Permutation test – trans-gene analysis (t-test + ash)
Yifan Zhou
6/16/2019
library(ashr)
library(ggplot2)
library(knitr)
library(kableExtra)
library(gridExtra)
options(stringsAsFactors = F)
wkdir <- '~/Downloads/ASoC/singlecell/'
load(paste0(wkdir,'data/cropseq_expression.Rd'))
nlocus <- colSums(exp.per.enhancer>0)
ncell_uniq <- rep(NA,nrow(exp.per.gRNA))
for (i in 1:nrow(exp.per.gRNA)){
glocus <- row.names(exp.per.gRNA)[i]
ncell_uniq[i]=sum(exp.per.gRNA[glocus,]>0 & nlocus==1)
}
tmp <- sapply(strsplit(row.names(exp.per.gRNA),split = '_'),
function(x){paste(x[1],x[2],sep = '_')})
names(ncell_uniq) <- tmpDE analysis on permutated sctransformed data
We still use the sctransformed Pearson residuals, and we filtered the genes to be those present in at least 20% of all cells (8117 in total).
For each target gRNA, we selected 2 groups of cells: cells that only contain the target gRNAs, and cells that only contain the negative control gRNAs. Then, we permute the group labels and conducted categorical regression of the Pearson residuals (against the 2 permuted conditions) to obtain the summary statistics. As before, 70 (gRNA vs neg ctrl) pairs were tested.
ashr meta-analysis on regression summary statistics
For each gene, we combined the 70 DE effect sizes and standard errors estimated in the previous step, and used an adaptive shrinkage method, ashr, to discern which effect sizes are truly non-zero (significant). Here, we used a cutoff of 0.1 for lfsr (the local false sign rate).
get.result <- function(betaest.mtx,lfsr.mtx){
tmp.gRNA <- colSums(lfsr.mtx<0.1)
tmp.gene <- rowSums(lfsr.mtx<0.1)
signif_genes <- row.names(betaest.mtx)[tmp.gene>0]
cat('There are',length(signif_genes),'significant genes in total.')
signif_genes.gRNA.lst <- list()
for(i in signif_genes){
gRNA.group <- colnames(betaest.mtx)[lfsr.mtx[i,]<0.1]
signif_genes.gRNA.lst[[i]] <- sapply(strsplit(gRNA.group,split = '_'),
function(x){paste(x[1],x[2],sep = '_')})
}
# Make table:
compact.genes.lst <- sapply(signif_genes.gRNA.lst,FUN = function(x){paste(x,collapse = ', ')})
compact.genes.df <- as.data.frame(compact.genes.lst)
tmp.indx <- order(rownames(compact.genes.df))
compact.genes.df <- data.frame(signif_gene=rownames(compact.genes.df)[tmp.indx],
gRNA_targets=compact.genes.df[tmp.indx,])
# Present table:
print(kable(compact.genes.df, caption = 'gRNA targets per significant gene') %>%
kable_styling() %>% scroll_box(width = "100%", height = "300px"))
num_gRNA_pergene <- sapply(signif_genes.gRNA.lst,FUN = function(x){length(x)})
### Significant genes per gRNA target
gRNA.signif_genes.lst <- list()
for (i in signif_genes){
cells <- signif_genes.gRNA.lst[[i]]
for (c in cells){
if (is.na(match(c, names(gRNA.signif_genes.lst)))){
gRNA.signif_genes.lst[[c]] <- i
} else {
gRNA.signif_genes.lst[[c]] <- c(gRNA.signif_genes.lst[[c]],i)
}
}
}
# Make table:
compact.gRNAs.lst <- sapply(gRNA.signif_genes.lst,FUN = function(x){paste(x,collapse = ', ')})
compact.gRNAs.df <- as.data.frame(compact.gRNAs.lst)
tmp.indx <- order(rownames(compact.gRNAs.df))
compact.gRNAs.df <- data.frame(gRNA_target=rownames(compact.gRNAs.df)[tmp.indx],
signif_genes=compact.gRNAs.df[tmp.indx,])
compact.gRNAs.df$gRNA_target <- sapply(strsplit(compact.gRNAs.df$gRNA_target,split = '_'),
function(x){paste(x[1],x[2],sep = '_')})
compact.gRNAs.df$num_cells <- ncell_uniq[compact.gRNAs.df$gRNA_target]
compact.gRNAs.df <- compact.gRNAs.df[,c(3,1,2)]
# Present table:
print(kable(compact.gRNAs.df, caption = 'Significant genes per gRNA target') %>%
kable_styling() %>% scroll_box(width = "100%", height = "300px"))
num_gene_pergRNA <- sapply(gRNA.signif_genes.lst,FUN = function(x){length(x)})
# Plots:
plot1 <- qplot(num_gRNA_pergene,geom = 'histogram',xlab = 'Number of gRNA targets per gene',ylab = 'Count',
main = paste('Distribution of', length(num_gRNA_pergene), 'significant genes'),bins=10)
plot2 <- qplot(num_gene_pergRNA,geom = 'histogram',xlab = 'Number of DE genes per gRNA target',
ylab = 'Count', main = paste('Distribution of', length(num_gene_pergRNA), 'gRNAs'),bins=20)
plot3 <- qplot(compact.gRNAs.df$num_cells,num_gene_pergRNA[compact.gRNAs.df$gRNA_target],
xlab = 'Number of cells in the target condition',ylab = 'Number of significant genes')
grid.arrange(plot1,plot2,plot3,ncol=2)
}Permutation 1
Here are all the significant genes (\(lfsr<0.1\)) and the gRNA groups where they are differentially expressed:
load(paste0(wkdir,'data/perm.ashr_estimation01.Rdata'))
get.result(betaest.mtx,lfsr.mtx)| signif_gene | gRNA_targets |
|---|---|
| ABAT | RERE_2 |
| ABHD14A | pos_SNAP91 |
| ABR | miR137_3 |
| AC004696.2 | SETD1A_1st |
| ACAT1 | KCTD13_3 |
| ACBD3 | pos_SNAP91 |
| ADAT1 | PCDHA123_3 |
| ADK | PPP1R16B_1 |
| AEN | SETD1A_1st |
| AFMID | miR137_3, LINC00637_2 |
| AGO3 | TRANK1_1 |
| AHCTF1 | UBE2Q2P1_2 |
| AKAP12 | CHRNA3_3 |
| AKAP8L | SETD1A_1st |
| AKIRIN1 | SETD1A_1st |
| ALS2 | KCTD13_2 |
| AMOTL1 | UBE2Q2P1_2 |
| AMPH | LOC100507431_2 |
| ANAPC13 | SETD1A_1st |
| ANKIB1 | PBRM1_3 |
| ANXA2 | CHRNA3_3 |
| AP1S1 | PPP1R16B_2 |
| APBB1 | LINC00637_1 |
| AR | SETD1A_1st |
| ARF4 | SETD1A_1st, NGEF_1 |
| ARHGEF1 | CHRNA3_3 |
| ARID3A | SETD1A_1st |
| ARL4C | NGEF_2 |
| ARMC1 | PPP1R16B_2 |
| ARMCX2 | VPS45_1 |
| ARPP19 | PCDHA123_3 |
| ATG4D | SETD1A_1st |
| ATXN1L | GALNT10_3 |
| BAALC | GALNT10_1 |
| BBS4 | UBE2Q2P1_1 |
| BCAP31 | SETD1A_1st |
| BTAF1 | LOC105376975_3 |
| BTG1 | pos_SNAP91 |
| BTG2 | pos_SNAP91 |
| BYSL | SETD1A_1st |
| C15orf61 | KMT2E_1 |
| C16orf52 | UBE2Q2P1_2 |
| C16orf70 | CHRNA3_3 |
| C17orf75 | UBE2Q2P1_3 |
| C18orf21 | PBRM1_3 |
| C19orf47 | UBE2Q2P1_2 |
| C1orf43 | SETD1A_1st |
| C5orf15 | RERE_3 |
| C9orf114 | KMT2E_2 |
| C9orf9 | BAG5_3 |
| CAB39L | pos_SNAP91 |
| CALD1 | CHRNA3_3 |
| CAMK2G | SETD1A_1st |
| CAND1 | GALNT10_1 |
| CAP1 | CHRNA3_3 |
| CAPZB | LOC105376975_1 |
| CARD19 | NGEF_1 |
| CASP6 | KCTD13_3 |
| CAV2 | CHRNA3_3 |
| CBY1 | DPYD_2 |
| CCNB1 | KMT2E_3 |
| CCND3 | PBRM1_3 |
| CCT4 | NAB2_2 |
| CDC5L | TCF4-ITF2_2 |
| CDH11 | NGEF_2 |
| CDK20 | SETD1A_1st |
| CDK5RAP3 | LINC00637_1 |
| CDKN1C | pos_SNAP91 |
| CEP85L | NGEF_2 |
| CHM | UBE2Q2P1_1 |
| CITED4 | NGEF_2 |
| CKB | LINC00637_3 |
| CLTB | KCTD13_2 |
| CNPY3 | TCF4-ITF2_1 |
| CNTLN | KMT2E_2 |
| COL6A1 | LOC105376975_3 |
| COPZ1 | LOC100507431_2 |
| CRTAP | TRANK1_1 |
| CSRP2 | NGEF_2 |
| CTDSPL2 | PCDHA123_2 |
| CTU1 | SETD1A_1st, NGEF_2 |
| CYCS | SETD1A_1st |
| DAD1 | SETD1A_1st |
| DCAF6 | VPS45_1 |
| DDX28 | KCTD13_3 |
| DDX31 | BCL11B_1 |
| DESI2 | NAB2_3 |
| DHCR7 | SETD1A_1st |
| DLL1 | pos_SNAP91 |
| DLST | SETD1A_1st |
| DMAP1 | SETD1A_1st |
| DNAJC25 | SETD1A_1st |
| DNPEP | PCDHA123_3 |
| DSEL | CHRNA3_3 |
| DTNBP1 | NAB2_2 |
| DUSP16 | TRANK1_1 |
| DUSP18 | NGEF_2 |
| EAF1 | SETD1A_1st, NGEF_2 |
| ECHS1 | LOC105376975_1 |
| EEF1AKMT4 | NGEF_1 |
| EHBP1 | pos_SNAP91 |
| EHD4 | PBRM1_1, PBRM1_3 |
| ELOVL4 | CHRNA3_1 |
| EMC7 | SETD1A_1st, NGEF_2 |
| EMP3 | CHRNA3_3 |
| ERBIN | PBRM1_2 |
| ERCC4 | SETD1A_1st |
| EXT1 | BCL11B_3 |
| EZH2 | CHRNA3_1 |
| FAM199X | CHRNA3_3 |
| FAM46A | DPYD_3 |
| FAM50A | PBRM1_3 |
| FAM98A | NAB2_1 |
| FANCE | KMT2E_3, UBE2Q2P1_3 |
| FDFT1 | SETD1A_1st |
| FGF19 | TRANK1_1 |
| FIZ1 | NGEF_2 |
| FJX1 | NGEF_2 |
| FLAD1 | VPS45_3 |
| FLYWCH1 | UBE2Q2P1_1 |
| FN1 | NGEF_2 |
| FOXRED1 | PBRM1_1 |
| FUOM | CHRNA3_3 |
| FZD2 | TCF4-ITF2_1 |
| GADD45G | pos_SNAP91 |
| GALNT2 | UBE2Q2P1_1 |
| GAN | PBRM1_3 |
| GAS2L1 | NGEF_2 |
| GCC2 | VPS45_1 |
| GDF15 | SETD1A_1st |
| GLE1 | NGEF_2 |
| GLRX3 | SETD1A_1st |
| GLT25D1 | BAG5_1 |
| GNPDA2 | miR137_1 |
| GOLGA3 | LOC105376975_3 |
| GOLGA4 | PBRM1_2 |
| GPCPD1 | LOC105376975_2 |
| GRN | SETD1A_1st |
| GTPBP4 | UBE2Q2P1_2 |
| HACD1 | CHRNA3_3 |
| HDAC1 | KMT2E_3 |
| HDAC8 | RERE_2 |
| HECTD4 | LOC100507431_1 |
| HES6 | pos_SNAP91 |
| HGSNAT | PBRM1_2 |
| HIST1H1C | SETD1A_1st |
| HIST3H2A | NGEF_2 |
| HJURP | CHRNA3_1 |
| HLA-E | NGEF_2 |
| HMGCS1 | SETD1A_1st |
| HOXB3 | BCL11B_3 |
| HS3ST3B1 | pos_SNAP91 |
| HSCB | NAB2_1 |
| HSD17B11 | LOC100507431_1 |
| HSD17B7 | SETD1A_1st |
| HSF2 | KMT2E_2 |
| HSPB1 | BCL11B_3, KCTD13_2, PBRM1_1 |
| IDI1 | SETD1A_1st |
| IK | SETD1A_1st |
| IL6ST | GALNT10_3 |
| ILKAP | CHRNA3_3 |
| INSIG1 | SETD1A_1st |
| INTS6 | KCTD13_1 |
| IQGAP2 | miR137_3 |
| ISG15 | LOC105376975_3 |
| JUN | TRANK1_1 |
| KCNC1 | PCDHA123_2 |
| KDM4B | CHRNA3_3 |
| KEAP1 | PBRM1_3 |
| KIAA0100 | PBRM1_3 |
| KIF1BP | SETD1A_1st |
| KIF23 | CHRNA3_1 |
| KIFC3 | CHRNA3_3 |
| KLHL12 | KMT2E_2 |
| KLHL7 | LOC105376975_1 |
| KMT5A | CHRNA3_1 |
| KRT18 | SETD1A_1st |
| LAMA1 | LOC105376975_3, VPS45_2 |
| LAMC1 | PBRM1_2 |
| LAMTOR1 | NGEF_2 |
| LARP1B | NAB2_2 |
| LARP4B | PBRM1_2 |
| LEMD2 | PBRM1_1 |
| LGALS3 | CHRNA3_3 |
| LMBR1L | LINC00637_3 |
| LMNA | NGEF_1 |
| LOXL2 | TCF4-ITF2_3 |
| LRCH3 | BCL11B_3 |
| LTBP1 | pos_SNAP91 |
| LYPD1 | NGEF_2 |
| MAGEH1 | SETD1A_1st |
| MAPK1 | BCL11B_2 |
| MAPK7 | VPS45_3 |
| MED10 | SETD1A_1st |
| MED14 | miR137_3 |
| MED20 | UBE2Q2P1_1 |
| MED26 | PBRM1_1 |
| MEG3 | UBE2Q2P1_1 |
| MELK | CHRNA3_1 |
| METAP1 | LOC105376975_3 |
| METTL12 | NGEF_2 |
| METTL14 | RERE_3 |
| MFSD5 | GALNT10_2 |
| MIR99AHG | BCL11B_1 |
| MIS18BP1 | CHRNA3_1 |
| MKNK2 | GALNT10_3 |
| MLLT4 | GALNT10_1 |
| MMGT1 | UBE2Q2P1_1 |
| MOB2 | SETD1A_1st |
| MOCS3 | NGEF_2 |
| MPDZ | TRANK1_1 |
| MPHOSPH10 | RERE_2 |
| MPHOSPH8 | NGEF_2 |
