA_pergene[gene]
zscore <- as.numeric(beta.mtx[gene,])/as.numeric(se.mtx[gene,])
zscore.df <- data.frame(score=zscore)
zscore.plot <- ggplot(zscore.df,aes(zscore)) + geom_histogram(aes(y=..density..),bins = 20) +
labs(x = 'z-score from regression',y='frequency',title = paste0(gene,' (',num,')')) +
stat_function(fun = dnorm, args = list(mean = 0, sd = 1),col=2)
return(zscore.plot)
}
plot1 <- plot_zscore('ARAF')
plot2 <- plot_zscore('CWF19L2')
plot3 <- plot_zscore('THG1L')
plot4 <- plot_zscore('ZNF48')
grid.arrange(plot1,plot2,plot3,plot4,ncol=2)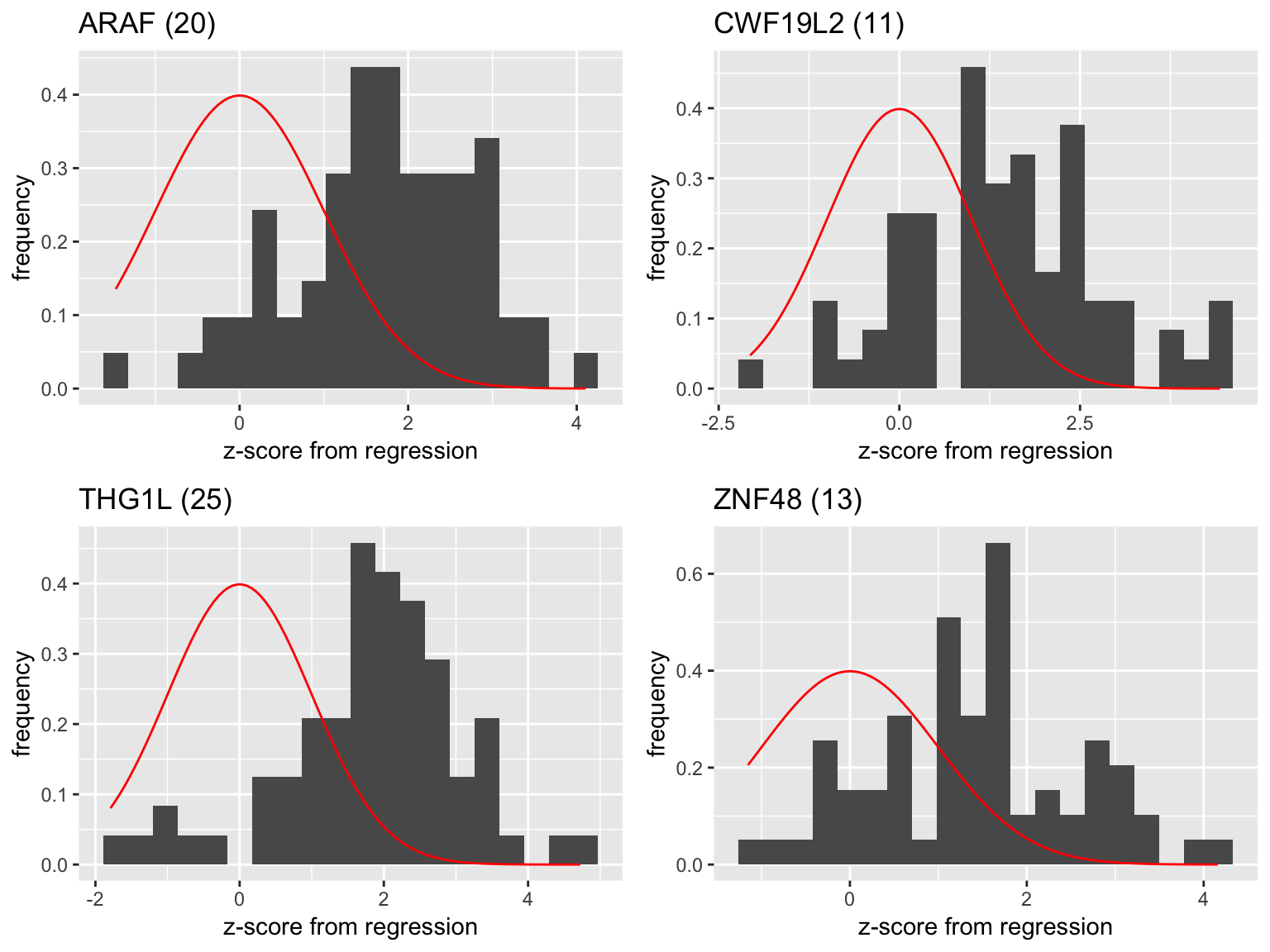
- Genes with low frequency of appearance
plot1 <- plot_zscore('A2M')
plot2 <- plot_zscore('SUMO3')
plot3 <- plot_zscore('DNAJC9')
plot4 <- plot_zscore('APC')
grid.arrange(plot1,plot2,plot3,plot4,ncol=2)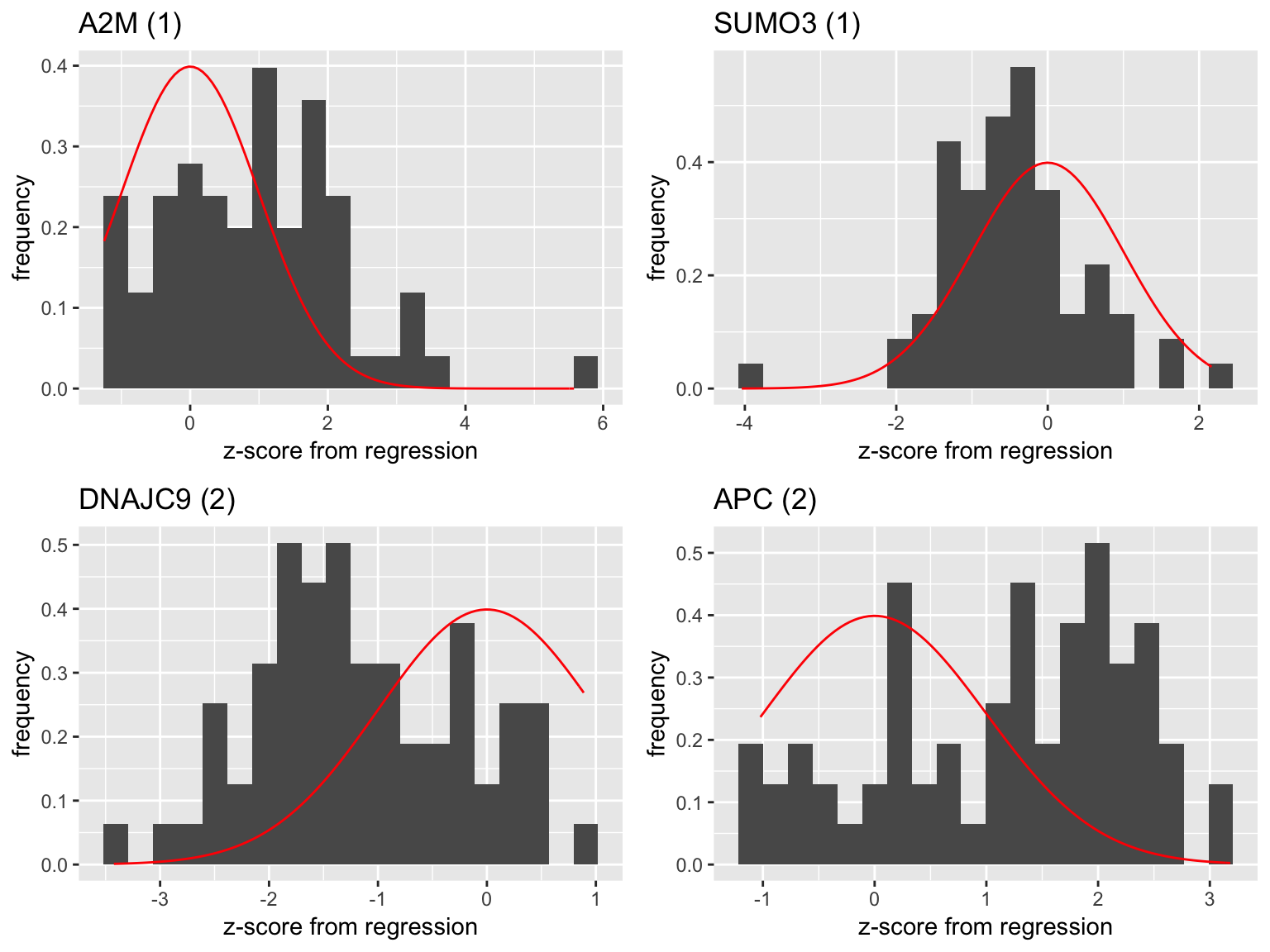
Significant genes per gRNA target
gRNA.signif_genes.lst <- list()
for (i in signif_genes){
cells <- signif_genes.gRNA.lst[[i]]
for (c in cells){
if (is.na(match(c, names(gRNA.signif_genes.lst)))){
gRNA.signif_genes.lst[[c]] <- i
} else {
gRNA.signif_genes.lst[[c]] <- c(gRNA.signif_genes.lst[[c]],i)
}
}
}
# Make table:
compact.gRNAs.lst <- sapply(gRNA.signif_genes.lst,FUN = function(x){paste(x,collapse = ', ')})
compact.gRNAs.df <- as.data.frame(compact.gRNAs.lst)
tmp.indx <- order(rownames(compact.gRNAs.df))
compact.gRNAs.df <- data.frame(gRNA_target=rownames(compact.gRNAs.df)[tmp.indx],
signif_genes=compact.gRNAs.df[tmp.indx,])
compact.gRNAs.df$gRNA_target <- sapply(strsplit(compact.gRNAs.df$gRNA_target,split = '_'), function(x){paste(x[1],x[2],sep = '_')})
compact.gRNAs.df$num_cells <- ncell_uniq[compact.gRNAs.df$gRNA_target]
compact.gRNAs.df <- compact.gRNAs.df[,c(3,1,2)]
# Present table:
kable(compact.gRNAs.df) %>%
kable_styling() %>%
scroll_box(width = "100%", height = "400px")| num_cells | gRNA_target | signif_genes |
|---|---|---|
| 36 | BAG5_1 | MPZL1, NCK2, NSUN2, THG1L, NUB1, PVT1, TCF7L2, CWF19L2, CPPED1, HEXIM1, TPGS1, CEP250, SPECC1L, ARAF, MED12 |
| 33 | BAG5_2 | RNF115, PIGM, THG1L, SEMA3A, PLPP5, BAG1, ZNF48, MED13, NDUFAF5, ARFGAP1, ARAF |
| 31 | BAG5_3 | QRICH1, THG1L, CWF19L2, FOXM1, RACGAP1, CDKN3, CPPED1, PRR11, CEP250, NRIP1 |
| 13 | BCL11B_1 | FAM228B, FNDC4, SNRNP27, QRICH1, ERMARD, PVT1, KAT6B, CWF19L2, TSPAN31, MOK, TMEM159, PRR11, RP11-231E4.4, PAXBP1, ARMCX6 |
| 33 | BCL11B_2 | RGS5, MPZL1, SKP2, VOPP1, SEMA3A, PVT1, SNAPC3, UBAP1, NINJ1, DNAJC9, TCF7L2, ASRGL1, CWF19L2, SLC2A3, PHLDA1, TFDP1, CAPN15, CDC6, ZNF24, GLT25D1, RANBP1, SNU13 |
| 13 | BCL11B_3 | PRKAB2, INPP5F, ZNF48, GGA3 |
| 3 | CHRNA3_1 | SEPN1, RGS5, ZNF496, PREPL, FBXW11, AC084018.1, BTBD7, RNF111, ABHD17A, MT-ND4 |
| 23 | CHRNA3_2 | MPZL1, HADHB, KLHL5, TRIO, PLPP1, CCDC136, TCF7L2, PRKAR1A, CBX4, FMR1 |
| 13 | CHRNA3_3 | C2orf49, RRP9, PVT1, CBWD1, SPTLC1, NUP98, DHRS7, ISCA2, BTBD1, CPPED1, PRR11, MARK4, HPS4, ARAF |
| 37 | DPYD_1 | FAM228B, MARCH7, QRICH1, DHFR, APC, HSD17B4, RHNO1, GINS2, ZNF24, MPST, ARMCX6, RBM41, LDOC1 |
| 68 | DPYD_2 | PLPP3, BCL10, RGS5, CDCA7, CLPTM1L, PGGT1B, THG1L, SHPRH, CASD1, CLN8, NOLC1, FXYD6, DERA, KCTD10, ZNF48, LRRC45, DOHH, GCAT, SCML1, ARAF |
| 28 | DPYD_3 | TRIT1, PLPP3, C2orf49, ITGA6, PITRM1, MAX, ZNF48, PRR11, ZNF24, PAXBP1, EIF2S3 |
| 38 | GALNT10_1 | CCDC50, SEMA3A, CCDC136, GRHPR, ASRGL1, BUD13, METAP2, KCTD10, SAMD4A, TRMT61A, TMEM101, PRKAR1A, GCAT, ARAF |
| 37 | GALNT10_2 | ST7L, RGS5, TTC31, THG1L, FZD3, RACGAP1, SAMD4A, NR2F2, MIS12, GLT25D1, ZNF787 |
| 50 | GALNT10_3 | FAM228B, ZNF512, TRIP12, QRICH1, GRIA1, MSRA, PVT1, KCTD10, PSMA6, EML1, ARAF, ARMCX1, RBM41 |
| 30 | KCTD13_1 | PLPP3, ADGRL2, SNX27, TRIP12, PLXNB1, QRICH1, NDUFAF4, SHPRH, TCF7L2, DHRS7, MAPK8IP3, ZNF48, CWC25, CEP250, CPNE1 |
| 32 | KCTD13_2 | SNX27, NCK2, NINJ1 |
| 15 | KCTD13_3 | DBT, IPO9, FAM228B, SNRNP27, QRICH1, GOLGB1, MAP3K4, TLE4, LATS2, GEMIN2, CHTF18, SMG1, NDUFAF5, PAXBP1 |
| 11 | KMT2E_1 | CNRIP1, QRICH1, CNPY4, NR6A1, EXOC6, ARAF |
| 19 | KMT2E_2 | NSUN2, SLC25A46, APC, EZR, MAD1L1, SEMA3A, TEAD1, COQ7, LPIN2 |
| 18 | KMT2E_3 | C1orf56, FAM228B, THG1L, LIN28B, EIF5AL1, SAMD4A, ARAF |
| 45 | LINC00637_1 | STXBP3, PSMA5, ARNT, PRCC, RCOR3, SMAD1, THG1L, C7orf73, FAM69B, PPIF, MALAT1, PKMYT1, SH2B1, ARAF |
| 34 | LINC00637_2 | PRCC, QRICH1, SRD5A1, TRIO, C7orf73, PVT1, SOX21, LPIN2 |
| 53 | LINC00637_3 | LSM10, RNF115, PRCC, FAM228B, C2orf49, THG1L, NDUFAF4, SEMA3A, SLC37A3, PPIF, RACGAP1, SAMD4A, GLT25D1, MCM5, ARAF, STK26 |
| 15 | LOC100507431_1 | FAM228B, ITGA6, ARAF, ARMCX6 |
| 29 | LOC100507431_2 | LAPTM4A, FAM228B, SEPT11, PGGT1B, SPOCK1, SEMA3A, NSMAF, E2F5, SDC2, CWF19L2, NDFIP2, DHRS7, ARFGAP1, ARAF, MED12, ARMCX6 |
| 49 | LOC100507431_3 | DNAJC16, PLPP3, GTF3C2, STT3B, QRICH1, THG1L, UBAP1, SLC38A1, MPHOSPH6 |
| 35 | LOC105376975_1 | PLPP3, QRICH1, ISY1, CHAMP1, SEC11A, TMEM50B |
| 50 | LOC105376975_2 | ATPIF1, PLPP3, MARCH7, TRIP12, QRICH1, THG1L, MAP3K4, NCOA4, EIF5AL1, ATAD1, KCTD10, NR2F2, MAPK8IP3, ZNF48, NAT9, ZNF24, SLC44A2, ZNF738, CECR5, MCM5, AIFM1 |