| MTCH1 | SETD1A_1st |
| MTFP1 | KCTD13_1 |
| MTMR14 | PBRM1_1 |
| MUT | SETD1A_1st |
| MUTYH | CHRNA3_1 |
| MVD | SETD1A_1st |
| MVK | SETD1A_1st |
| MYO9B | NGEF_2 |
| NAA20 | NAB2_2 |
| NAMPT | miR137_3 |
| NDUFAF7 | pos_SNAP91 |
| NECAP1 | SETD1A_1st |
| NFYC | UBE2Q2P1_3 |
| NKAIN4 | PPP1R16B_1 |
| NKIRAS2 | SETD1A_1st, NGEF_2 |
| NOA1 | PCDHA123_3 |
| NOL7 | BCL11B_1 |
| NQO1 | PBRM1_3 |
| NRBP1 | KCTD13_1 |
| NUDT2 | NGEF_2 |
| NUP188 | TCF4-ITF2_3 |
| OLMALINC | SETD1A_1st |
| OTULIN | SETD1A_1st, NGEF_2 |
| PAIP2 | pos_SNAP91 |
| PAK1IP1 | BAG5_1 |
| PAPSS1 | LOC105376975_1 |
| PBDC1 | LOC100507431_2 |
| PCGF2 | LOC100507431_2 |
| PCMT1 | SETD1A_1st |
| PDCD6IP | pos_SNAP91 |
| PDE9A | TRANK1_2 |
| PDPR | STAT6_2 |
| PDRG1 | SETD1A_1st |
| PGM1 | SETD1A_1st |
| PIAS3 | PBRM1_2 |
| PIDD1 | SETD1A_1st |
| PIGP | NGEF_1 |
| PIK3C2B | pos_SNAP91 |
| PLCD3 | PBRM1_1 |
| PLEKHA5 | NGEF_2 |
| PLPP1 | CHRNA3_3 |
| PMM2 | miR137_1 |
| PNKD | UBE2Q2P1_3 |
| PNO1 | PBRM1_3 |
| POC5 | CHRNA3_1 |
| POFUT2 | SETD1A_1st |
| POLR2A | NGEF_1 |
| POLR3A | DPYD_2 |
| POP4 | NGEF_2 |
| PPIL3 | SETD1A_1st |
| PPP1R1A | TRANK1_1 |
| PPP2R5D | LOC105376975_3 |
| PPP2R5E | STAT6_3 |
| PRAG1 | SETD1A_1st |
| PRELID3B | UBE2Q2P1_3 |
| PREPL | BCL11B_3 |
| PRMT7 | DPYD_3 |
| PSEN1 | NGEF_2 |
| PTRF | CHRNA3_3 |
| PUM3 | NGEF_2 |
| RAB28 | UBE2Q2P1_1 |
| RAD23A | SETD1A_1st |
| RAI1 | STAT6_1 |
| RAPGEF2 | UBE2Q2P1_3 |
| RARA | NGEF_1 |
| RASSF1 | SETD1A_1st |
| RBFOX2 | SETD1A_1st |
| RBMS1 | NGEF_2 |
| RGS19 | PCDHA123_3 |
| RHBDD3 | STAT6_3 |
| RHOC | KCTD13_2 |
| RIMKLB | CHRNA3_1 |
| RNF14 | KMT2E_2 |
| RNF167 | SETD1A_1st |
| RNF7 | UBE2Q2P1_1 |
| RNPEP | LINC00637_3 |
| RP11-32K4.2 | pos_SNAP91 |
| RP11-357C3.3 | KMT2E_1 |
| RP11-539L10.3 | PBRM1_1 |
| RP11-701H24.2 | TRANK1_1 |
| RPF2 | CHRNA3_3 |
| RPL12 | TCF4-ITF2_2 |
| RPL32 | KCTD13_2 |
| RPL6 | KCTD13_2 |
| RPL7 | KCTD13_2 |
| RPL7A | KCTD13_2 |
| RPP40 | NGEF_2 |
| RPS11 | KCTD13_2 |
| RPS27 | KCTD13_2 |
| RPUSD2 | DPYD_2 |
| RRBP1 | PBRM1_2 |
| RRP9 | SETD1A_1st |
| S100A10 | PBRM1_1 |
| SEC13 | SETD1A_1st |
| SEC61G | SETD1A_1st |
| SEL1L | TCF4-ITF2_3 |
| SERINC3 | pos_SNAP91 |
| SERTAD1 | SETD1A_1st |
| SERTAD3 | TCF4-ITF2_3 |
| SFXN4 | PPP1R16B_2 |
| SH3GLB1 | SETD1A_1st |
| SLC12A2 | UBE2Q2P1_2, PBRM1_1 |
| SLC16A14 | UBE2Q2P1_2 |
| SLC25A4 | CHRNA3_3 |
| SLC26A11 | GALNT10_2 |
| SLC38A7 | NGEF_2 |
| SLX4IP | SETD1A_1st |
| SMAD5 | RERE_3 |
| SMARCC1 | CHRNA3_1 |
| SMG5 | BCL11B_1 |
| SMG6 | UBE2Q2P1_2 |
| SNHG11 | CHRNA3_3 |
| SNU13 | CHRNA3_3, UBE2Q2P1_2 |
| SOBP | KCTD13_1 |
| SORT1 | RERE_3, TRANK1_1 |
| SOX3 | pos_SNAP91 |
| SOX4 | pos_SNAP91 |
| SP1 | KCTD13_3 |
| SP2 | VPS45_3 |
| SPATS2L | CHRNA3_3, UBE2Q2P1_2 |
| SQLE | SETD1A_1st |
| SSR1 | SETD1A_1st, PPP1R16B_3 |
| STK17A | SETD1A_1st |
| SUGP2 | pos_SNAP91 |
| SUMO1 | RERE_2 |
| SURF1 | BCL11B_2 |
| SWAP70 | KCTD13_2 |
| TACC1 | PBRM1_1 |
| TAF6L | LOC100507431_1 |
| TBKBP1 | CHRNA3_3 |
| TBL1X | CHRNA3_1 |
| TBP | LOC100507431_1 |
| TCAIM | PBRM1_1 |
| TDRD3 | LINC00637_3 |
| TENM2 | LINC00637_2 |
| TGDS | SETD1A_1st |
| THAP12 | CHRNA3_1 |
| THAP7 | RERE_1 |
| THEM6 | PBRM1_2 |
| THOC2 | NAB2_1 |
| TIAL1 | VPS45_3 |
| TIMP2 | PBRM1_2 |
| TLK2 | CHRNA3_2 |
| TMED2 | SETD1A_1st |
| TMEM106B | pos_SNAP91 |
| TMEM126B | PBRM1_1 |
| TMEM208 | PBRM1_2 |
| TMEM63B | VPS45_1 |
| TMEM99 | SETD1A_1st, NGEF_2 |
| TMX2 | SETD1A_1st |
| TNIK | UBE2Q2P1_2 |
| TOGARAM1 | KMT2E_3 |
| TOMM70 | TRANK1_1 |
| TOR1AIP2 | BCL11B_3 |
| TPM1 | CHRNA3_3 |
| TPST2 | PBRM1_1 |
| TRAF3IP1 | NGEF_2 |
| TRMU | CHRNA3_3 |
| TSEN54 | SETD1A_1st |
| TSPAN12 | pos_SNAP91 |
| TTC33 | pos_SNAP91 |
| TTK | KMT2E_3 |
| TTLL1 | BCL11B_2 |
| TUBA1C | NGEF_1 |
| TUSC2 | GALNT10_2 |
| TXNRD2 | UBE2Q2P1_3 |
| UBE2K | UBE2Q2P1_1 |
| UBE4B | TRANK1_1 |
| UBR2 | NGEF_2 |
| UFC1 | VPS45_1 |
| UFM1 | SETD1A_1st |
| UIMC1 | NGEF_2 |
| UNC119 | KCTD13_1, PCDHA123_3 |
| UNK | SETD1A_1st |
| UPF1 | NAB2_3 |
| VAT1 | CHRNA3_3 |
| VPS26B | miR137_2 |
| VPS8 | pos_SNAP91 |
| WBP2 | KCTD13_2 |
| WDR26 | BAG5_2 |
| WDR89 | pos_SNAP91 |
| WFIKKN1 | PBRM1_3 |
| WSCD1 | CHRNA3_1 |
| XRCC2 | KMT2E_2 |
| XRCC3 | SETD1A_1st |
| YAE1D1 | NGEF_1 |
| YWHAE | LOC100507431_2 |
| ZBTB20 | KCTD13_2 |
| ZC3H18 | GALNT10_3 |
| ZC3H7B | UBE2Q2P1_2 |
| ZDHHC4 | NGEF_2 |
| ZFAND2A | SETD1A_1st, NGEF_2 |
| ZFPL1 | SETD1A_1st, NGEF_1 |
| ZFYVE19 | NGEF_2 |
| ZNF140 | SETD1A_1st |
| ZNF174 | pos_SNAP91 |
| ZNF362 | pos_SNAP91 |
| ZNF524 | KCTD13_3 |
| ZNF573 | NGEF_1 |
| ZNF655 | UBE2Q2P1_2 |
| ZNF93 | NAB2_2 |
| ZNHIT2 | NGEF_1 |
| ZNRF3 | TRANK1_1 |
| ZSCAN18 | SETD1A_1st |
| ZYX | CHRNA3_3 |
| num_cells | gRNA_target | signif_genes |
|---|---|---|
| 36 | BAG5_1 | PAK1IP1, GLT25D1 |
| 33 | BAG5_2 | WDR26 |
| 31 | BAG5_3 | C9orf9 |
| 13 | BCL11B_1 | SMG5, NOL7, DDX31, MIR99AHG |
| 33 | BCL11B_2 | SURF1, MAPK1, TTLL1 |
| 13 | BCL11B_3 | TOR1AIP2, PREPL, LRCH3, HSPB1, EXT1, HOXB3 |
| 3 | CHRNA3_1 | MUTYH, HJURP, SMARCC1, POC5, ELOVL4, EZH2, MELK, THAP12, RIMKLB, KMT5A, MIS18BP1, KIF23, WSCD1, TBL1X |
| 23 | CHRNA3_2 | TLK2 |
| 13 | CHRNA3_3 | CAP1, SPATS2L, ILKAP, SLC25A4, PLPP1, RPF2, AKAP12, CAV2, CALD1, ZYX, HACD1, FUOM, LGALS3, ANXA2, TPM1, KIFC3, C16orf70, PTRF, VAT1, TBKBP1, DSEL, KDM4B, ARHGEF1, EMP3, SNHG11, SNU13, TRMU, FAM199X |
| 68 | DPYD_2 | POLR3A, RPUSD2, CBY1 |
| 28 | DPYD_3 | FAM46A, PRMT7 |
| 38 | GALNT10_1 | MLLT4, BAALC, CAND1 |
| 37 | GALNT10_2 | TUSC2, MFSD5, SLC26A11 |
| 50 | GALNT10_3 | IL6ST, ATXN1L, ZC3H18, MKNK2 |
| 30 | KCTD13_1 | NRBP1, SOBP, INTS6, UNC119, MTFP1 |
| 32 | KCTD13_2 | RHOC, RPS27, ALS2, RPL32, ZBTB20, CLTB, HSPB1, RPL7, RPL7A, SWAP70, RPL6, WBP2, RPS11 |
| 15 | KCTD13_3 | CASP6, ACAT1, SP1, DDX28, ZNF524 |
| 11 | KMT2E_1 | C15orf61, RP11-357C3.3 |
| 19 | KMT2E_2 | KLHL12, RNF14, HSF2, XRCC2, CNTLN, C9orf114 |
| 18 | KMT2E_3 | HDAC1, CCNB1, FANCE, TTK, TOGARAM1 |
| 45 | LINC00637_1 | APBB1, CDK5RAP3 |
| 34 | LINC00637_2 | TENM2, AFMID |
| 53 | LINC00637_3 | RNPEP, LMBR1L, TDRD3, CKB |
| 15 | LOC100507431_1 | HSD17B11, TBP, TAF6L, HECTD4 |
| 29 | LOC100507431_2 | AMPH, COPZ1, YWHAE, PCGF2, PBDC1 |
| 35 | LOC105376975_1 | CAPZB, PAPSS1, KLHL7, ECHS1 |
| 50 | LOC105376975_2 | GPCPD1 |
| 8 | LOC105376975_3 | ISG15, METAP1, PPP2R5D, BTAF1, GOLGA3, LAMA1, COL6A1 |
| 44 | miR137_1 | GNPDA2, PMM2 |
| 30 | miR137_2 | VPS26B |
| 54 | miR137_3 | IQGAP2, NAMPT, ABR, AFMID, MED14 |
| 36 | NAB2_1 | FAM98A, HSCB, THOC2 |
| 24 | NAB2_2 | CCT4, LARP1B, DTNBP1, ZNF93, NAA20 |
| 25 | NAB2_3 | DESI2, UPF1 |
| 11 | NGEF_1 | LMNA, ARF4, EEF1AKMT4, YAE1D1, CARD19, ZFPL1, ZNHIT2, TUBA1C, POLR2A, RARA, ZNF573, PIGP |
| 2 | NGEF_2 | CITED4, HIST3H2A, LYPD1, RBMS1, FN1, ARL4C, TRAF3IP1, EAF1, OTULIN, UIMC1, RPP40, HLA-E, UBR2, CEP85L, ZFAND2A, ZDHHC4, PUM3, NUDT2, GLE1, FJX1, METTL12, LAMTOR1, PLEKHA5, CSRP2, MPHOSPH8, PSEN1, EMC7, ZFYVE19, SLC38A7, CDH11, TMEM99, NKIRAS2, MYO9B, POP4, CTU1, FIZ1, MOCS3, GAS2L1, DUSP18 |
| 4 | PBRM1_1 | S100A10, MTMR14, TCAIM, RP11-539L10.3, SLC12A2, LEMD2, HSPB1, TACC1, TMEM126B, FOXRED1, EHD4, PLCD3, MED26, TPST2 |
| 12 | PBRM1_2 | PIAS3, LAMC1, GOLGA4, ERBIN, HGSNAT, THEM6, LARP4B, TMEM208, TIMP2, RRBP1 |
| 4 | PBRM1_3 | PNO1, CCND3, ANKIB1, EHD4, WFIKKN1, NQO1, GAN, KIAA0100, C18orf21, KEAP1, FAM50A |
| 64 | PCDHA123_2 | KCNC1, CTDSPL2 |
| 62 | PCDHA123_3 | DNPEP, NOA1, ARPP19, ADAT1, UNC119, RGS19 |
| 3 | pos_SNAP91 | ZNF362, BTG2, PIK3C2B, ACBD3, LTBP1, NDUFAF7, EHBP1, HES6, PDCD6IP, ABHD14A, VPS8, TTC33, PAIP2, SOX4, DLL1, TMEM106B, TSPAN12, RP11-32K4.2, GADD45G, CDKN1C, BTG1, CAB39L, WDR89, ZNF174, HS3ST3B1, SUGP2, SERINC3, SOX3 |
| 44 | PPP1R16B_1 | ADK, NKAIN4 |
| 31 | PPP1R16B_2 | AP1S1, ARMC1, SFXN4 |
| 35 | PPP1R16B_3 | SSR1 |
| 48 | RERE_1 | THAP7 |
| 46 | RERE_2 | MPHOSPH10, SUMO1, ABAT, HDAC8 |
| 25 | RERE_3 | SORT1, METTL14, C5orf15, SMAD5 |
| 23 | SETD1A_1st | AKIRIN1, DMAP1, PGM1, SH3GLB1, C1orf43, HSD17B7, PPIL3, SEC13, EAF1, RASSF1, RRP9, ARF4, ANAPC13, MED10, OTULIN, HMGCS1, IK, SSR1, HIST1H1C, MTCH1, BYSL, MUT, PCMT1, ZFAND2A, CYCS, STK17A, SEC61G, INSIG1, PRAG1, FDFT1, SQLE, CDK20, DNAJC25, IDI1, KIF1BP, CAMK2G, OLMALINC, GLRX3, PIDD1, MOB2, TMX2, ZFPL1, DHCR7, NECAP1, KRT18, MVK, TMED2, ZNF140, UFM1, TGDS, DAD1, DLST, XRCC3, EMC7, AEN, ERCC4, MVD, RNF167, TMEM99, NKIRAS2, GRN, TSEN54, UNK, ARID3A, ATG4D, RAD23A, AKAP8L, GDF15, SERTAD1, CTU1, AC004696.2, ZSCAN18, SLX4IP, PDRG1, POFUT2, RBFOX2, MAGEH1, AR, BCAP31 |
| 24 | STAT6_1 | RAI1 |
| 38 | STAT6_2 | PDPR |
| 53 | STAT6_3 | PPP2R5E, RHBDD3 |
| 50 | TCF4-ITF2_1 | CNPY3, FZD2 |
| 45 | TCF4-ITF2_2 | CDC5L, RPL12 |
| 12 | TCF4-ITF2_3 | LOXL2, NUP188, SEL1L, SERTAD3 |
| 3 | TRANK1_1 | UBE4B, AGO3, JUN, SORT1, CRTAP, TOMM70, MPDZ, FGF19, DUSP16, PPP1R1A, RP11-701H24.2, ZNRF3 |
| 19 | TRANK1_2 | PDE9A |
| 11 | UBE2Q2P1_1 | GALNT2, RNF7, RAB28, UBE2K, MED20, MEG3, BBS4, FLYWCH1, CHM, MMGT1 |
| 8 | UBE2Q2P1_2 | AHCTF1, SPATS2L, SLC16A14, TNIK, SLC12A2, ZNF655, GTPBP4, AMOTL1, C16orf52, SMG6, C19orf47, ZC3H7B, SNU13 |
| 11 | UBE2Q2P1_3 | NFYC, PNKD, RAPGEF2, FANCE, C17orf75, PRELID3B, TXNRD2 |
| 14 | VPS45_1 | UFC1, DCAF6, GCC2, TMEM63B, ARMCX2 |
| 79 | VPS45_2 | LAMA1 |
| 9 | VPS45_3 | FLAD1, TIAL1, MAPK7, SP2 |
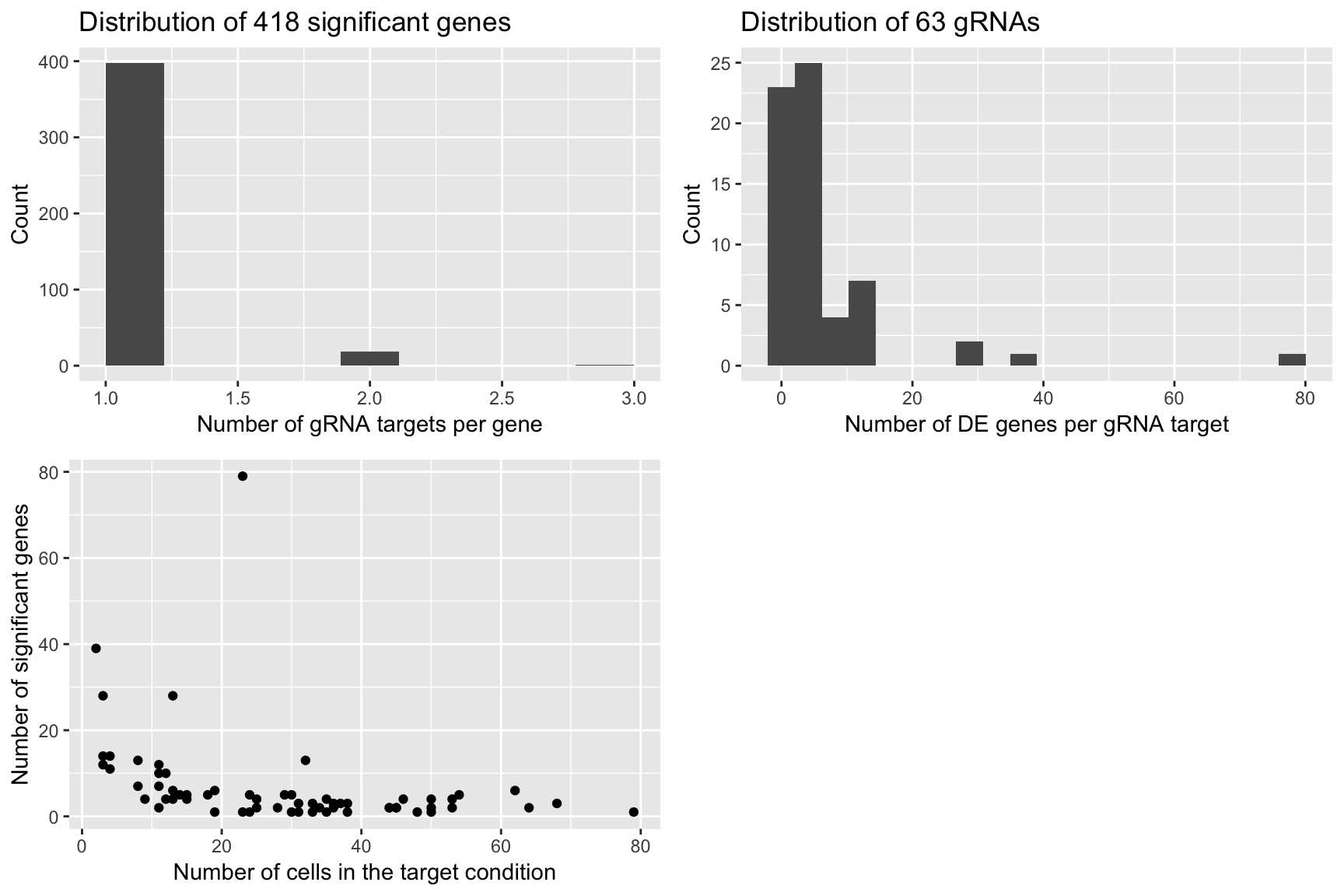
- We notice that gRNA targets with a large number of significant genes are the ones with relatively few cells (compared to the 111 negative control cells).
Permutation 2
load(paste0(wkdir,'data/perm.ashr_estimation02.Rdata'))
get.result(betaest.mtx,lfsr.mtx)| signif_gene | gRNA_targets |
|---|---|
| AAAS | NGEF_2 |
| ABL1 | GALNT10_3 |
| AC068491.1 | pos_SNAP91 |
| AC104655.3 | KCTD13_3 |
| ACBD5 | RERE_2, pos_SNAP91 |
| ACSL3 | LOC105376975_3 |
| ACTR2 | miR137_3 |
| ADO | PBRM1_3 |
| AFTPH | CHRNA3_1 |
| AHSA1 | UBE2Q2P1_1 |
| AKAP8 | UBE2Q2P1_1 |
| ALG5 | NAB2_2 |
| ANLN | CHRNA3_1 |
| ARF4 | UBE2Q2P1_1 |
| ARHGAP21 | CHRNA3_1 |
| ARHGEF17 | miR137_1 |
| ARID3A | UBE2Q2P1_1 |
| ARID5B | TRANK1_1 |
| ARL8B | UBE2Q2P1_1 |
| ARMC1 | PBRM1_3 |
| ARPC1B | LOC105376975_3 |
| ASXL1 | pos_SNAP91 |
| ATAD2 | TRANK1_1 |
| ATP11C | pos_SNAP91 |
| ATP5G1 | VPS45_2 |
| ATP5G3 | UBE2Q2P1_2 |
| AUNIP | BAG5_2 |
| AURKB | PBRM1_2 |
| B3GLCT | PBRM1_3 |
| BACE2 | miR137_2 |
| BAIAP2-AS1 | CHRNA3_1 |
| BAZ2A | LINC00637_1 |
| BAZ2B | SETD1A_1st |
| BEX1 | TRANK1_1 |
| BLOC1S2 | TRANK1_1 |
| BNIP3 | UBE2Q2P1_1 |
| BOK | UBE2Q2P1_3 |
| BUB1 | KCTD13_3 |
| BUB3 | PBRM1_2 |
| C11orf68 | UBE2Q2P1_1 |
| C11orf83 | LOC105376975_3 |
| C17orf75 | TCF4-ITF2_3 |
| C1GALT1C1 | TCF4-ITF2_2 |
| C7orf55 | BAG5_3 |
| C9orf40 | PCDHA123_1 |
| CABIN1 | TCF4-ITF2_1 |
| CAP2 | CHRNA3_1 |
| CASC3 | pos_SNAP91 |
| CASK | UBE2Q2P1_1 |
| CAV1 | NAB2_3 |
| CCDC184 | CHRNA3_1 |
| CCDC80 | pos_SNAP91 |
| CCNF | LOC100507431_1 |
| CCNJL | CHRNA3_3 |
| CD200 | NGEF_2 |
| CD24 | LINC00637_1 |
| CDC25A | TRANK1_1 |
| CDC25B | VPS45_1 |
| CDC34 | UBE2Q2P1_1 |
| CDK12 | SETD1A_1st |
| CDKN2A | PPP1R16B_3 |
| CENPM | TCF4-ITF2_1 |
| CENPV | TCF4-ITF2_1 |
| CEP250 | pos_SNAP91 |
| CEP350 | LOC105376975_3 |
| CHCHD2 | TRANK1_1 |
| CLDND1 | NAB2_3 |
| CLNS1A | CHRNA3_3 |
| CLOCK | PBRM1_1 |
| CLPTM1L | NGEF_1 |
| CMTM8 | CHRNA3_1 |
| CNRIP1 | NGEF_2 |
| COPS7B | miR137_3 |
| COQ5 | TCF4-ITF2_2 |
| CPD | TCF4-ITF2_2 |
| CPSF4 | pos_SNAP91 |
| CPXM1 | KMT2E_2 |
| CRCP | TCF4-ITF2_3 |
| CREB3 | UBE2Q2P1_1 |
| CRELD2 | VPS45_2 |
| CRTC1 | CHRNA3_1 |
| CRYBB2 | pos_SNAP91 |
| CTB-31O20.2 | UBE2Q2P1_1 |
| CXXC1 | RERE_1 |
| CYCS | UBE2Q2P1_1 |
| DCAF10 | TCF4-ITF2_1 |
| DCBLD2 | LOC100507431_2 |
| DDX6 | CHRNA3_1 |
| DHRS12 | UBE2Q2P1_2 |
| DKC1 | TRANK1_1 |
| DLGAP5 | KCTD13_2 |
| DLL3 | SETD1A_1st |
| DNAJB1 | RERE_3 |
| DNAJB11 | NGEF_2 |
| DNAJB2 | UBE2Q2P1_1 |
| DPP8 | pos_SNAP91 |
| DSCC1 | TRANK1_1 |
| DSEL | RERE_2 |
| DTX3 | NGEF_2 |
| DUSP14 | UBE2Q2P1_1 |
| DUSP3 | UBE2Q2P1_1 |
| DYNC2H1 | CHRNA3_1 |
| EDA2R | KCTD13_1 |
| EIF4A1 | pos_SNAP91 |
| ELMO1 | NGEF_2 |
| ELP5 | PBRM1_3 |
| EMD | TRANK1_1 |
| ENKD1 | UBE2Q2P1_1 |
| ENTPD6 | CHRNA3_3 |
| EPHA2 | UBE2Q2P1_1 |
| EPN2 | LOC105376975_3 |
| ESCO1 | LINC00637_1 |
| EXTL3 | TRANK1_1, PBRM1_3 |
| FAM134C | STAT6_1 |
| FAM20B | pos_SNAP91 |
| FAM46A | TCF4-ITF2_3 |
| FANCB | PBRM1_3 |
| FBRS | pos_SNAP91 |
| FBXO3 | BCL11B_3 |
| FGFR1 | UBE2Q2P1_2 |
| FN1 | BCL11B_3 |
| FOSL1 | UBE2Q2P1_1 |
| FSD1L | LOC105376975_2 |
| GATS | NGEF_2 |
| GBA2 | NGEF_2 |
| GJA1 | BCL11B_3 |
| GNAI1 | STAT6_2 |
| GNB1 | NAB2_3 |
| GOLGA7 | UBE2Q2P1_1 |
| GPS1 | VPS45_2 |
| GSTM3 | BCL11B_1 |
| GSTM4 | BCL11B_3 |
| HARS2 | CHRNA3_1 |
| HAT1 | VPS45_2 |
| HAUS8 | NAB2_3 |
| HDAC2 | TCF4-ITF2_1 |
| HIC2 | PCDHA123_1 |
| HIRA | SETD1A_1st |
| HKR1 | LOC105376975_3 |
| HNRNPA1 | KCTD13_1 |
| HSD17B11 | CHRNA3_1 |
| HSP90AB1 | LOC105376975_2 |
| IFT122 | RERE_1 |
| IFT27 | LINC00637_3 |
| IK | NAB2_2 |
| INTS14 | NAB2_2 |
| IP6K1 | UBE2Q2P1_1 |
| ITFG2 | PCDHA123_2 |
| KAZN | DPYD_1 |
| KCTD9 | KMT2E_2 |
| KDM5B | SETD1A_1st |
| KHNYN | UBE2Q2P1_1 |
| KLHDC10 | pos_SNAP91 |
| KLHL25 | CHRNA3_1 |
| KLHL5 | TRANK1_2 |
| KRTCAP2 | STAT6_3 |
| LAMTOR1 | BCL11B_3 |
| LDHB | UBE2Q2P1_1 |
| LETMD1 | PBRM1_1 |
| LINC00467 | BCL11B_3, UBE2Q2P1_2 |
| LMNA | UBE2Q2P1_1 |
| LRIF1 | UBE2Q2P1_1 |
| LSAMP | VPS45_2 |
| LTBP4 | KMT2E_1 |
| MACF1 | pos_SNAP91 |
| MAFF | UBE2Q2P1_1 |
| MAP2K3 | pos_SNAP91 |
| MAPKAP1 | UBE2Q2P1_1 |
| MAPKAPK2 | pos_SNAP91 |
| MDM1 | UBE2Q2P1_3 |
| MED29 | TCF4-ITF2_3 |
| METTL2B | NAB2_1 |
| MFSD13A | PCDHA123_3 |
| MGAT2 | PCDHA123_1 |
| MICU2 | VPS45_3 |
| MIR99AHG | SETD1A_1st |
| MOGS | PCDHA123_3 |
| MOK | KMT2E_2 |
| MORF4L1 | NAB2_2 |
| MORN4 | PBRM1_3 |
| MRPS18C | DPYD_2 |
| MT1X | pos_SNAP91 |
| MTERF1 | UBE2Q2P1_3 |
| MTIF2 | DPYD_1 |
| MYC | pos_SNAP91 |
| MZT2B | BCL11B_2 |
| NAA35 | DPYD_2 |
| NANS | KCTD13_1 |
| NAPG | NGEF_2 |
| NCBP1 | pos_SNAP91 |
| NDUFS4 | GALNT10_2 |
| NDUFS8 | UBE2Q2P1_1 |
| NEAT1 | pos_SNAP91 |
| NEK9 | PCDHA123_3 |
| NEU1 | CHRNA3_1, UBE2Q2P1_3 |
| NINJ1 | TRANK1_1 |
| NKAIN3 | TCF4-ITF2_1 |
| NMU | TCF4-ITF2_1 |
| NSFL1C | TRANK1_2 |
| NSUN2 | BCL11B_3 |
| NTN1 | NGEF_2 |
| NUMB | pos_SNAP91 |
| NUP214 | BCL11B_1 |
| OBFC1 | LOC105376975_3 |
| ORAI1 | TRANK1_2, LINC00637_1 |
| PAAF1 | UBE2Q2P1_1 |
| PAK2 | KCTD13_3 |
| PARP16 | NGEF_1 |
| PCGF1 | LOC105376975_3 |
| PCYT2 | STAT6_2 |
| PDCD2L | NGEF_2 |
| PFDN4 | NAB2_2 |
| PGM3 | CHRNA3_1 |
| PHC2 | BAG5_2 |
| PHF12 | PBRM1_2 |
| PHF23 | LINC00637_1 |
| PIAS3 | LOC105376975_3 |
| PJA1 | VPS45_3 |
| PLEKHG1 | NGEF_2 |
| PLXDC2 | UBE2Q2P1_2 |
| PNKP | TRANK1_3 |
| POLR3F | NAB2_3 |
| POR | PPP1R16B_2 |
| PPFIA3 | PCDHA123_1 |
| PPIP5K2 | NGEF_2 |
| PPME1 | UBE2Q2P1_1 |
| PPP1CB | NAB2_3 |
| PPP1R8 | PPP1R16B_1 |
| PPP1R9A | CHRNA3_1 |
| PPP2CB | UBE2Q2P1_1 |
| PRDX5 | KCTD13_1 |
| PRKCSH | PBRM1_1 |
| PRPF39 | UBE2Q2P1_2 |
| PRR14 | KMT2E_3 |
| PRTG | PBRM1_3 |
| PSMC1 | NAB2_2 |
| PSMC3 | NGEF_2 |
| PTPN1 | pos_SNAP91 |
| PTRF | UBE2Q2P1_1, pos_SNAP91 |
| QSER1 | pos_SNAP91 |
| R3HDM1 | GALNT10_1 |
| RABGGTB | PCDHA123_1 |
| RABL6 | NAB2_2, DPYD_1 |
| RAPGEF1 | PBRM1_1 |
| RBAK-RBAKDN | TRANK1_1 |
| RCOR2 | DPYD_3 |
| RELA | PPP1R16B_2 |
| RGMA | PBRM1_2 |
| RIMKLB | UBE2Q2P1_3 |
| RITA1 | PBRM1_3 |
| RNF14 | TCF4-ITF2_3 |
| RNF141 | NGEF_2 |
| RNF168 | TRANK1_1 |
| RP11-14N7.2 | pos_SNAP91 |
| RP11-448A19.1 | GALNT10_3 |
| RP11-566K11.6 | PBRM1_1 |
| RP11-5C23.3 | miR137_1 |
| RP11-701H24.2 | NGEF_2 |
| RP13-104F24.2 | VPS45_1 |
| RP13-942N8.1 | LOC105376975_1 |
| RPL17 | LOC100507431_3 |
| RPS23 | LOC105376975_2 |
| RPS9 | LOC105376975_2 |
| RPUSD3 | DPYD_2 |
| RRAGD | LOC105376975_1 |
| RRBP1 | pos_SNAP91 |
| RRM2B | TRANK1_1, TCF4-ITF2_3 |
| RRP1 | BCL11B_1 |
| RRP7A | TRANK1_1 |
| RWDD2B | BCL11B_3 |
| SAC3D1 | TRANK1_1 |
| SDF2L1 | NGEF_2 |
| SEC31A | UBE2Q2P1_1 |
| SERPINH1 | LOC105376975_3 |
| SETD9 | VPS45_1 |
| SF3B2 | UBE2Q2P1_1 |
| SFR1 | miR137_3 |
| SHC1 | pos_SNAP91 |
| SIGIRR | miR137_2 |
| SIN3A | TCF4-ITF2_3 |
| SLC12A4 | UBE2Q2P1_1 |
| SLC19A1 | TRANK1_2 |
| SLC25A14 | SETD1A_1st |
| SLC2A3 | BCL11B_3 |
| SLC30A9 | KCTD13_2, PPP1R16B_1 |
| SLC38A7 | RERE_2 |
| SLC39A9 | pos_SNAP91 |
| SMAD4 | CHRNA3_1 |
| SMOC1 | pos_SNAP91 |
| SMURF2 | pos_SNAP91 |
| SMYD3 | NAB2_3 |
| SNHG11 | UBE2Q2P1_1 |
| SNU13 | TRANK1_1 |
| SPTY2D1 | UBE2Q2P1_1 |