| 8 | LOC105376975_3 | ACBD3, RP11-46C20.1, SPOCK1, THG1L, FBXO9, SDC2, MAPK8IP3, ZNF134, CRYBB2 |
| 44 | miR137_1 | PLPP3, PSMA5, FAM228B, TMEM165, HSD17B4, THG1L, NINJ1, PPIF, CWF19L2, SOX21, NR2F2, CLCN7, PKMYT1, ZNF48, CDC6, PRR11, APMAP, ARAF, MED12, ARMCX6, STK26 |
| 30 | miR137_2 | PRCC, RCOR3, HSD17B4, MAP3K4, RBAK-RBAKDN, SEMA3A, PVT1, IKBKAP, ASRGL1, KCTD10, COMMD6, CPPED1 |
| 54 | miR137_3 | RGS5, RCOR3, FAM228B, GPN1, RND3, STT3B, ZNF660, PLXNB1, GRIA1, DST, RBAK-RBAKDN, SEMA3A, CWF19L2, SLC2A3, GTF3A, SAMD4A, SMG1, PRR11, ZNF787, ARMCX6 |
| 36 | NAB2_1 | PSMA5, PRCC, GTF3C2, MARCH7, ISY1, SEMA3A, SOX21, ROMO1, ARAF |
| 24 | NAB2_2 | SNX27, PRCC, GTF3C2, FZD7, GAP43, THG1L, ETV1, PVT1, PPP2R5E, IFI27L1, TMEM101, ZNF134, ARFGAP1, ARAF, ARMCX6 |
| 25 | NAB2_3 | EVA1B, PKN2, RBAK-RBAKDN, CWF19L2, PPP2R5E, CPPED1, PRR11, HCCS, PAK3 |
| 11 | NGEF_1 | PLPP3, EIF2AK2, TEX10, KMT5A, CPPED1, CNTROB, PRR11, TTC39C, ARAF |
| 2 | NGEF_2 | DEPDC1, PSRC1, C1orf131, CENPA, C2orf69, SGO2, FANCD2, SRD5A3, IPO11, TTK, CCDC126, KBTBD2, REEP4, CDCA2, BBIP1, MKI67, MSANTD2, CDCA3, EMG1, TROAP, SKA3, BORA, MIS18BP1, ARHGAP11A, PIF1, MYO9A, NPRL3, CCNF, RP11-343C2.12, GLOD4, AURKB, HEXIM1, NDC80, NOTCH3, FAM83D, NHSL2 |
| 4 | PBRM1_1 | MYCL, JUN, PSRC1, ZNF687, NUF2, ASPM, NEK2, AHCTF1, PPP2R3A, ECT2, PTTG1, NUDCD2, CCDC167, RTN4IP1, RBAK-RBAKDN, RP11-339B21.12, NCAPD3, UNC119B, MZT1, PLK1, ZNF48, PAPD5, SKA2, NDC80, CDC25B, TPX2, CHEK2 |
| 12 | PBRM1_2 | RGS5, ITGA6, QRICH1, INTU, THG1L, CCDC136, TLE4, EFCAB11, PRC1, CPPED1, ZNF48, TMEM101, MED13, C19orf47, ZNF134 |
| 4 | PBRM1_3 | RGS5, MPZL1, CMBL, TRIO, SPARC, FAM193B, ANKMY2, TPM2, SCD, SESN3, HEBP1, NDN, CASC4, TPPP3, ARHGEF9 |
| 53 | PCDHA123_1 | TMEM69, C1orf56, RGS5, C2orf49, NCK2, QRICH1, SSR3, GRIA1, THG1L, PDLIM2, UBAP1, ASRGL1, BUD13, NUP58, MAPK8IP3, GOSR1, TMEM101, SUMO2, ARMCX6 |
| 64 | PCDHA123_2 | PRCC, MPZL1, WDR26, GTF3C2, MARCH7, CDCA7, PDE6D, SEPT2, ZNF660, QRICH1, GRSF1, CLPTM1L, MSRA, NINJ1, KCTD10, SAMD4A, DVL2, PRKAR1A, IMPA2, MCM5 |
| 62 | PCDHA123_3 | NRDC, QRICH1, GNL3, NCAPG, CNPY4, MSRA, CWF19L2, KCTD10, PTPN11, SMG1, TMEM101, IGF2BP1, GPM6B, MED12 |
| 3 | pos_SNAP91 | RSRP1, ADGRL2, RNF115, RGS5, FLNB, GUF1, SMAD1, THG1L, TCF19, LIN28B, AMPH, PVT1, FAM69B, NCOA4, NUP98, HYOU1, USP12, NHLRC3, AKAP11, EDC3, CPPED1, NME2, TIMP2, LPIN2, LRP3, MGME1, RP11-357C3.3, UPRT, FMR1, MT-CO2, MT-CYB |