| SSBP2 | CHRNA3_1 |
| STARD10 | CHRNA3_1 |
| STARD3NL | BCL11B_3 |
| STX3 | UBE2Q2P1_1 |
| SUPT6H | pos_SNAP91 |
| SUPT7L | TRANK1_1 |
| SUV39H1 | NGEF_2 |
| SVIL | pos_SNAP91 |
| SVIP | LOC105376975_1 |
| SYS1 | CHRNA3_1 |
| TACC2 | UBE2Q2P1_3 |
| TADA3 | DPYD_2 |
| TALDO1 | TCF4-ITF2_1 |
| TCEAL4 | BCL11B_3 |
| TERF1 | NAB2_3 |
| TEX264 | UBE2Q2P1_1 |
| TFPI2 | TRANK1_1 |
| THBS3 | RERE_3, UBE2Q2P1_1 |
| THYN1 | KCTD13_2 |
| TIPRL | PCDHA123_1 |
| TKFC | PBRM1_3 |
| TLDC1 | SETD1A_1st |
| TMED5 | PPP1R16B_2 |
| TMEM170B | CHRNA3_1 |
| TMEM194A | PPP1R16B_2 |
| TMEM223 | KMT2E_2 |
| TMEM230 | NAB2_2 |
| TMEM245 | LOC105376975_3 |
| TMEM259 | UBE2Q2P1_1 |
| TMEM41B | LOC105376975_3 |
| TMEM50B | NGEF_2 |
| TMTC4 | pos_SNAP91 |
| TNFRSF12A | UBE2Q2P1_1 |
| TNIK | TRANK1_1 |
| TOB1 | VPS45_1 |
| TPD52L2 | UBE2Q2P1_1 |
| TRIM69 | SETD1A_1st |
| TSSC4 | pos_SNAP91 |
| TUBA1B | BAG5_1 |
| TXNRD2 | TRANK1_1 |
| TYW5 | NGEF_2 |
| UBE2C | PBRM1_2 |
| UFL1 | BCL11B_1, NGEF_2 |
| UFM1 | UBE2Q2P1_1 |
| UNC119 | KMT2E_1 |
| UNC13B | PBRM1_3 |
| UQCR11 | BCL11B_1 |
| UROD | VPS45_2 |
| UTP14A | KMT2E_1 |
| VCPKMT | CHRNA3_1 |
| VDAC1 | BAG5_1 |
| VPS52 | UBE2Q2P1_2 |
| WASF2 | pos_SNAP91 |
| WASL | SETD1A_1st |
| WDR20 | UBE2Q2P1_1 |
| WDR45 | NAB2_2 |
| WFIKKN1 | PBRM1_1 |
| YDJC | NAB2_3 |
| YKT6 | UBE2Q2P1_1 |
| ZBTB40 | UBE2Q2P1_1 |
| ZBTB43 | BCL11B_3 |
| ZFAND2A | KCTD13_1 |
| ZFC3H1 | TCF4-ITF2_3 |
| ZMIZ2 | NGEF_2 |
| ZNF12 | UBE2Q2P1_1 |
| ZNF124 | CHRNA3_3, SETD1A_1st, STAT6_2 |
| ZNF138 | BCL11B_2 |
| ZNF146 | miR137_2 |
| ZNF263 | SETD1A_1st |
| ZNF264 | TRANK1_1 |
| ZNF35 | KCTD13_2 |
| ZNF480 | SETD1A_1st |
| ZNF573 | PBRM1_3 |
| ZNF581 | STAT6_2 |
| ZNF649 | PBRM1_2 |
| ZNF655 | TRANK1_1 |
| ZNF711 | NAB2_1 |
| ZNF738 | BCL11B_2 |
| ZNF789 | TRANK1_1 |
| ZYX | RERE_3 |
| num_cells | gRNA_target | signif_genes |
|---|---|---|
| 36 | BAG5_1 | VDAC1, TUBA1B |
| 33 | BAG5_2 | AUNIP, PHC2 |
| 31 | BAG5_3 | C7orf55 |
| 13 | BCL11B_1 | GSTM3, UFL1, NUP214, UQCR11, RRP1 |
| 33 | BCL11B_2 | MZT2B, ZNF138, ZNF738 |
| 13 | BCL11B_3 | GSTM4, LINC00467, FN1, NSUN2, GJA1, STARD3NL, ZBTB43, FBXO3, LAMTOR1, SLC2A3, RWDD2B, TCEAL4 |
| 3 | CHRNA3_1 | AFTPH, CMTM8, HSD17B11, SSBP2, HARS2, TMEM170B, CAP2, NEU1, PGM3, ANLN, PPP1R9A, ARHGAP21, STARD10, DYNC2H1, DDX6, CCDC184, VCPKMT, KLHL25, BAIAP2-AS1, SMAD4, CRTC1, SYS1 |
| 13 | CHRNA3_3 | ZNF124, CCNJL, CLNS1A, ENTPD6 |
| 37 | DPYD_1 | KAZN, MTIF2, RABL6 |
| 68 | DPYD_2 | TADA3, RPUSD3, MRPS18C, NAA35 |
| 28 | DPYD_3 | RCOR2 |
| 38 | GALNT10_1 | R3HDM1 |
| 37 | GALNT10_2 | NDUFS4 |
| 50 | GALNT10_3 | RP11-448A19.1, ABL1 |
| 30 | KCTD13_1 | ZFAND2A, NANS, PRDX5, HNRNPA1, EDA2R |
| 32 | KCTD13_2 | ZNF35, SLC30A9, THYN1, DLGAP5 |
| 15 | KCTD13_3 | AC104655.3, BUB1, PAK2 |
| 11 | KMT2E_1 | UNC119, LTBP4, UTP14A |
| 19 | KMT2E_2 | KCTD9, TMEM223, MOK, CPXM1 |
| 18 | KMT2E_3 | PRR14 |
| 45 | LINC00637_1 | CD24, BAZ2A, ORAI1, PHF23, ESCO1 |
| 53 | LINC00637_3 | IFT27 |
| 15 | LOC100507431_1 | CCNF |
| 29 | LOC100507431_2 | DCBLD2 |
| 49 | LOC100507431_3 | RPL17 |
| 35 | LOC105376975_1 | RRAGD, SVIP, RP13-942N8.1 |
| 50 | LOC105376975_2 | RPS23, HSP90AB1, FSD1L, RPS9 |
| 8 | LOC105376975_3 | PIAS3, CEP350, PCGF1, ACSL3, ARPC1B, TMEM245, OBFC1, TMEM41B, C11orf83, SERPINH1, EPN2, HKR1 |
| 44 | miR137_1 | RP11-5C23.3, ARHGEF17 |
| 30 | miR137_2 | SIGIRR, ZNF146, BACE2 |
| 54 | miR137_3 | ACTR2, COPS7B, SFR1 |
| 36 | NAB2_1 | METTL2B, ZNF711 |
| 24 | NAB2_2 | IK, RABL6, ALG5, PSMC1, INTS14, MORF4L1, TMEM230, PFDN4, WDR45 |
| 25 | NAB2_3 | GNB1, SMYD3, PPP1CB, CLDND1, CAV1, TERF1, HAUS8, POLR3F, YDJC |
| 11 | NGEF_1 | CLPTM1L, PARP16 |
| 2 | NGEF_2 | CNRIP1, TYW5, CD200, DNAJB11, PPIP5K2, UFL1, PLEKHG1, ELMO1, ZMIZ2, GATS, GBA2, RNF141, PSMC3, AAAS, DTX3, RP11-701H24.2, NTN1, NAPG, PDCD2L, TMEM50B, SDF2L1, SUV39H1 |
| 4 | PBRM1_1 | CLOCK, RAPGEF1, LETMD1, WFIKKN1, RP11-566K11.6, PRKCSH |
| 12 | PBRM1_2 | BUB3, RGMA, AURKB, PHF12, ZNF649, UBE2C |
| 4 | PBRM1_3 | EXTL3, ARMC1, UNC13B, ADO, MORN4, TKFC, RITA1, B3GLCT, PRTG, ELP5, ZNF573, FANCB |
| 53 | PCDHA123_1 | RABGGTB, TIPRL, C9orf40, MGAT2, PPFIA3, HIC2 |
| 64 | PCDHA123_2 | ITFG2 |
| 62 | PCDHA123_3 | MOGS, MFSD13A, NEK9 |
| 3 | pos_SNAP91 | WASF2, MACF1, RP11-14N7.2, SHC1, FAM20B, MAPKAPK2, AC068491.1, CCDC80, CPSF4, KLHDC10, MYC, NCBP1, ACBD5, SVIL, TSSC4, QSER1, NEAT1, TMTC4, SLC39A9, SMOC1, NUMB, DPP8, FBRS, MT1X, EIF4A1, MAP2K3, SUPT6H, CASC3, PTRF, SMURF2, RRBP1, ASXL1, CEP250, PTPN1, CRYBB2, ATP11C |
| 44 | PPP1R16B_1 | PPP1R8, SLC30A9 |
| 31 | PPP1R16B_2 | TMED5, POR, RELA, TMEM194A |
| 35 | PPP1R16B_3 | CDKN2A |
| 48 | RERE_1 | IFT122, CXXC1 |
| 46 | RERE_2 | ACBD5, SLC38A7, DSEL |
| 25 | RERE_3 | THBS3, ZYX, DNAJB1 |
| 23 | SETD1A_1st | KDM5B, ZNF124, BAZ2B, WASL, TRIM69, ZNF263, TLDC1, CDK12, DLL3, ZNF480, MIR99AHG, HIRA, SLC25A14 |
| 24 | STAT6_1 | FAM134C |
| 38 | STAT6_2 | ZNF124, GNAI1, PCYT2, ZNF581 |
| 53 | STAT6_3 | KRTCAP2 |
| 50 | TCF4-ITF2_1 | NMU, HDAC2, NKAIN3, DCAF10, TALDO1, CENPV, CABIN1, CENPM |
| 45 | TCF4-ITF2_2 | COQ5, CPD, C1GALT1C1 |
| 12 | TCF4-ITF2_3 | RNF14, FAM46A, CRCP, RRM2B, ZFC3H1, SIN3A, C17orf75, MED29 |
| 3 | TRANK1_1 | SUPT7L, CDC25A, TNIK, RNF168, RBAK-RBAKDN, CHCHD2, TFPI2, ZNF789, ZNF655, EXTL3, RRM2B, DSCC1, ATAD2, NINJ1, ARID5B, BLOC1S2, SAC3D1, ZNF264, TXNRD2, SNU13, RRP7A, BEX1, EMD, DKC1 |
| 19 | TRANK1_2 | KLHL5, ORAI1, NSFL1C, SLC19A1 |
| 65 | TRANK1_3 | PNKP |
| 11 | UBE2Q2P1_1 | EPHA2, ZBTB40, LRIF1, THBS3, LMNA, DNAJB2, ARL8B, IP6K1, TEX264, ARF4, SEC31A, ZNF12, CYCS, YKT6, PPP2CB, GOLGA7, CREB3, MAPKAP1, BNIP3, SPTY2D1, STX3, FOSL1, C11orf68, SF3B2, NDUFS8, PAAF1, PPME1, LDHB, UFM1, KHNYN, AHSA1, WDR20, TNFRSF12A, ENKD1, SLC12A4, DUSP14, PTRF, DUSP3, CDC34, ARID3A, TMEM259, CTB-31O20.2, AKAP8, SNHG11, TPD52L2, MAFF, CASK |
| 8 | UBE2Q2P1_2 | LINC00467, ATP5G3, VPS52, FGFR1, PLXDC2, DHRS12, PRPF39 |
| 11 | UBE2Q2P1_3 | BOK, NEU1, MTERF1, TACC2, RIMKLB, MDM1 |
| 14 | VPS45_1 | SETD9, TOB1, RP13-104F24.2, CDC25B |
| 79 | VPS45_2 | UROD, HAT1, LSAMP, ATP5G1, GPS1, CRELD2 |
| 9 | VPS45_3 | MICU2, PJA1 |
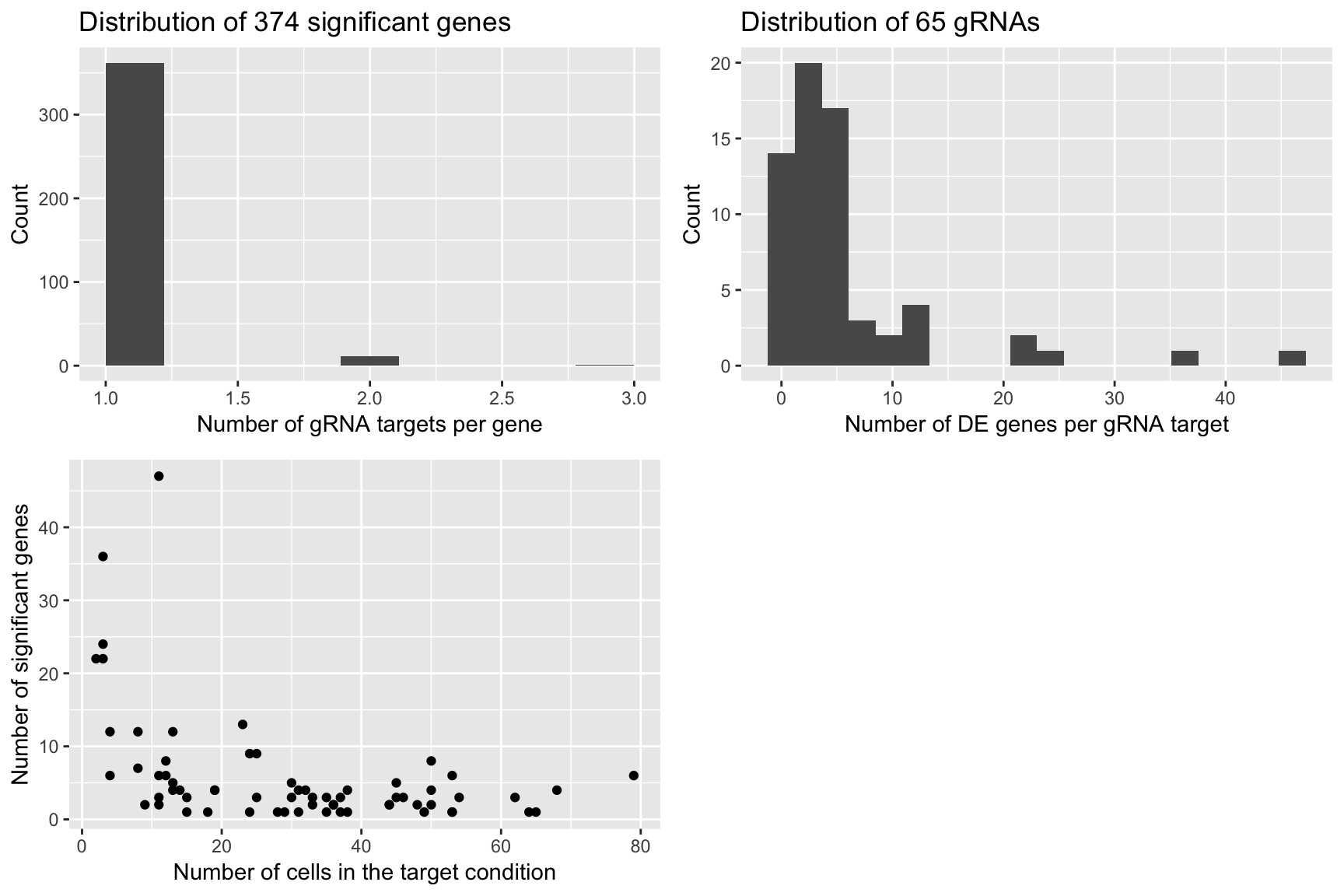
Permutation 3
load(paste0(wkdir,'data/perm.ashr_estimation03.Rdata'))
get.result(betaest.mtx,lfsr.mtx)| signif_gene | gRNA_targets |
|---|---|
| ABCF1 | UBE2Q2P1_2 |
| ACAP2 | CHRNA3_1 |
| ACVR1B | KMT2E_2 |
| ADAT1 | PBRM1_3 |
| ADD2 | UBE2Q2P1_2 |
| ADH5 | UBE2Q2P1_1 |
| ADPRM | PCDHA123_2 |
| AGGF1 | LOC105376975_3, SETD1A_1st |
| AIP | NAB2_2 |
| AKAP12 | CHRNA3_1 |
| ANKRD11 | KMT2E_1 |
| ANTXR1 | PBRM1_3 |
| APOM | NGEF_2 |
| ARL4A | TRANK1_1 |
| ASH1L | miR137_1 |
| ASPM | LOC105376975_1 |
| ASTN2 | BCL11B_2 |
| ATAD2B | pos_SNAP91 |
| ATP5G1 | BAG5_1 |
| AXL | PBRM1_2 |
| BEX2 | CHRNA3_1 |
| BLOC1S6 | LINC00637_3 |
| BMPR1A | NAB2_2 |
| BOC | LOC105376975_1 |
| BOD1L1 | PBRM1_3 |
| BTG3 | miR137_3 |
| BUB3 | LOC105376975_1 |
| C12orf76 | VPS45_1 |
| C16orf72 | KMT2E_3 |
| C2orf76 | NAB2_2 |
| C4orf27 | miR137_3 |
| C5orf45 | LOC105376975_3 |
| C9orf3 | NGEF_2 |
| C9orf9 | pos_SNAP91 |
| CARM1 | KCTD13_2 |
| CARS | TCF4-ITF2_3 |
| CASD1 | NGEF_2 |
| CBL | KMT2E_2 |
| CBX2 | SETD1A_1st |
| CBX6 | PBRM1_2 |
| CCDC115 | STAT6_3 |
| CCDC157 | BCL11B_1 |
| CDK5RAP3 | NAB2_2 |
| CDKN1C | PBRM1_3 |
| CELF2 | PCDHA123_1, PBRM1_2 |
| CENPH | NAB2_3 |
| CENPW | NGEF_2 |
| CEP70 | pos_SNAP91 |
| CHST10 | PCDHA123_1 |
| CLPX | VPS45_1 |
| COA1 | BAG5_1 |
| COG3 | BAG5_1, UBE2Q2P1_1 |
| COMMD3 | KMT2E_1 |
| COPG2 | PPP1R16B_3 |
| CREBL2 | VPS45_3 |
| CSRNP3 | KMT2E_2 |
| CTB-175P5.4 | TRANK1_3 |
| CTD-2037K23.2 | TRANK1_1 |
| CYB5D2 | PBRM1_3 |
| DDX10 | LOC100507431_3 |
| DEPDC1 | BAG5_3, LOC100507431_2 |
| DIAPH3 | pos_SNAP91 |
| DIP2C | KMT2E_1 |
| DLG3 | pos_SNAP91 |
| DLL1 | PBRM1_3 |
| DNAJB9 | DPYD_1 |
| DNAJC18 | UBE2Q2P1_1 |
| DPY19L3 | SETD1A_1st |
| DRAM1 | TRANK1_1 |
| DUSP12 | KCTD13_3 |
| DYRK2 | RERE_1 |
| EEF1AKMT1 | RERE_2 |
| EEF1B2 | SETD1A_1st |
| EFR3B | STAT6_1 |
| EGLN2 | PBRM1_1 |
| EI24 | BAG5_1 |
| EIF2D | NGEF_2 |
| EIF4H | NAB2_3 |
| EPB41L4A-AS1 | UBE2Q2P1_1 |
| EPHA4 | BCL11B_3 |
| ERCC8 | DPYD_2 |
| ETAA1 | STAT6_3 |
| EXOSC2 | LOC105376975_3 |
| EYA3 | NAB2_2, LINC00637_3 |
| FABP5 | BAG5_1 |
| FAM107B | KMT2E_2 |
| FAM122A | TRANK1_3 |
| FAM136A | BAG5_1 |
| FAM195A | KMT2E_3 |
| FAM234A | PBRM1_1 |
| FAM64A | PPP1R16B_1 |
| FBRSL1 | BCL11B_2 |
| FGD4 | UBE2Q2P1_1 |
| FKBP5 | LOC105376975_3 |
| FMNL2 | SETD1A_1st |
| FOCAD | TRANK1_1 |
| FRG1 | miR137_3 |
| FRYL | RERE_1, DPYD_3 |
| FRZB | SETD1A_1st |
| FUNDC1 | UBE2Q2P1_2 |
| FYN | RERE_1 |
| GADD45A | VPS45_3 |
| GALE | TRANK1_3 |
| GALT | STAT6_3 |
| GAS5 | VPS45_1 |
| GCLM | CHRNA3_3 |
| GIN1 | KMT2E_1 |
| GJC1 | LOC105376975_3 |
| GPC6 | SETD1A_1st |
| GSTCD | PBRM1_1 |
| GTSE1 | LOC105376975_1 |
| HARS2 | KMT2E_1 |
| HAUS8 | LOC105376975_3, SETD1A_1st |
| HGSNAT | LOC105376975_3 |
| ID2 | NGEF_2 |
| IGFBP4 | GALNT10_3 |
| ILK | CHRNA3_2 |
| INPP5E | SETD1A_1st |
| IQSEC1 | PBRM1_1 |
| IRS2 | CHRNA3_1 |
| IVD | pos_SNAP91 |
| KATNAL1 | PBRM1_1 |
| KHNYN | KCTD13_1 |
| KMT2B | PCDHA123_1 |
| LAS1L | miR137_2 |
| LDB1 | SETD1A_1st |
| LRP1 | BCL11B_1, PBRM1_3 |
| LRP2 | NAB2_3 |
| LRRC75A | BAG5_2 |
| LRRC75A-AS1 | VPS45_1 |
| LTN1 | TRANK1_1 |
| MAFF | CHRNA3_1 |
| MAP4K4 | BCL11B_3 |
| MCM6 | UBE2Q2P1_3 |
| MCOLN1 | NGEF_2 |
| MDM2 | CHRNA3_1 |
| METTL15 | TRANK1_1 |
| MFAP1 | PBRM1_1 |
| MGAT2 | TRANK1_2 |
| MIB2 | TRANK1_1 |
| MMP15 | DPYD_3 |
| MON1B | SETD1A_1st |
| MPHOSPH6 | TRANK1_1 |
| MRPL28 | miR137_2 |
| MTFP1 | PBRM1_1 |
| MTR | NGEF_2 |
| MYCBP2 | NAB2_3 |
| NCAM1 | KMT2E_2 |
| NCAPD2 | BAG5_3 |
| NCAPG | NAB2_3 |
| NCK1 | NAB2_2 |
| NCKAP5 | TRANK1_1 |
| NCOR2 | KMT2E_3 |
| NECTIN2 | miR137_2 |
| NFRKB | KMT2E_1 |
| NOL12 | GALNT10_2 |
| NPPC | TRANK1_1 |
| NR2F1 | NGEF_2 |
| NRG1 | PBRM1_1 |
| NSMCE4A | miR137_3 |
| NT5DC2 | BAG5_1 |
| NUDCD1 | PCDHA123_3 |
| NUP160 | SETD1A_1st |
| NUP188 | UBE2Q2P1_3 |
| OFD1 | BCL11B_3 |
| PACRGL | CHRNA3_1 |
| PAF1 | LINC00637_3, PBRM1_3 |
| PALMD | PBRM1_1 |
| PAX6 | NAB2_2 |
| PCGF6 | RERE_1, LOC100507431_1 |
| PDHX | miR137_2 |
| PDIA5 | GALNT10_2 |
| PDZD8 | CHRNA3_1 |
| PEX13 | UBE2Q2P1_2 |
| PHYH | PPP1R16B_3 |
| PIN4 | UBE2Q2P1_2 |
| PITPNA | KCTD13_2 |
| PJA1 | pos_SNAP91 |
| PKD1 | pos_SNAP91 |
| PKIB | TRANK1_2 |
| PNN | CHRNA3_1, miR137_3 |
| PPP1R12A | CHRNA3_1 |
| PPP1R7 | TCF4-ITF2_3 |
| PPRC1 | UBE2Q2P1_2 |
| PRKACB | KMT2E_3 |
| PROX1 | PBRM1_2 |
| PRTFDC1 | NGEF_2 |
| PSME4 | LOC100507431_3 |
| PTP4A2 | CHRNA3_1 |
| PTPN9 | KCTD13_3 |
| PYGO2 | PCDHA123_2 |
| RAB33A | TRANK1_1 |
| RABEP2 | BAG5_2 |
| RAP2B | UBE2Q2P1_2 |
| RASSF4 | KMT2E_3, PBRM1_3 |
| RBM26 | PCDHA123_1 |
| RBM4 | SETD1A_1st |
| RBM45 | pos_SNAP91 |
| REV1 | BCL11B_3 |
| RFLNB | KMT2E_1 |
| RIC8B | BCL11B_2, NAB2_2 |
| RNASEH2A | LOC100507431_3 |
| RNASEH2C | LINC00637_3 |
| RNF165 | UBE2Q2P1_1 |
| RNF169 | UBE2Q2P1_2 |
| RNF44 | BCL11B_2 |
| RNFT2 | LOC105376975_3 |
| RP11-12G12.7 | BCL11B_1 |
| RP11-15H20.6 | NGEF_2 |
| RP11-182L21.6 | UBE2Q2P1_3 |
| RP11-186B7.4 | VPS45_1 |
| RP11-448A19.1 | UBE2Q2P1_3 |
| RP11-477B16.5 | PBRM1_1 |
| RP3-395M20.12 | NGEF_2 |
| RQCD1 | SETD1A_1st |
| RRP1 | LOC100507431_3 |
| SCAF8 | NAB2_2 |
| SCAP | LINC00637_2 |
| SCARA3 | SETD1A_1st |
| SDHAF3 | BAG5_1 |
| SEC24C | KMT2E_3 |
| SEC61A2 | BAG5_1 |
| SEC62 | miR137_2 |
| SEMA3A | KCTD13_1 |
| SEPN1 | TRANK1_1 |
| SERPING1 | pos_SNAP91 |
| SESN2 | LOC100507431_1 |
| SETD2 | TCF4-ITF2_3 |
| SFXN1 | PBRM1_1 |
| SFXN2 | TRANK1_2 |
| SGSM2 | LOC105376975_2 |
| SHMT1 | LOC105376975_3 |
| SKA3 | SETD1A_1st |
| SLC25A19 | SETD1A_1st |
| SLC29A4 | pos_SNAP91 |
| SLC30A1 | TRANK1_1 |
| SLC39A6 | DPYD_3 |
| SLC41A3 | STAT6_3 |
| SMAD2 | LINC00637_1 |
| SMARCB1 | BCL11B_1 |
| SNAP23 | RERE_2 |
| SNHG8 | KCTD13_1 |
| SNRNP27 | LOC100507431_3 |
| SNX9 | CHRNA3_1 |
| SOCS7 | TCF4-ITF2_3 |
| SOX3 | LINC00637_3 |
| SOX4 | PBRM1_3 |
| SPG21 | PCDHA123_1 |
| SPP1 | CHRNA3_1 |
| SPTAN1 | CHRNA3_1 |
| SRP68 | miR137_3 |
| SRPRA | PPP1R16B_2 |
| SSFA2 | KMT2E_2 |
| STAM | PBRM1_3, pos_SNAP91 |
| STK38 | PBRM1_3, PPP1R16B_3 |
| STK4 | UBE2Q2P1_3 |
| STRADA | pos_SNAP91 |
| STX12 | miR137_3 |
| STX6 | VPS45_1 |
| SUGP2 | PBRM1_3 |
| SYT17 | NAB2_2 |
| TAF1 | SETD1A_1st, pos_SNAP91 |
| TBCC | BCL11B_2 |
| TCAF1 | UBE2Q2P1_2 |
| TCEAL4 | RERE_2 |
| TCF7L1 | BAG5_3 |
| TEAD2 | LINC00637_3 |
| TFPT | LINC00637_1 |
| TGDS | NAB2_3 |
| TMEM18 | TCF4-ITF2_1 |
| TMEM199 | TCF4-ITF2_3 |
| TRDMT1 | NAB2_1 |
| TRIM11 | KCTD13_3 |
| TRIM24 | STAT6_3 |
| TRIP12 | PCDHA123_1 |
| TRMT10C | CHRNA3_1 |
| TSPAN18 | GALNT10_2 |
| TTC1 | KCTD13_3 |
| UBLCP1 | RERE_3 |
| UBN2 | TCF4-ITF2_1 |
| UTP11 | miR137_2 |
| VPS33B | TRANK1_1 |
| VSIG10 | pos_SNAP91 |
| WDR12 | TCF4-ITF2_3 |
| WWC2 | VPS45_1 |
| WWP2 | SETD1A_1st |
| XBP1 | LOC100507431_1 |
| XPR1 | UBE2Q2P1_2 |
| YY1 | miR137_2 |
| ZBTB40 | NGEF_2 |
| ZBTB45 | LOC105376975_3, SETD1A_1st |
| ZBTB5 | NGEF_2 |
| ZDHHC2 | BAG5_1 |
| ZFPL1 | PCDHA123_2 |
| ZNF136 | UBE2Q2P1_1 |
| ZNF174 | PBRM1_3 |
| ZNF205 | miR137_3 |
| ZNF444 | miR137_1 |
| ZNF516 | BAG5_2 |
| ZNF567 | TRANK1_1 |
| ZNF75A | SETD1A_1st |
| ZNF84 | TRANK1_1 |
| num_cells | gRNA_target | signif_genes |
|---|---|---|
| 36 | BAG5_1 | FAM136A, NT5DC2, COA1, SDHAF3, ZDHHC2, FABP5, SEC61A2, EI24, COG3, ATP5G1 |
| 33 | BAG5_2 | RABEP2, LRRC75A, ZNF516 |
| 31 | BAG5_3 | DEPDC1, TCF7L1, NCAPD2 |
| 13 | BCL11B_1 | LRP1, RP11-12G12.7, SMARCB1, CCDC157 |
| 33 | BCL11B_2 | RNF44, TBCC, ASTN2, RIC8B, FBRSL1 |
| 13 | BCL11B_3 | REV1, MAP4K4, EPHA4, OFD1 |
| 3 | CHRNA3_1 | PTP4A2, TRMT10C, ACAP2, PACRGL, SPP1, AKAP12, SNX9, SPTAN1, PDZD8, MDM2, PPP1R12A, IRS2, PNN, MAFF, BEX2 |
| 23 | CHRNA3_2 | ILK |
| 13 | CHRNA3_3 | GCLM |
| 37 | DPYD_1 | DNAJB9 |