| 44 | PPP1R16B_1 | PRCC, GTF3C2, STAMBP, STT3B, PLXNB1, TRIO, SKP2, VOPP1, CCDC136, KCTD10, TFDP1, SMG1, TMX3, ZNF787, GCAT, WDR45 |
| 31 | PPP1R16B_2 | ZNF512, ATF2, TLE4, AMN1, MAPK8IP3, PAPD5, SLC44A2, CEP250 |
| 35 | PPP1R16B_3 | RPS6KA1, DBI, PLXNB1, THG1L, TRIM24, GTF2F2, NR2F2, MAPK8IP3, ZNF205, SMG1, IRX5, MRPL45, TMEM101, SNAPC2, C19orf47 |
| 48 | RERE_1 | PLPP3, PKN2, PSMA5, SNX27, TROVE2, RPE, PLXNB1, NCAPG, NINJ1, DNAJC9, PAPD5, GEMIN4, NDC80, ZNF24, ARAF |
| 46 | RERE_2 | PLPP3, STT3B, PLXNB1, QRICH1, KDELR2, TRIM24, USMG5, KCTD10, GPATCH2L, ZNF48, NDUFAF5, TMEM50B |
| 25 | RERE_3 | OMA1, FAM228B, HSD17B4, THG1L, UBE3D, PAXBP1 |
| 23 | SETD1A_1st | PLPP3, MSTO1, PRCC, RGS5, MPZL1, FAM228B, GTF3C2, BIRC6, USP34, TMSB10, PLXNB1, RAB6B, GRSF1, THG1L, MMS22L, MAP3K4, ANLN, SEMA3A, BRAF, MAF1, SNAPC3, UBAP1, NINJ1, SHOC2, TCF7L2, ASRGL1, CCDC84, KCTD10, STX2, NDFIP2, TRIM9, GPATCH2L, MGRN1, MMP2, MPRIP, SCPEP1, LPIN2, DLGAP1, PARD6G, POLR2E, POFUT1, MED12 |
| 24 | STAT6_1 | PLPP3, BCL10, RAB28, PVT1, NUP98, SOX21, KCTD2 |
| 38 | STAT6_2 | PLPP3, MPZL1, QRICH1, SEC61A1, DHFR, CCNH, CYFIP2, CCDC136, KCTD10, TFDP1, IMPA2, STK26 |
| 53 | STAT6_3 | PLPP3, MPZL1, PLXNB1, QRICH1, THG1L, SEMA3A, C7orf73, FZD3, CWF19L2, CCDC84, LPIN2, ZNF787 |
| 50 | TCF4-ITF2_1 | SNX27, FAM228B, QRICH1, SCD5, TRIO, CAMSAP1, EIF5AL1, HMBS, MAX, TRMT61A, CLK3, HEXIM1, ARFGAP1, ARAF |
| 45 | TCF4-ITF2_2 | ATPIF1, RNF115, DNAH14, THG1L, TDP2, MAP3K4, E2F5, CWF19L2, KCTD10, HECTD4, ZNF48, MCM5, ARMCX6 |
| 12 | TCF4-ITF2_3 | MMS22L, E2F5, NCOA4, EIF5AL1, CCDC84, CHTF18, PRR11, ZNF134, LMF2, DDX3Y |
| 3 | TRANK1_1 | CLCN6, UBXN11, ATG4C, METTL18, CEP170, CKAP2L, FAM13B, IGF2BP3, INIP, EEF2KMT, C16orf72, PYCR1, NRIP1, MED12 |
| 19 | TRANK1_2 | MANEAL, FZD7, THG1L, FZD3, ALDH1B1, ZNF24, ZNF134 |
| 65 | TRANK1_3 | PSMA5, CDCA7, TAMM41, FAM105A, NUP155, THG1L, KCTD10, SOX21, PSMA6, CLK3, RPUSD1, PKMYT1, ZNF48 |
| 11 | UBE2Q2P1_1 | RGS5, RP11-75C9.1, SERPINH1 |
| 8 | UBE2Q2P1_2 | RPS6KA1, TTC32, GOLGB1, PPP1R2, AFF4, SPOCK1, USP42, RUFY2, NUP98, MALAT1, ATP5B, FRS2, IDH3A, TERF2IP, FAM134C, KCTD2, KDM4B, ZNF444, LTN1 |
| 11 | UBE2Q2P1_3 | NAV1, CCNH, THG1L, MAP3K4, ACER3, AMN1, PAN3, RHBDL3, ARAF |
| 14 | VPS45_1 | FAM228B, TRIP12, ISY1, TRIO, PLPP1, NEDD9, WDR11, IFITM3, TTC17, A2M, SAMD4A, JOSD1 |
| 79 | VPS45_2 | DNAJC16, HSPG2, SNX27, FAM228B, QRICH1, THG1L, MSRA, SNAPC3, NINJ1, SLF2, TRMT112, MAP3K11, KCTD10, PSMA6, PAQR4, ZNF48, C17orf80, ZNF134, ARFGAP1, SUMO3, TRMT2A, ARAF |