| 68 | DPYD_2 | ERCC8 |
| 28 | DPYD_3 | FRYL, MMP15, SLC39A6 |
| 37 | GALNT10_2 | PDIA5, TSPAN18, NOL12 |
| 50 | GALNT10_3 | IGFBP4 |
| 30 | KCTD13_1 | SNHG8, SEMA3A, KHNYN |
| 32 | KCTD13_2 | PITPNA, CARM1 |
| 15 | KCTD13_3 | DUSP12, TRIM11, TTC1, PTPN9 |
| 11 | KMT2E_1 | GIN1, HARS2, DIP2C, COMMD3, NFRKB, ANKRD11, RFLNB |
| 19 | KMT2E_2 | CSRNP3, SSFA2, FAM107B, NCAM1, CBL, ACVR1B |
| 18 | KMT2E_3 | PRKACB, RASSF4, SEC24C, NCOR2, FAM195A, C16orf72 |
| 45 | LINC00637_1 | SMAD2, TFPT |
| 34 | LINC00637_2 | SCAP |
| 53 | LINC00637_3 | EYA3, RNASEH2C, BLOC1S6, PAF1, TEAD2, SOX3 |
| 15 | LOC100507431_1 | SESN2, PCGF6, XBP1 |
| 29 | LOC100507431_2 | DEPDC1 |
| 49 | LOC100507431_3 | PSME4, SNRNP27, DDX10, RNASEH2A, RRP1 |
| 35 | LOC105376975_1 | ASPM, BOC, BUB3, GTSE1 |
| 50 | LOC105376975_2 | SGSM2 |
| 8 | LOC105376975_3 | AGGF1, C5orf45, FKBP5, HGSNAT, EXOSC2, RNFT2, SHMT1, GJC1, HAUS8, ZBTB45 |
| 44 | miR137_1 | ASH1L, ZNF444 |
| 30 | miR137_2 | UTP11, SEC62, PDHX, YY1, MRPL28, NECTIN2, LAS1L |
| 54 | miR137_3 | STX12, C4orf27, FRG1, NSMCE4A, PNN, ZNF205, SRP68, BTG3 |
| 36 | NAB2_1 | TRDMT1 |
| 24 | NAB2_2 | EYA3, C2orf76, NCK1, SCAF8, BMPR1A, PAX6, AIP, RIC8B, SYT17, CDK5RAP3 |
| 25 | NAB2_3 | LRP2, NCAPG, CENPH, EIF4H, MYCBP2, TGDS |
| 2 | NGEF_2 | RP3-395M20.12, ZBTB40, EIF2D, MTR, ID2, NR2F1, APOM, CENPW, CASD1, ZBTB5, C9orf3, PRTFDC1, MCOLN1, RP11-15H20.6 |
| 4 | PBRM1_1 | PALMD, IQSEC1, GSTCD, SFXN1, NRG1, KATNAL1, RP11-477B16.5, MFAP1, FAM234A, EGLN2, MTFP1 |
| 12 | PBRM1_2 | PROX1, CELF2, AXL, CBX6 |
| 4 | PBRM1_3 | ANTXR1, BOD1L1, SOX4, STK38, DLL1, STAM, RASSF4, CDKN1C, LRP1, ZNF174, ADAT1, CYB5D2, SUGP2, PAF1 |
| 53 | PCDHA123_1 | CHST10, TRIP12, CELF2, RBM26, SPG21, KMT2B |
| 64 | PCDHA123_2 | PYGO2, ZFPL1, ADPRM |
| 62 | PCDHA123_3 | NUDCD1 |
| 3 | pos_SNAP91 | ATAD2B, RBM45, CEP70, SLC29A4, C9orf9, STAM, SERPING1, VSIG10, DIAPH3, IVD, PKD1, STRADA, PJA1, DLG3, TAF1 |
| 44 | PPP1R16B_1 | FAM64A |
| 31 | PPP1R16B_2 | SRPRA |
| 35 | PPP1R16B_3 | STK38, COPG2, PHYH |
| 48 | RERE_1 | FRYL, FYN, PCGF6, DYRK2 |
| 46 | RERE_2 | EEF1AKMT1, SNAP23, TCEAL4 |
| 25 | RERE_3 | UBLCP1 |
| 23 | SETD1A_1st | FMNL2, FRZB, EEF1B2, RQCD1, AGGF1, SCARA3, INPP5E, LDB1, NUP160, RBM4, SKA3, GPC6, ZNF75A, WWP2, MON1B, SLC25A19, CBX2, HAUS8, DPY19L3, ZBTB45, TAF1 |
| 24 | STAT6_1 | EFR3B |
| 53 | STAT6_3 | ETAA1, CCDC115, SLC41A3, TRIM24, GALT |
| 50 | TCF4-ITF2_1 | TMEM18, UBN2 |
| 12 | TCF4-ITF2_3 | WDR12, PPP1R7, SETD2, CARS, TMEM199, SOCS7 |
| 3 | TRANK1_1 | MIB2, SEPN1, SLC30A1, NCKAP5, NPPC, CTD-2037K23.2, ARL4A, FOCAD, METTL15, DRAM1, ZNF84, VPS33B, MPHOSPH6, ZNF567, LTN1, RAB33A |
| 19 | TRANK1_2 | PKIB, SFXN2, MGAT2 |
| 65 | TRANK1_3 | GALE, FAM122A, CTB-175P5.4 |
| 11 | UBE2Q2P1_1 | ADH5, EPB41L4A-AS1, DNAJC18, FGD4, COG3, RNF165, ZNF136 |
| 8 | UBE2Q2P1_2 | XPR1, PEX13, ADD2, RAP2B, ABCF1, TCAF1, PPRC1, RNF169, FUNDC1, PIN4 |
| 11 | UBE2Q2P1_3 | MCM6, RP11-448A19.1, NUP188, RP11-182L21.6, STK4 |
| 14 | VPS45_1 | GAS5, STX6, WWC2, C12orf76, CLPX, RP11-186B7.4, LRRC75A-AS1 |
| 9 | VPS45_3 | GADD45A, CREBL2 |
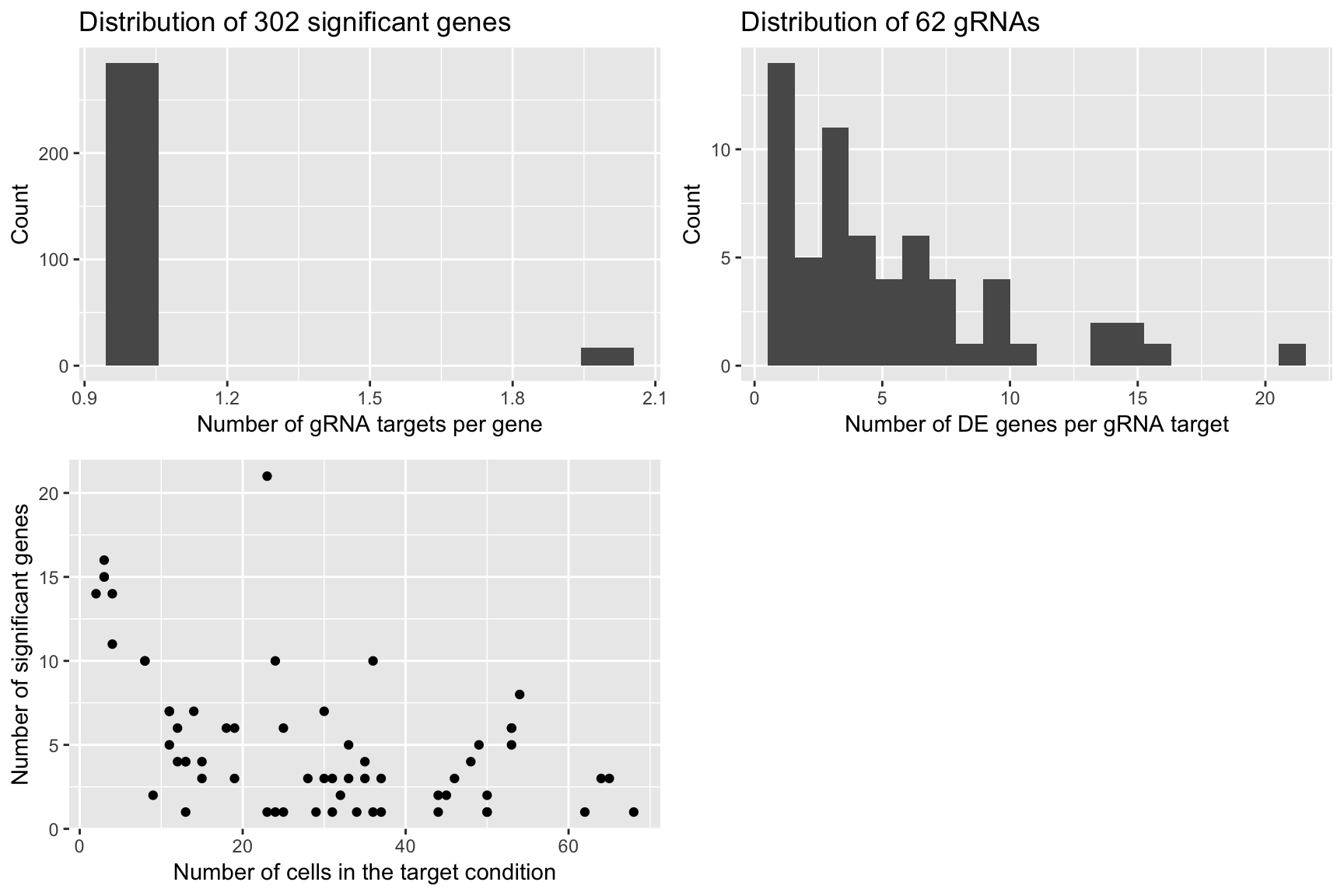
Permutation 4
load(paste0(wkdir,'data/perm.ashr_estimation04.Rdata'))
get.result(betaest.mtx,lfsr.mtx)| signif_gene | gRNA_targets |
|---|---|
| AAMP | PPP1R16B_1 |
| ACAP3 | NGEF_2 |
| ACSL4 | NGEF_2 |
| ACSS2 | KMT2E_3, TRANK1_1 |
| AGBL5 | VPS45_2 |
| AHCYL1 | NAB2_3 |
| AJUBA | LOC105376975_2 |
| AKAP9 | CHRNA3_3 |
| AMDHD2 | UBE2Q2P1_3 |
| ANKMY2 | UBE2Q2P1_1 |
| ANLN | SETD1A_1st |
| APBB2 | NGEF_2 |
| ARHGAP32 | KCTD13_3 |
| ARHGAP5 | LOC105376975_3 |
| ARNT2 | KCTD13_2 |
| ASPM | SETD1A_1st |
| ASRGL1 | CHRNA3_1 |
| ATG5 | PPP1R16B_1 |
| ATOH8 | NGEF_2 |
| ATXN10 | PBRM1_3 |
| ATXN3 | NGEF_2 |
| AURKA | SETD1A_1st |
| B4GALNT4 | NGEF_2 |
| B4GALT4 | VPS45_1 |
| BASP1 | NGEF_2 |
| BICD2 | CHRNA3_1 |
| BORCS6 | NGEF_2 |
| BRAP | PBRM1_1 |
| BTBD1 | KCTD13_3 |
| BUB3 | SETD1A_1st, VPS45_1 |
| C14orf1 | TRANK1_1 |
| C19orf47 | KMT2E_1 |
| C21orf62 | TRANK1_2 |
| C2orf49 | CHRNA3_1 |
| C9orf142 | DPYD_3 |
| CAMK1D | PBRM1_1 |
| CASC5 | SETD1A_1st |
| CASP7 | LOC105376975_3 |
| CBFA2T2 | NGEF_2 |
| CBR1 | VPS45_3 |
| CCDC138 | VPS45_2 |
| CCDC140 | CHRNA3_2 |
| CCDC174 | KMT2E_3, UBE2Q2P1_1 |
| CCDC189 | CHRNA3_1 |
| CCDC91 | STAT6_1 |
| CCNC | RERE_3 |
| CCNI | NGEF_2 |
| CCNY | KMT2E_1 |
| CDC37 | LOC105376975_2 |
| CDCA3 | SETD1A_1st, pos_SNAP91 |
| CDCA8 | SETD1A_1st |
| CDYL | RERE_2 |
| CENPB | KMT2E_2 |
| CENPE | SETD1A_1st |
| CENPF | SETD1A_1st |
| CEP41 | NAB2_1, pos_SNAP91 |
| CFL2 | PCDHA123_2 |
| CHCHD7 | LOC105376975_2 |
| CHPT1 | NGEF_1 |
| CHST12 | UBE2Q2P1_2 |
| CLCN3 | KCTD13_1 |
| CLGN | GALNT10_1 |
| CLINT1 | BAG5_3 |
| CMTM8 | UBE2Q2P1_1 |
| CNN1 | KCTD13_1 |
| COG8 | NGEF_2 |
| CORO1B | NGEF_2 |
| CTD-2001E22.2 | UBE2Q2P1_1 |
| CTD-2568A17.3 | PBRM1_3 |
| CWF19L1 | PBRM1_3 |
| DCAF10 | PPP1R16B_2 |
| DCLK1 | NGEF_2 |
| DCTN5 | NGEF_2 |
| DCX | NGEF_2 |
| DDIT4 | UBE2Q2P1_2 |
| DDT | KCTD13_1, LINC00637_1 |
| DFFA | pos_SNAP91 |
| DFNA5 | pos_SNAP91 |
| DHX16 | LOC105376975_3 |
| DHX40 | miR137_3 |
| DIS3L2 | LOC105376975_3, NAB2_3 |
| DLL3 | NGEF_2 |
| DMTF1 | NGEF_2 |
| DNAAF5 | LINC00637_2 |
| DNAJC18 | NGEF_1 |
| DOLK | NGEF_2 |
| DPY19L3 | pos_SNAP91 |
| DTWD1 | TCF4-ITF2_2 |
| DUS3L | pos_SNAP91 |
| DYNC1H1 | NGEF_2 |
| DYRK1B | NGEF_2 |
| EED | PBRM1_3 |
| EFCAB14 | LOC105376975_3 |
| EFNB3 | BCL11B_1 |
| ELAVL2 | NGEF_2 |
| EMC2 | RERE_1 |
| ENO2 | NGEF_2 |
| EPB41L2 | PBRM1_3 |
| EPHA4 | NGEF_2 |
| EPS8L1 | NGEF_2 |
| ERICH1 | STAT6_1 |
| ERP44 | SETD1A_1st |
| ETV4 | LINC00637_2 |
| EXD3 | BCL11B_3, NGEF_2 |
| EXOC2 | NAB2_3 |
| FAM127A | NGEF_2 |
| FAM131A | RERE_2 |
| FAM174A | GALNT10_1 |
| FAM58A | STAT6_1 |
| FAM83D | SETD1A_1st |
| FASN | TRANK1_1 |
| FASTKD5 | NGEF_2 |
| FBXO16 | LOC105376975_1 |
| FNBP1L | NGEF_2 |
| FST | pos_SNAP91 |
| FTX | NGEF_2 |
| FUCA2 | DPYD_2 |
| FZD9 | GALNT10_3 |
| GAP43 | NGEF_2 |
| GATB | STAT6_2 |
| GDAP1L1 | NGEF_2 |
| GDI1 | NGEF_2 |
| GFRA1 | KCTD13_2 |
| GGA2 | NGEF_2 |
| GGT7 | NGEF_2 |
| GJC1 | KMT2E_2 |
| GLRX | NGEF_2 |
| GMFB | NGEF_1 |
| GNG3 | NGEF_2 |
| GON4L | KMT2E_3 |
| GORAB | LOC105376975_3 |
| GPHN | CHRNA3_2 |
| GPKOW | NAB2_1 |
| GTSE1 | SETD1A_1st |
| HDHD2 | TRANK1_2 |
| HES1 | CHRNA3_1 |
| HNRNPF | GALNT10_2 |
| IDH3A | PBRM1_3 |
| IL13RA2 | UBE2Q2P1_2 |
| IMPDH1 | CHRNA3_3 |
| IP6K2 | NGEF_2 |
| IQSEC1 | NGEF_2 |
| IRGQ | NGEF_2 |
| JUNB | NGEF_2 |
| KDM5B | NGEF_2 |
| KDM6B | NGEF_2 |
| KIAA0895L | NGEF_2 |
| KIAA1715 | NGEF_2 |
| KIF11 | DPYD_3 |
| KIF1BP | STAT6_1 |
| KIF20A | SETD1A_1st |
| KIF2C | SETD1A_1st |
| KLHL8 | PBRM1_1 |
| KPNA2 | SETD1A_1st |
| LARP7 | LOC100507431_1 |
| LGR4 | GALNT10_3, NGEF_1 |
| LMBR1L | NGEF_2 |
| LPIN2 | BCL11B_1 |
| LRP1 | CHRNA3_1 |
| LRRC57 | UBE2Q2P1_3 |
| LRRC8D | TRANK1_1 |
| MAP3K4 | PBRM1_1 |
| MAP6 | NGEF_2 |