| 9 | VPS45_3 | WDR26, FAM228B, PREPL, TSC1, TPT1-AS1, DHRS7, SPATA5L1, CPPED1, KIF3B, ARMCX2, FMR1 |
num_gene_pergRNA <- sapply(gRNA.signif_genes.lst,FUN = function(x){length(x)})
hist(num_gene_pergRNA,xlab = 'Number of DE genes per gRNA target',ylab = 'Count',
main = 'Distribution of 70 gRNAs')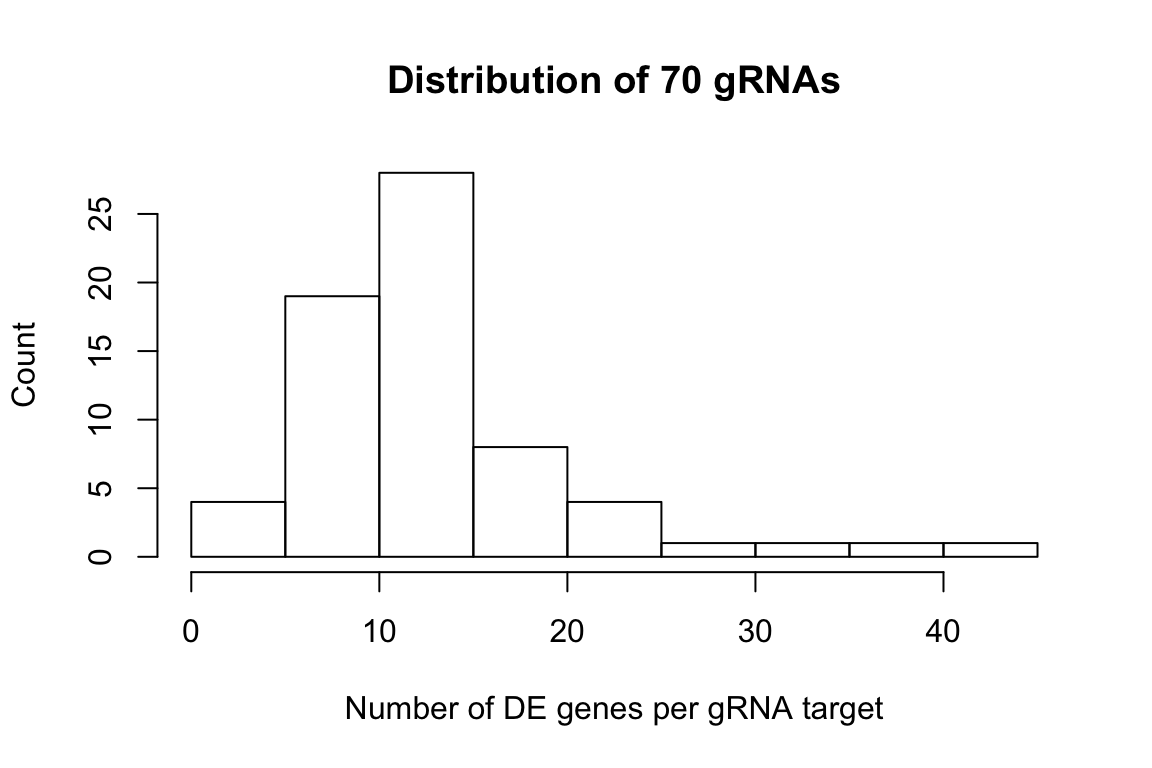
GO over-representation analysis
Gene ontology enrichment of the 462 significant genes (out of ~30,000 genes in total):
- Enrichment based on biological processes
gene_info <- read.delim(paste0(wkdir,'data/genes.tsv'),header = F,
sep = '\t',col.names = c('ID','name'))
gene_info$ID <- sapply(strsplit(gene_info$ID,split = '[.]'),
FUN = function(x){x[[1]]})
tmp.indx <- match(signif_genes,gene_info$name)
ensembl.signif_genes <- gene_info$ID[tmp.indx]
library(org.Hs.eg.db)
library(clusterProfiler)
ego.bp.signif_genes <- enrichGO(gene = ensembl.signif_genes,
universe = gene_info$ID,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "BP",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05,
readable = TRUE)
barplot(ego.bp.signif_genes, drop=TRUE, showCategory=6)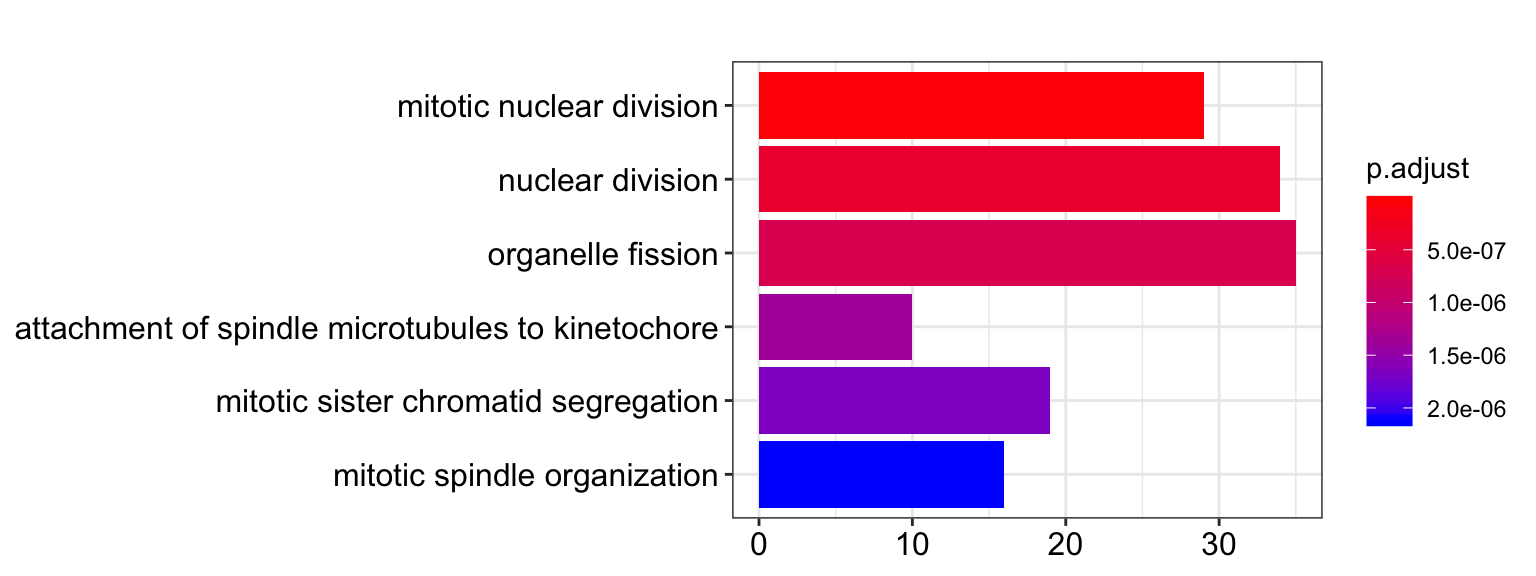
kable(head(ego.bp.signif_genes),row.names = F) %>%
kable_styling() %>%
scroll_box(width = "100%", height = "300px")| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|
| GO:0140014 | mitotic nuclear division | 29/405 | 277/17444 | 0 | 0.0e+00 | 0.0e+00 | PSRC1/MSTO1/NUF2/NEK2/NCAPG/APC/PTTG1/TTK/MAD1L1/ANLN/SLF2/MKI67/NCAPD3/RACGAP1/MZT1/BORA/CHAMP1/PRC1/CCNF/PKMYT1/PLK1/SH2B1/MIS12/AURKB/CDC6/NDC80/TPX2/KIF3B/CHEK2 | 29 |
| GO:0000280 | nuclear division | 34/405 | 418/17444 | 0 | 4.0e-07 | 4.0e-07 | PSRC1/MSTO1/NUF2/ASPM/NEK2/SGO2/FANCD2/NCAPG/APC/PTTG1/TTK/MAD1L1/ANLN/SLF2/MKI67/NCAPD3/RACGAP1/MZT1/BORA/CHAMP1/PRC1/CCNF/PKMYT1/PLK1/SH2B1/MIS12/AURKB/CDC6/PRKAR1A/NDC80/CDC25B/TPX2/KIF3B/CHEK2 | 34 |
| GO:0048285 | organelle fission | 35/405 | 457/17444 | 0 | 7.0e-07 | 7.0e-07 | PSRC1/MSTO1/NUF2/ASPM/NEK2/SGO2/FANCD2/NCAPG/SLC25A46/APC/PTTG1/TTK/MAD1L1/ANLN/SLF2/MKI67/NCAPD3/RACGAP1/MZT1/BORA/CHAMP1/PRC1/CCNF/PKMYT1/PLK1/SH2B1/MIS12/AURKB/CDC6/PRKAR1A/NDC80/CDC25B/TPX2/KIF3B/CHEK2 | 35 |
| GO:0008608 | attachment of spindle microtubules to kinetochore | 10/405 | 32/17444 | 0 | 1.4e-06 | 1.3e-06 | NUF2/NEK2/ECT2/APC/MAD1L1/RACGAP1/CHAMP1/MIS12/AURKB/NDC80 | 10 |
| GO:0000070 | mitotic sister chromatid segregation | 19/405 | 152/17444 | 0 | 1.7e-06 | 1.6e-06 | PSRC1/MSTO1/NUF2/NEK2/NCAPG/APC/PTTG1/TTK/MAD1L1/SLF2/NCAPD3/RACGAP1/CHAMP1/PRC1/PLK1/MIS12/AURKB/CDC6/NDC80 | 19 |