| MAPRE3 | NGEF_2 |
| MBD1 | LOC100507431_3 |
| MCM5 | pos_SNAP91 |
| MECP2 | BCL11B_1 |
| MED9 | VPS45_1, pos_SNAP91 |
| MEG3 | NGEF_2 |
| METAP1D | GALNT10_2 |
| MICAL1 | KMT2E_1 |
| MKI67 | SETD1A_1st |
| MKNK1 | UBE2Q2P1_3 |
| MLYCD | BCL11B_1 |
| MNAT1 | PBRM1_1 |
| MRPL19 | pos_SNAP91 |
| MRPS21 | PPP1R16B_3 |
| MT1X | miR137_2 |
| MTCH2 | GALNT10_2 |
| MTHFD2 | DPYD_1 |
| MXI1 | NGEF_2 |
| MYO6 | STAT6_3 |
| NAA38 | UBE2Q2P1_3 |
| NAF1 | KMT2E_3 |
| NAV2 | CHRNA3_2 |
| NBAS | pos_SNAP91 |
| NCAM1 | NGEF_2 |
| NCK1 | BCL11B_2 |
| NCOA6 | UBE2Q2P1_3 |
| NDUFAF6 | PBRM1_3 |
| NEFM | NGEF_2 |
| NELFB | NAB2_2 |
| NGRN | miR137_1 |
| NHLRC3 | NGEF_2 |
| NHSL2 | VPS45_3 |
| NIT1 | PPP1R16B_1 |
| NME6 | RERE_1 |
| NMNAT1 | PPP1R16B_2 |
| NONO | RERE_2 |
| NR6A1 | PBRM1_1 |
| NREP | NGEF_2 |
| NSG1 | NGEF_2 |
| NUDT11 | PPP1R16B_3 |
| NUDT22 | GALNT10_3 |
| NUP133 | TCF4-ITF2_3 |
| NUP214 | KCTD13_2 |
| ORAOV1 | PBRM1_3 |
| ORMDL2 | NGEF_1 |
| PAAF1 | NGEF_2 |
| PAFAH1B1 | KCTD13_1 |
| PAIP2 | SETD1A_1st |
| PANK1 | KMT2E_1 |
| PCGF3 | NGEF_2 |
| PDCD11 | PBRM1_3 |
| PDCD2L | TRANK1_1 |
| PDE12 | NGEF_2 |
| PDE7A | RERE_3 |
| PDE9A | VPS45_3 |
| PDS5B | SETD1A_1st |
| PFDN2 | LOC105376975_2 |
| PGAM5 | TRANK1_1 |
| PGM1 | TRANK1_2 |
| PHF21A | NGEF_2 |
| PHRF1 | PBRM1_3 |
| PIF1 | SETD1A_1st |
| PIM1 | TRANK1_3 |
| PKIA | NGEF_2 |
| PLPP1 | CHRNA3_1 |
| PLSCR1 | PBRM1_3 |
| PNP | BCL11B_2 |
| POC5 | STAT6_1 |
| POU3F3 | TRANK1_3 |
| PPIC | UBE2Q2P1_1 |
| PPP1R16A | KMT2E_3 |
| PPP2R2B | CHRNA3_3 |
| PQLC2 | pos_SNAP91 |
| PRKCI | LOC100507431_1 |
| PRKD3 | CHRNA3_1 |
| PRRC1 | RERE_1 |
| PSMA1 | BCL11B_1 |
| PSMB3 | GALNT10_3 |
| PSMC3IP | PBRM1_3 |
| PTGR1 | KMT2E_3 |
| RAB4A | LOC100507431_3 |
| RABL3 | PBRM1_3 |
| RANBP9 | BCL11B_3 |
| RAP2A | PCDHA123_3 |
| RAPGEF1 | UBE2Q2P1_1 |
| RASSF2 | KMT2E_2 |
| RASSF8 | CHRNA3_3 |
| RBFOX2 | NGEF_2 |
| RBM38 | GALNT10_2 |
| RBM7 | GALNT10_2 |
| RBX1 | PBRM1_3 |
| REEP4 | RERE_1 |
| RNF175 | UBE2Q2P1_2 |
| ROBO3 | NGEF_2 |
| RP11-220I1.1 | miR137_2 |
| RP11-446H18.3 | PBRM1_3 |
| RPL39L | UBE2Q2P1_2 |
| RPUSD3 | STAT6_2 |
| RRM2B | PBRM1_3 |
| RRN3 | CHRNA3_3 |
| RRP7A | PBRM1_3 |
| RUFY3 | NGEF_2 |
| SAC3D1 | PBRM1_3 |
| SARS | VPS45_1 |
| SARS2 | LOC100507431_3 |
| SASH1 | KMT2E_1 |
| SDCCAG3 | CHRNA3_1 |
| SDCCAG8 | NGEF_2 |
| SEMA4C | pos_SNAP91 |
| SEPW1 | CHRNA3_1 |
| SERAC1 | PPP1R16B_3 |
| SETD2 | NAB2_3 |
| SFR1 | STAT6_1 |
| SFT2D3 | PBRM1_3 |
| SFXN5 | PBRM1_1 |
| SGTB | TRANK1_2 |
| SHOC2 | PCDHA123_1 |
| SIMC1 | pos_SNAP91 |
| SKAP2 | LINC00637_3 |
| SLC35A3 | UBE2Q2P1_3 |
| SLC6A8 | NAB2_3, NGEF_2 |
| SLC9A3 | PBRM1_3 |
| SMIM4 | CHRNA3_2 |
| SMOC1 | SETD1A_1st, NGEF_2 |
| SNAPC2 | LOC105376975_3 |
| SNRNP200 | RERE_1 |
| SNUPN | LOC100507431_2 |
| SORT1 | PBRM1_3 |
| SOX11 | PPP1R16B_2 |
| SP1 | CHRNA3_1 |
| SPA17 | RERE_1 |
| SPAG5 | SETD1A_1st |
| SPATA6 | BCL11B_1 |
| SPIN3 | NGEF_2 |
| ST20 | CHRNA3_1 |
| ST3GAL3 | LOC105376975_3 |
| STK33 | TRANK1_1 |
| STMN2 | NGEF_2 |
| STXBP3 | TRANK1_1 |
| SUB1 | PBRM1_3 |
| SUPT5H | miR137_1 |
| SYNE2 | PBRM1_1 |
| TAGLN2 | PBRM1_3 |
| TARDBP | PPP1R16B_3 |
| TBC1D15 | NAB2_3 |
| TBC1D23 | BCL11B_1 |
| TCF12 | PPP1R16B_3 |
| TENM2 | LOC100507431_1 |
| TEX261 | NAB2_1 |
| TEX9 | pos_SNAP91 |
| THBS3 | NGEF_2 |
| TIMELESS | CHRNA3_2 |
| TMEM126A | NAB2_1 |
| TMEM38B | PPP1R16B_1 |
| TMEM43 | LOC105376975_2 |
| TMEM57 | NGEF_2 |
| TOP2A | SETD1A_1st |
| TOP3B | TRANK1_1 |
| TPM2 | CHRNA3_3 |
| TPRG1L | PPP1R16B_3 |
| TRIM16 | BCL11B_2 |
| TRIM32 | pos_SNAP91 |
| TRIM69 | TRANK1_2 |
| TSHZ2 | CHRNA3_1 |
| TTC28 | NGEF_2 |
| TTLL4 | RERE_3 |
| TUBA1A | NGEF_2 |
| TUBB2B | NGEF_2 |
| UACA | NGEF_1 |
| UBE2C | SETD1A_1st |
| UBE2H | NGEF_2 |
| UBE4A | NGEF_2 |
| USP22 | NGEF_2 |
| USP42 | PBRM1_2 |
| UTP3 | NGEF_2 |
| VMP1 | CHRNA3_1 |
| VPS37B | BAG5_3 |
| WDR55 | KMT2E_3 |
| WRB | LOC105376975_1 |
| WWC2 | STAT6_2 |
| WWP2 | pos_SNAP91 |
| XRN2 | PBRM1_1 |
| Z83851.1 | PPP1R16B_3 |
| ZBTB22 | UBE2Q2P1_3 |
| ZC2HC1A | NGEF_2 |
| ZC3H4 | LOC100507431_3 |
| ZNF408 | NAB2_1 |
| ZNF48 | miR137_2 |
| ZNF573 | KMT2E_3 |
| ZNF670 | RERE_3 |
| ZNF691 | BCL11B_1 |
| ZNF827 | CHRNA3_3 |
| ZRANB1 | DPYD_1 |
| ZW10 | PBRM1_1 |
| num_cells | gRNA_target | signif_genes |
|---|---|---|
| 31 | BAG5_3 | CLINT1, VPS37B |
| 13 | BCL11B_1 | ZNF691, SPATA6, TBC1D23, PSMA1, MLYCD, EFNB3, LPIN2, MECP2 |
| 33 | BCL11B_2 | NCK1, PNP, TRIM16 |
| 13 | BCL11B_3 | RANBP9, EXD3 |
| 3 | CHRNA3_1 | PRKD3, C2orf49, HES1, PLPP1, BICD2, SDCCAG3, ASRGL1, SP1, LRP1, ST20, CCDC189, VMP1, SEPW1, TSHZ2 |
| 23 | CHRNA3_2 | CCDC140, SMIM4, NAV2, TIMELESS, GPHN |
| 13 | CHRNA3_3 | ZNF827, PPP2R2B, AKAP9, IMPDH1, TPM2, RASSF8, RRN3 |
| 37 | DPYD_1 | MTHFD2, ZRANB1 |
| 68 | DPYD_2 | FUCA2 |
| 28 | DPYD_3 | C9orf142, KIF11 |
| 38 | GALNT10_1 | CLGN, FAM174A |
| 37 | GALNT10_2 | METAP1D, HNRNPF, MTCH2, RBM7, RBM38 |
| 50 | GALNT10_3 | FZD9, LGR4, NUDT22, PSMB3 |
| 30 | KCTD13_1 | CLCN3, PAFAH1B1, CNN1, DDT |
| 32 | KCTD13_2 | NUP214, GFRA1, ARNT2 |
| 15 | KCTD13_3 | ARHGAP32, BTBD1 |
| 11 | KMT2E_1 | MICAL1, SASH1, CCNY, PANK1, C19orf47 |
| 19 | KMT2E_2 | GJC1, CENPB, RASSF2 |
| 18 | KMT2E_3 | GON4L, CCDC174, NAF1, WDR55, PPP1R16A, PTGR1, ZNF573, ACSS2 |
| 45 | LINC00637_1 | DDT |
| 34 | LINC00637_2 | DNAAF5, ETV4 |
| 53 | LINC00637_3 | SKAP2 |
| 15 | LOC100507431_1 | PRKCI, LARP7, TENM2 |
| 29 | LOC100507431_2 | SNUPN |
| 49 | LOC100507431_3 | RAB4A, MBD1, SARS2, ZC3H4 |
| 35 | LOC105376975_1 | FBXO16, WRB |
| 50 | LOC105376975_2 | PFDN2, TMEM43, CHCHD7, AJUBA, CDC37 |
| 8 | LOC105376975_3 | ST3GAL3, EFCAB14, GORAB, DIS3L2, DHX16, CASP7, ARHGAP5, SNAPC2 |
| 44 | miR137_1 | NGRN, SUPT5H |
| 30 | miR137_2 | RP11-220I1.1, ZNF48, MT1X |
| 54 | miR137_3 | DHX40 |
| 36 | NAB2_1 | TEX261, CEP41, ZNF408, TMEM126A, GPKOW |
| 24 | NAB2_2 | NELFB |
| 25 | NAB2_3 | AHCYL1, DIS3L2, SETD2, EXOC2, TBC1D15, SLC6A8 |
| 11 | NGEF_1 | DNAJC18, LGR4, ORMDL2, CHPT1, GMFB, UACA |
| 2 | NGEF_2 | ACAP3, TMEM57, FNBP1L, THBS3, KDM5B, SDCCAG8, MAPRE3, ATOH8, KIAA1715, EPHA4, IQSEC1, IP6K2, PDE12, GAP43, PCGF3, NSG1, APBB2, UTP3, RUFY3, CCNI, BASP1, GLRX, NREP, TUBB2B, DMTF1, UBE2H, NEFM, PKIA, ZC2HC1A, STMN2, ELAVL2, DOLK, EXD3, MXI1, B4GALNT4, PHF21A, GNG3, CORO1B, PAAF1, MAP6, NCAM1, UBE4A, ROBO3, ENO2, LMBR1L, TUBA1A, DCLK1, NHLRC3, SMOC1, ATXN3, MEG3, DYNC1H1, GGA2, DCTN5, KIAA0895L, COG8, KDM6B, BORCS6, USP22, JUNB, DLL3, DYRK1B, IRGQ, EPS8L1, FASTKD5, CBFA2T2, GGT7, GDAP1L1, TTC28, RBFOX2, SPIN3, FTX, ACSL4, DCX, FAM127A, SLC6A8, GDI1 |
| 4 | PBRM1_1 | SFXN5, KLHL8, MAP3K4, NR6A1, CAMK1D, ZW10, BRAP, MNAT1, SYNE2, XRN2 |
| 12 | PBRM1_2 | USP42 |
| 4 | PBRM1_3 | SORT1, TAGLN2, SFT2D3, RP11-446H18.3, RABL3, PLSCR1, SLC9A3, SUB1, EPB41L2, NDUFAF6, RRM2B, CWF19L1, PDCD11, PHRF1, SAC3D1, ORAOV1, EED, IDH3A, PSMC3IP, CTD-2568A17.3, RBX1, RRP7A, ATXN10 |