| GO:0007052 | mitotic spindle organization | 16/405 | 108/17444 | 0 | 2.1e-06 | 2.0e-06 | PSRC1/MSTO1/NUF2/NEK2/TTK/RACGAP1/MZT1/BORA/EML1/PRC1/PLK1/AURKB/NDC80/TPX2/KIF3B/CHEK2 | 16 |
- Enrichment based on cellular components
ego.cc.signif_genes <- enrichGO(gene = ensembl.signif_genes,
universe = gene_info$ID,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "CC",
pAdjustMethod = "BH",
pvalueCutoff = 0.01,
qvalueCutoff = 0.05,
readable = TRUE)
barplot(ego.cc.signif_genes, drop=TRUE, showCategory=6)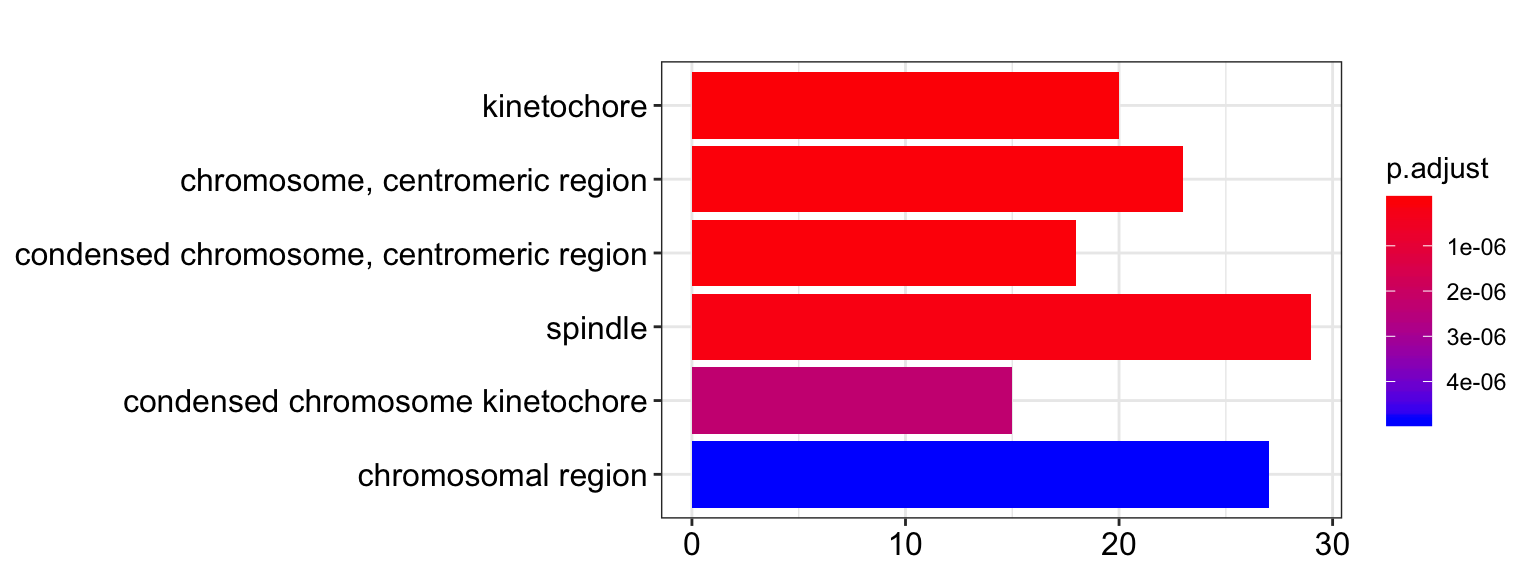
kable(head(ego.cc.signif_genes),row.names = F) %>%
kable_styling() %>%
scroll_box(width = "100%", height = "300px")| ID | Description | GeneRatio | BgRatio | pvalue | p.adjust | qvalue | geneID | Count |
|---|---|---|---|---|---|---|---|---|
| GO:0000776 | kinetochore | 20/435 | 134/18503 | 0e+00 | 0.0e+00 | 0.0e+00 | NUF2/NEK2/AHCTF1/CENPA/SGO2/SEPT2/APC/NUDCD2/TTK/MAD1L1/NUP98/SKA3/CHAMP1/MIS18BP1/PLK1/MIS12/AURKB/SKA2/NDC80/SUMO3 | 20 |
| GO:0000775 | chromosome, centromeric region | 23/435 | 195/18503 | 0e+00 | 1.0e-07 | 0.0e+00 | NUF2/NEK2/AHCTF1/CENPA/SGO2/SEPT2/NCAPG/APC/NUDCD2/TTK/MAD1L1/NUP98/NCAPD3/SKA3/CHAMP1/MIS18BP1/PLK1/MIS12/AURKB/SKA2/NDC80/SUMO3/FMR1 | 23 |
| GO:0000779 | condensed chromosome, centromeric region | 18/435 | 118/18503 | 0e+00 | 1.0e-07 | 0.0e+00 | NUF2/NEK2/AHCTF1/CENPA/SGO2/SEPT2/NCAPG/NUDCD2/MAD1L1/NCAPD3/SKA3/CHAMP1/MIS18BP1/PLK1/MIS12/AURKB/SKA2/NDC80 | 18 |
| GO:0005819 | spindle | 29/435 | 333/18503 | 0e+00 | 2.0e-07 | 2.0e-07 | PSRC1/ASPM/NEK2/CEP170/BIRC6/CKAP2L/SEPT2/ECT2/NSUN2/NUDCD2/NEDD9/TTK/MAD1L1/RACGAP1/LATS2/SKA3/MZT1/CHAMP1/EML1/PRC1/PLK1/AURKB/CDC6/SKA2/CDC25B/TPX2/KIF3B/FAM83D/SPECC1L | 29 |
| GO:0000777 | condensed chromosome kinetochore | 15/435 | 105/18503 | 0e+00 | 2.3e-06 | 2.1e-06 | NUF2/NEK2/AHCTF1/CENPA/SGO2/SEPT2/NUDCD2/MAD1L1/SKA3/CHAMP1/MIS18BP1/PLK1/MIS12/SKA2/NDC80 | 15 |
| GO:0098687 | chromosomal region | 27/435 | 347/18503 | 1e-07 | 4.9e-06 | 4.5e-06 | NUF2/NEK2/AHCTF1/CENPA/SGO2/SEPT2/NCAPG/APC/NUDCD2/TTK/MAD1L1/NUP98/NCAPD3/SKA3/CHAMP1/MIS18BP1/PIF1/PLK1/TERF2IP/MIS12/AURKB/SKA2/NDC80/SUMO3/CHEK2/MCM5/FMR1 | 27 |
[Gene ontology enrichment of the 53 significant genes that appeared in > 3 gRNA conditions (out of ~30,000 genes in total) does not result in anything significant.]
Gene set enrichment analysis
with ordered geneList values being the number of gRNA groups each gene is differentially expressed in.
- Enrichment based on biological processes
gene.num <- sapply(signif_genes.gRNA.lst,FUN = function(x){length(x)})
names(gene.num) <- ensembl.signif_genes
gene.num <- gene.num[order(gene.num,decreasing = T)]
gsea.signif_genes <- gseGO(geneList = gene.num,
OrgDb = org.Hs.eg.db,
keyType = 'ENSEMBL',
ont = "BP",
nPerm = 1000,
minGSSize = 100,
maxGSSize = 500,
pvalueCutoff = 0.05,
verbose = FALSE)
dotplot(gsea.signif_genes)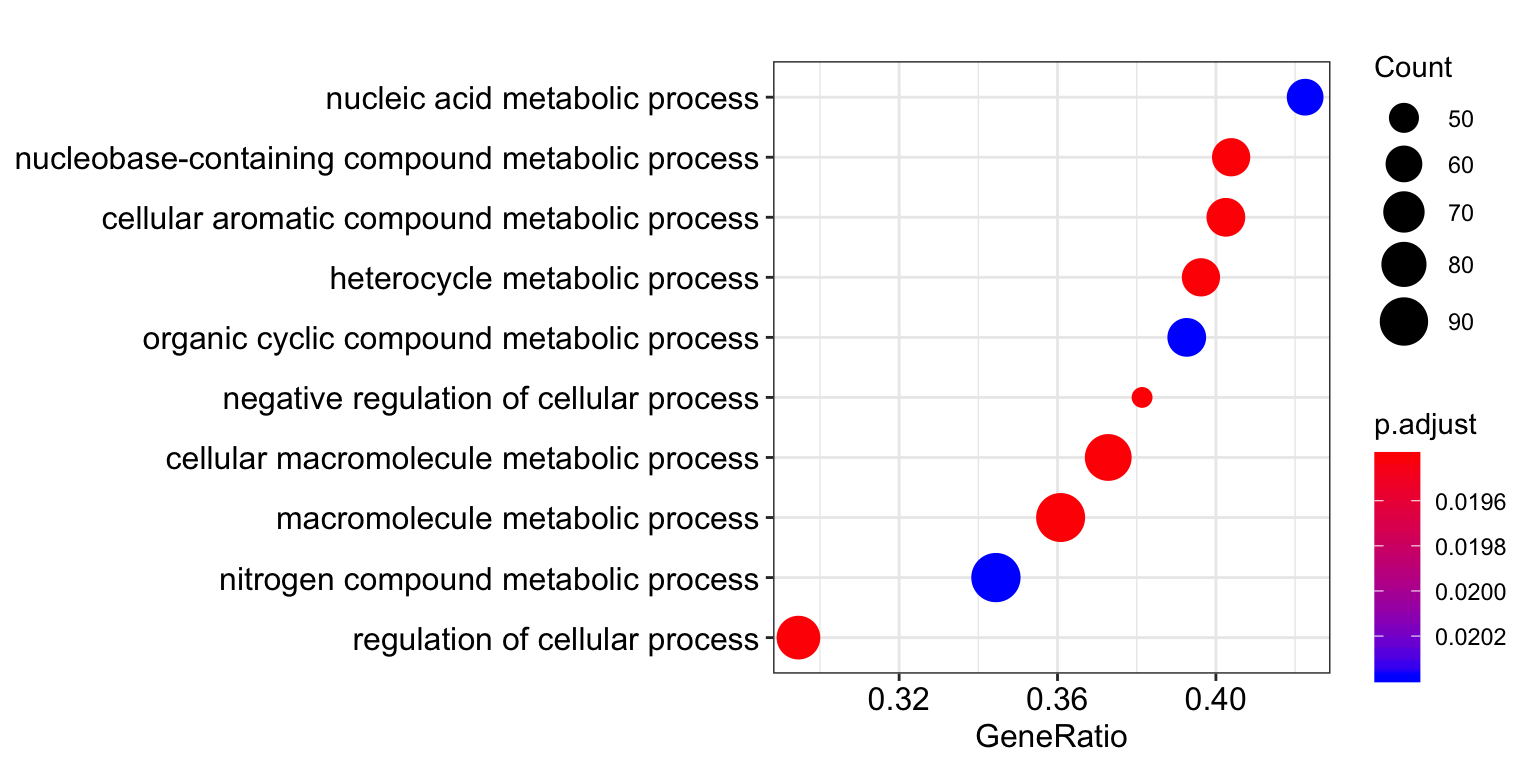
# convert the results back to gene names
convert <- function(x){
indx <- match(x,gene_info$ID)
names <- gene_info$name[indx]
return(paste(names,collapse = '/'))
}
tmp <- strsplit(gsea.signif_genes@result$core_enrichment,split = '/')
tmp1 <- sapply(tmp,FUN = convert)
gsea.signif_genes@result$core_enrichment <- tmp1
kable(head(gsea.signif_genes),row.names = F) %>%
kable_styling() %>%
scroll_box(width = "100%", height = "400px")| ID | Description | setSize | enrichmentScore | NES | pvalue | p.adjust | qvalues | rank | leading_edge | core_enrichment |
|---|---|---|---|---|---|---|---|---|---|---|
| GO:0043170 | macromolecule metabolic process | 255 | 0.5179999 | 1.391744 | 0.000999 | 0.0194092 | 0.0078117 | 131 | tags=36%, list=28%, signal=58% | THG1L/QRICH1/ARAF/PLPP3/KCTD10/ZNF48/SEMA3A/PRCC/CPPED1/SAMD4A/GTF3C2/TRIO/MAP3K4/MAPK8IP3/ZNF24/ZNF134/MED12/PSMA5/TCF7L2/ASRGL1/SOX21/SMG1/LPIN2/RNF115/MARCH7/TRIP12/STT3B/MSRA/UBAP1/EIF5AL1/NUP98/NR2F2/ZNF787/PAXBP1/MCM5/RCOR3/NCK2/ITGA6/CDCA7/ISY1/SPOCK1/GRIA1/E2F5/SNAPC3/TLE4/NCOA4/PPIF/TFDP1/PSMA6/PKMYT1/PAPD5/HEXIM1/PRKAR1A/NDUFAF5/STK26/FMR1/RPS6KA1/ATPIF1/BCL10/PKN2/PSRC1/ZNF512/PREPL/SNRNP27/FZD7/ZNF660/GOLGB1/GRSF1/SMAD1/NSUN2/SKP2/PLPP1/DHFR/CCNH/APC/PGGT1B/MMS22L/LIN28B/SHPRH/TRIM24/SDC2/BUD13/AMN1/NDFIP2/PPP2R5E/MAX/TRMT61A/CLK3/CHTF18/CDC6/MED13/KCTD2 |
| GO:0044260 | cellular macromolecule metabolic process | 228 | 0.5119208 | 1.378147 | 0.000999 | 0.0194092 | 0.0078117 | 131 | tags=37%, list=28%, signal=53% | QRICH1/ARAF/PLPP3/KCTD10/ZNF48/SEMA3A/CPPED1/SAMD4A/GTF3C2/TRIO/MAP3K4/MAPK8IP3/ZNF24/ZNF134/MED12/PSMA5/TCF7L2/SOX21/SMG1/LPIN2/RNF115/MARCH7/TRIP12/STT3B/MSRA/UBAP1/EIF5AL1/NUP98/NR2F2/ZNF787/PAXBP1/MCM5/RCOR3/NCK2/ITGA6/CDCA7/ISY1/SPOCK1/GRIA1/E2F5/SNAPC3/TLE4/NCOA4/PPIF/TFDP1/PSMA6/PKMYT1/PAPD5/HEXIM1/PRKAR1A/NDUFAF5/STK26/FMR1/RPS6KA1/ATPIF1/BCL10/PKN2/PSRC1/ZNF512/FZD7/ZNF660/GOLGB1/SMAD1/NSUN2/SKP2/PLPP1/DHFR/CCNH/APC/PGGT1B/MMS22L/LIN28B/SHPRH/TRIM24/SDC2/AMN1/NDFIP2/PPP2R5E/MAX/TRMT61A/CLK3/CHTF18/CDC6/MED13/KCTD2 |
| GO:0048523 | negative regulation of cellular process | 118 | 0.5499471 | 1.479355 | 0.000999 | 0.0194092 | 0.0078117 | 106 | tags=38%, list=23%, signal=39% | ARAF/PLPP3/RGS5/SEMA3A/PRCC/PRR11/PLXNB1/SAMD4A/TRIO/ZNF24/MED12/TCF7L2/SOX21/RNF115/MARCH7/TRIP12/NR2F2/PAXBP1/RCOR3/NCK2/ITGA6/SPOCK1/GRIA1/FZD3/E2F5/TLE4/PPIF/TFDP1/PKMYT1/PAPD5/HEXIM1/PRKAR1A/NDC80/STK26/FMR1/RPS6KA1/ATPIF1/BCL10/PSRC1/FZD7/SMAD1/NSUN2/PLPP1/DHFR/APC |
| GO:0006139 | nucleobase-containing compound metabolic process | 156 | 0.5164218 | 1.389733 | 0.001998 | 0.0194092 | 0.0078117 | 130 | tags=40%, list=28%, signal=44% | THG1L/QRICH1/PLPP3/ZNF48/PRCC/SAMD4A/GTF3C2/MAP3K4/ZNF24/ZNF134/MED12/TCF7L2/SOX21/SMG1/LPIN2/TRIP12/HSD17B4/NUP98/NR2F2/ZNF787/PAXBP1/MCM5/RCOR3/NCK2/ITGA6/CDCA7/ISY1/E2F5/SNAPC3/TLE4/NCOA4/PPIF/TFDP1/PSMA6/PAPD5/HEXIM1/PRKAR1A/FMR1/RPS6KA1/ATPIF1/BCL10/PSRC1/ZNF512/SNRNP27/FZD7/ZNF660/GOLGB1/GRSF1/SMAD1/NSUN2/DHFR/CCNH/MMS22L/LIN28B/SHPRH/TRIM24/BUD13/MAX/TRMT61A/CLK3/CHTF18/CDC6/MED13 |
| GO:0006725 | cellular aromatic compound metabolic process | 159 | 0.5171509 | 1.392743 | 0.001998 | 0.0194092 | 0.0078117 | 130 | tags=40%, list=28%, signal=44% | THG1L/QRICH1/PLPP3/ZNF48/PRCC/SAMD4A/GTF3C2/MAP3K4/ZNF24/ZNF134/MED12/TCF7L2/ASRGL1/SOX21/SMG1/LPIN2/TRIP12/HSD17B4/NUP98/NR2F2/ZNF787/PAXBP1/MCM5/RCOR3/NCK2/ITGA6/CDCA7/ISY1/E2F5/SNAPC3/TLE4/NCOA4/PPIF/TFDP1/PSMA6/PAPD5/HEXIM1/PRKAR1A/FMR1/RPS6KA1/ATPIF1/BCL10/PSRC1/ZNF512/SNRNP27/FZD7/ZNF660/GOLGB1/GRSF1/SMAD1/NSUN2/DHFR/CCNH/MMS22L/LIN28B/SHPRH/TRIM24/BUD13/MAX/TRMT61A/CLK3/CHTF18/CDC6/MED13 |
| GO:0046483 | heterocycle metabolic process | 159 | 0.5081597 | 1.368529 | 0.001998 | 0.0194092 | 0.0078117 | 130 | tags=40%, list=28%, signal=43% | THG1L/QRICH1/PLPP3/ZNF48/PRCC/SAMD4A/GTF3C2/MAP3K4/ZNF24/ZNF134/MED12/TCF7L2/SOX21/SMG1/LPIN2/TRIP12/HSD17B4/NUP98/NR2F2/ZNF787/PAXBP1/MCM5/RCOR3/NCK2/ITGA6/CDCA7/ISY1/E2F5/SNAPC3/TLE4/NCOA4/PPIF/TFDP1/PSMA6/PAPD5/HEXIM1/PRKAR1A/FMR1/RPS6KA1/ATPIF1/BCL10/PSRC1/ZNF512/SNRNP27/FZD7/ZNF660/GOLGB1/GRSF1/SMAD1/NSUN2/DHFR/CCNH/MMS22L/LIN28B/SHPRH/TRIM24/BUD13/MAX/TRMT61A/CLK3/CHTF18/CDC6/MED13 |
Permutation test
Please see this page for results of permutation tests.
Session Information
sessionInfo()R version 3.5.2 (2018-12-20)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] parallel stats4 stats graphics grDevices utils datasets
[8] methods base
other attached packages:
[1] clusterProfiler_3.10.1 org.Hs.eg.db_3.7.0 AnnotationDbi_1.44.0
[4] IRanges_2.16.0 S4Vectors_0.20.1 Biobase_2.42.0
[7] BiocGenerics_0.28.0 gridExtra_2.3 kableExtra_1.0.1
[10] knitr_1.22 ggplot2_3.1.0 ashr_2.2-32
loaded via a namespace (and not attached):
[1] enrichplot_1.2.0 bit64_0.9-7 progress_1.2.0
[4] doParallel_1.0.14 webshot_0.5.1 RColorBrewer_1.1-2
[7] httr_1.4.0 UpSetR_1.3.3 tools_3.5.2
[10] R6_2.4.0 DBI_1.0.0 lazyeval_0.2.1
[13] colorspace_1.4-0 withr_2.1.2 prettyunits_1.0.2
[16] tidyselect_0.2.5 bit_1.1-14 compiler_3.5.2
[19] rvest_0.3.2 xml2_1.2.0 triebeard_0.3.0
[22] labeling_0.3 scales_1.0.0 SQUAREM_2017.10-1
[25] readr_1.3.1 ggridges_0.5.1 mixsqp_0.1-97
[28] stringr_1.4.0 digest_0.6.18 rmarkdown_1.11
[31] DOSE_3.8.2 pscl_1.5.2 pkgconfig_2.0.2
[34] htmltools_0.3.6 highr_0.7 rlang_0.3.1
[37] rstudioapi_0.9.0 RSQLite_2.1.1 gridGraphics_0.3-0
[40] farver_1.1.0 jsonlite_1.6 BiocParallel_1.16.6
[43] GOSemSim_2.8.0 dplyr_0.8.0.1 magrittr_1.5
[46] ggplotify_0.0.3 GO.db_3.7.0 Matrix_1.2-16
[49] Rcpp_1.0.0 munsell_0.5.0 viridis_0.5.1
[52] stringi_1.3.1 yaml_2.2.0 ggraph_1.0.2
[55] MASS_7.3-51.1 plyr_1.8.4 qvalue_2.14.1
[58] grid_3.5.2 blob_1.1.1 ggrepel_0.8.0
[61] DO.db_2.9 crayon_1.3.4 lattice_0.20-38
[64] cowplot_0.9.4 splines_3.5.2 hms_0.4.2
[67] pillar_1.3.1 fgsea_1.8.0 igraph_1.2.4
[70] reshape2_1.4.3 codetools_0.2-16 fastmatch_1.1-0
[73] glue_1.3.0 evaluate_0.13 data.table_1.12.0
[76] urltools_1.7.2 tweenr_1.0.1 foreach_1.4.4
[79] tidyr_0.8.3 gtable_0.2.0 purrr_0.3.1
[82] polyclip_1.9-1 assertthat_0.2.0 xfun_0.5
[85] ggforce_0.2.1 europepmc_0.3 viridisLite_0.3.0
[88] truncnorm_1.0-8 tibble_2.0.1 rvcheck_0.1.3
[91] iterators_1.0.10 memoise_1.1.0