| 53 | PCDHA123_1 | SHOC2 |
| 64 | PCDHA123_2 | CFL2 |
| 62 | PCDHA123_3 | RAP2A |
| 3 | pos_SNAP91 | DFFA, PQLC2, NBAS, MRPL19, SEMA4C, FST, SIMC1, DFNA5, CEP41, TRIM32, CDCA3, TEX9, WWP2, MED9, DUS3L, DPY19L3, MCM5 |
| 44 | PPP1R16B_1 | NIT1, AAMP, ATG5, TMEM38B |
| 31 | PPP1R16B_2 | NMNAT1, SOX11, DCAF10 |
| 35 | PPP1R16B_3 | TPRG1L, TARDBP, MRPS21, SERAC1, TCF12, Z83851.1, NUDT11 |
| 48 | RERE_1 | SNRNP200, NME6, PRRC1, REEP4, EMC2, SPA17 |
| 46 | RERE_2 | FAM131A, CDYL, NONO |
| 25 | RERE_3 | ZNF670, TTLL4, CCNC, PDE7A |
| 23 | SETD1A_1st | CDCA8, KIF2C, ASPM, CENPF, CENPE, KIF20A, PAIP2, ANLN, ERP44, BUB3, MKI67, CDCA3, PDS5B, SMOC1, CASC5, PIF1, SPAG5, TOP2A, KPNA2, FAM83D, UBE2C, AURKA, GTSE1 |
| 24 | STAT6_1 | POC5, ERICH1, KIF1BP, SFR1, CCDC91, FAM58A |
| 38 | STAT6_2 | RPUSD3, GATB, WWC2 |
| 53 | STAT6_3 | MYO6 |
| 45 | TCF4-ITF2_2 | DTWD1 |
| 12 | TCF4-ITF2_3 | NUP133 |
| 3 | TRANK1_1 | LRRC8D, STXBP3, STK33, PGAM5, C14orf1, FASN, PDCD2L, ACSS2, TOP3B |
| 19 | TRANK1_2 | PGM1, SGTB, TRIM69, HDHD2, C21orf62 |
| 65 | TRANK1_3 | POU3F3, PIM1 |
| 11 | UBE2Q2P1_1 | CCDC174, CMTM8, CTD-2001E22.2, PPIC, ANKMY2, RAPGEF1 |
| 8 | UBE2Q2P1_2 | RPL39L, RNF175, CHST12, DDIT4, IL13RA2 |
| 11 | UBE2Q2P1_3 | MKNK1, SLC35A3, ZBTB22, LRRC57, AMDHD2, NAA38, NCOA6 |
| 14 | VPS45_1 | SARS, B4GALT4, BUB3, MED9 |
| 79 | VPS45_2 | AGBL5, CCDC138 |
| 9 | VPS45_3 | CBR1, PDE9A, NHSL2 |
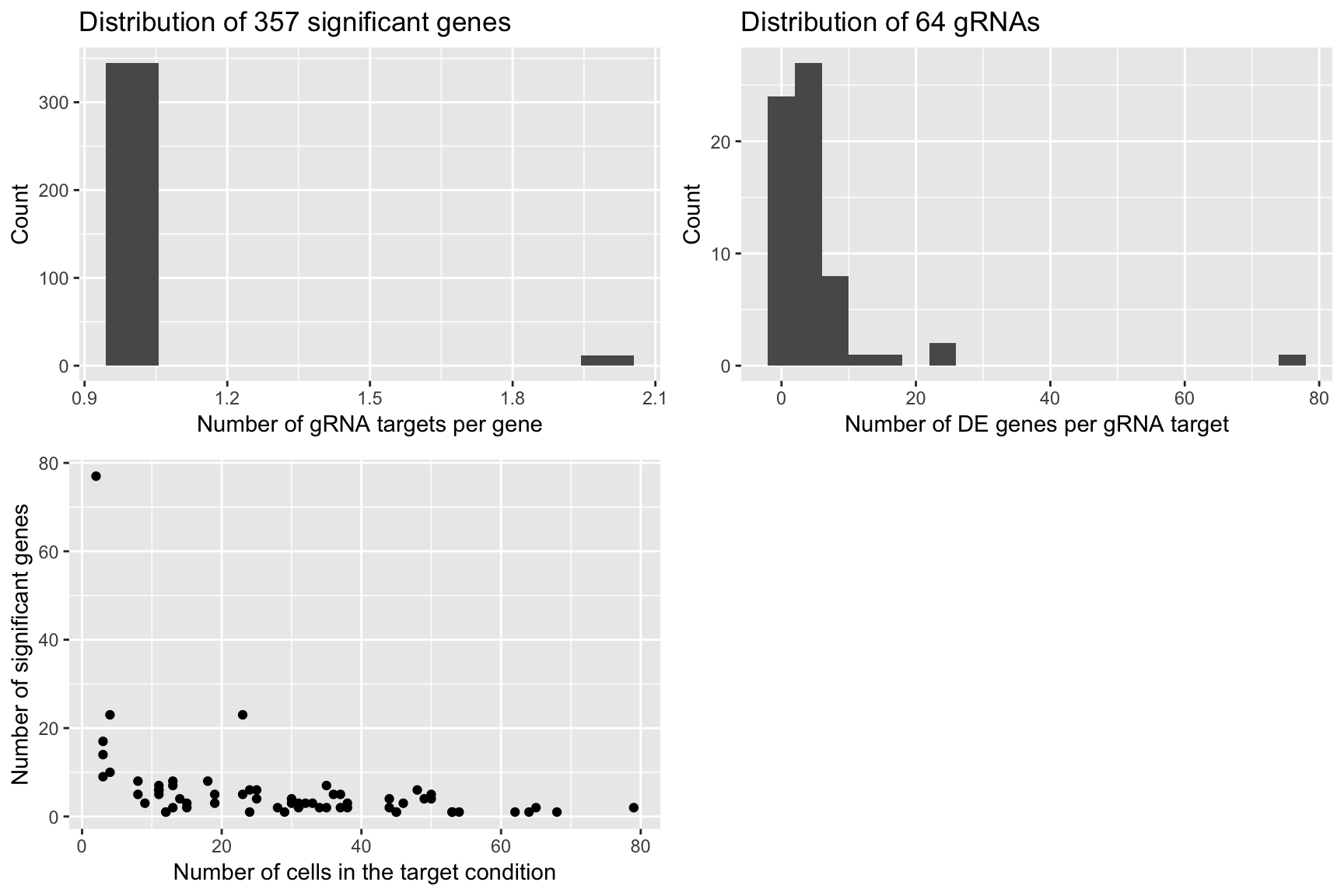
Permutation 5
load(paste0(wkdir,'data/perm.ashr_estimation05.Rdata'))
get.result(betaest.mtx,lfsr.mtx)| signif_gene | gRNA_targets |
|---|---|
| A2M | PBRM1_1 |
| ABHD17B | UBE2Q2P1_2 |
| AC004951.6 | VPS45_1 |
| AC092296.1 | NGEF_2 |
| ACSL4 | CHRNA3_1 |
| AEN | TRANK1_2 |
| AHCTF1 | pos_SNAP91 |
| AIMP1 | TRANK1_2 |
| AJUBA | LINC00637_1 |
| AKT3 | VPS45_3 |
| ALPL | PBRM1_1 |
| ALS2 | NGEF_2 |
| AMN1 | TRANK1_1 |
| AMOT | KCTD13_3 |
| AMOTL1 | LOC105376975_3 |
| ANKRD1 | VPS45_3, NGEF_1 |
| AP3M2 | LOC105376975_3 |
| AP4M1 | VPS45_1 |
| APPL2 | LOC100507431_1 |
| ARFGEF1 | miR137_2 |
| ARHGAP11A | KCTD13_1 |
| ARNT | NGEF_2 |
| ASAP1 | CHRNA3_2 |
| ASTN2 | TRANK1_2 |
| ATN1 | UBE2Q2P1_2 |
| ATP5SL | CHRNA3_3 |
| ATP6V0B | TRANK1_2 |
| ATP6V1D | TRANK1_2 |
| ATXN1L | pos_SNAP91 |
| AURKA | pos_SNAP91 |
| B4GALNT4 | CHRNA3_1 |
| B4GALT5 | PBRM1_2 |
| BBC3 | UBE2Q2P1_2 |
| BCAT1 | PBRM1_2 |
| BNIP3L | PCDHA123_1 |
| BORA | pos_SNAP91 |
| BRCA1 | KCTD13_3, TCF4-ITF2_1 |
| BZW1 | PBRM1_2 |
| C11orf68 | NGEF_1 |
| C12orf43 | LOC100507431_2 |
| C17orf89 | NGEF_2 |
| C19orf24 | TRANK1_2 |
| C19orf60 | PBRM1_1 |
| C1orf35 | KMT2E_2 |
| C8orf33 | NGEF_2 |
| C9orf72 | LOC105376975_1 |
| CAB39L | LOC100507431_1 |
| CAMK2N1 | CHRNA3_2, PBRM1_1 |
| CAMSAP1 | CHRNA3_1 |
| CAND1 | UBE2Q2P1_2 |
| CAPRIN1 | TCF4-ITF2_3 |
| CBR1 | PBRM1_3 |
| CCAR1 | NAB2_2 |
| CCDC126 | CHRNA3_1 |
| CCDC71L | PBRM1_1 |
| CCDC85C | CHRNA3_1, STAT6_2 |
| CCND2 | PBRM1_3, TCF4-ITF2_3 |
| CDC25A | KCTD13_3 |
| CDC42EP3 | NGEF_2 |
| CDCA8 | pos_SNAP91 |
| CDIP1 | CHRNA3_3 |
| CDK12 | UBE2Q2P1_2 |
| CDK19 | CHRNA3_1 |
| CDKN2AIP | LOC105376975_1 |
| CEBPD | DPYD_2 |
| CEPT1 | CHRNA3_1, NGEF_2 |
| CHEK2 | STAT6_2 |
| CHPF | KCTD13_2 |
| CKAP2 | pos_SNAP91 |
| CLIP3 | UBE2Q2P1_2 |
| CLN5 | RERE_2 |
| CLUH | PBRM1_3 |
| CMAS | LINC00637_1 |
| CMIP | UBE2Q2P1_2 |
| COG8 | CHRNA3_1 |
| COMMD9 | RERE_3 |
| COPS8 | LINC00637_3 |
| CSNK2A2 | PBRM1_1 |
| CTD-2270L9.4 | NAB2_2 |
| CTDNEP1 | BCL11B_2 |
| CTNNBIP1 | PPP1R16B_1 |
| CXorf56 | CHRNA3_1 |
| CYB5D2 | TCF4-ITF2_3 |
| CYP20A1 | BCL11B_2 |
| CYTOR | NGEF_1 |
| DAGLB | TCF4-ITF2_3 |
| DAP3 | BCL11B_2 |
| DAZAP1 | PBRM1_2 |
| DCAF6 | VPS45_3 |
| DCK | UBE2Q2P1_3 |
| DCLK2 | LOC100507431_1 |
| DCTN1 | CHRNA3_1 |
| DCTN3 | miR137_2 |
| DCX | CHRNA3_1 |
| DDI2 | GALNT10_2 |
| DHRS4L2 | KMT2E_1 |
| DHX34 | CHRNA3_1, NGEF_2 |
| DIAPH3 | LOC100507431_1 |
| DLG3 | pos_SNAP91 |
| DNAH14 | CHRNA3_1 |
| DNAJB2 | TRANK1_2 |
| DNAL1 | pos_SNAP91 |
| DNMT3A | NGEF_2 |
| DPP3 | SETD1A_1st |
| DRAXIN | LOC100507431_2 |
| DYNC2H1 | PPP1R16B_3 |
| EDA2R | NGEF_2 |
| EFS | LOC105376975_3 |
| EIF2AK2 | NAB2_2, STAT6_3 |
| ELMOD2 | NGEF_2 |
| ENKD1 | miR137_2 |
| ENO2 | CHRNA3_1 |
| EPHB4 | TRANK1_1 |
| ERCC3 | VPS45_1 |
| ETV1 | STAT6_3 |
| FAAP100 | TCF4-ITF2_3 |
| FAM110A | pos_SNAP91 |
| FAM122B | SETD1A_1st |
| FAM136A | LOC100507431_3 |
| FAM216A | NGEF_2 |
| FAM60A | LOC105376975_1 |
| FAT1 | KCTD13_2 |
| FBXO45 | TCF4-ITF2_3 |
| FBXW2 | KMT2E_3 |
| FGFR1OP2 | NGEF_2 |
| FIGN | UBE2Q2P1_2 |
| FIZ1 | LOC105376975_3 |
| FNDC3A | RERE_2 |
| FOXP1 | CHRNA3_2 |
| FRZB | pos_SNAP91 |
| FXYD6 | LINC00637_1 |
| FZD10 | KMT2E_1 |
| FZR1 | pos_SNAP91 |
| GABPB1 | CHRNA3_3 |
| GALNT2 | DPYD_2 |
| GAP43 | CHRNA3_1 |
| GATC | UBE2Q2P1_2 |
| GBA2 | KMT2E_3 |
| GDAP1L1 | CHRNA3_1 |
| GFER | miR137_2 |
| GLA | PBRM1_3 |
| GLI4 | VPS45_1 |
| GLTP | VPS45_2 |
| GNA12 | PPP1R16B_3 |
| GNB5 | NAB2_1 |
| GNG3 | CHRNA3_1 |
| GNG5 | STAT6_1 |
| GOLGA4 | CHRNA3_3 |
| GOLGA8B | KMT2E_1 |
| GPATCH8 | GALNT10_2 |
| HAGH | BCL11B_1 |
| HDLBP | NGEF_1 |
| HECTD1 | GALNT10_3 |
| HIRA | NGEF_2 |
| HIST1H1C | LOC105376975_1 |
| HPS5 | NAB2_1 |
| HSD11B1L | PBRM1_1 |
| IFNAR1 | NGEF_2 |
| IFT122 | LINC00637_1 |
| IGFBP2 | VPS45_3 |
| IKBIP | UBE2Q2P1_1 |
| IRX2 | NGEF_2 |
| ISCA2 | TRANK1_2 |
| ISCU | TCF4-ITF2_1 |
| ITPK1 | LOC105376975_3 |
| JRK | UBE2Q2P1_2 |
| KCNG1 | BAG5_1 |
| KDSR | LOC105376975_1 |
| KEAP1 | CHRNA3_1 |
| KIF15 | TRANK1_1 |
| KIF20A | pos_SNAP91 |
| KIF4A | pos_SNAP91 |
| KLHL20 | TCF4-ITF2_3 |
| KLHL7 | NGEF_1 |
| KMT5A | pos_SNAP91 |
| KRR1 | NAB2_2 |
| LAMP2 | pos_SNAP91 |
| LATS1 | PCDHA123_2 |
| LRIF1 | TRANK1_2 |
| LRRCC1 | pos_SNAP91 |
| LSS | UBE2Q2P1_1 |
| LTBP1 | TRANK1_1 |
| MAP6 | CHRNA3_2 |
| MAPK14 | CHRNA3_2 |
| MAPK8 | DPYD_2 |
| MASTL | CHRNA3_1 |
| MCFD2 | DPYD_2 |
| MED21 | pos_SNAP91 |
| MEG8 | NGEF_2 |
| METAP1 | LINC00637_2 |
| MGEA5 | LOC100507431_2 |
| MGST2 | TCF4-ITF2_3 |
| MIR99AHG | NGEF_2 |
| MKI67 | pos_SNAP91 |
| MLLT4 | CHRNA3_2 |
| MOB4 | NAB2_2 |
| MRPL20 | CHRNA3_2, LOC105376975_1 |
| MRPL35 | LOC100507431_1 |
| MRPL40 | NGEF_2 |
| MRPL46 | DPYD_3 |
| MRPL52 | NGEF_2 |
| MRPS18B | CHRNA3_3 |
| MSL1 | pos_SNAP91 |
| MT1X | PBRM1_3 |
| MTG2 | NGEF_2 |
| MTHFSD | LOC105376975_1 |
| MTUS1 | PBRM1_3 |
| MTX1 | TCF4-ITF2_1 |
| MXD3 | pos_SNAP91 |
| MYC | PBRM1_3 |
| MYCBP | NAB2_1 |
| NAPG | CHRNA3_1 |
| NDUFB1 | STAT6_1 |
| NDUFV1 | LINC00637_3 |
| NHP2 | NGEF_2 |
| NINJ1 | BCL11B_3 |
| NNAT | CHRNA3_1 |
| NOVA2 | UBE2Q2P1_2 |
| NR2F1 | miR137_2 |
| NUAK2 | UBE2Q2P1_2 |
| NUDT3 | NGEF_2 |
| NUF2 | pos_SNAP91 |
| NUP153 | pos_SNAP91 |
| NUP98 | CHRNA3_1 |
| NUS1 | NGEF_2 |
| OFD1 | KMT2E_2 |
| OLMALINC | UBE2Q2P1_1 |
| OMA1 | PBRM1_3 |
| ORC1 | PBRM1_2 |
| OTUD4 | UBE2Q2P1_2 |
| PARVA | UBE2Q2P1_1 |
| PAXBP1 | NAB2_3, TRANK1_1, PPP1R16B_2 |
| PCNX1 | CHRNA3_3 |
| PDCD6 | UBE2Q2P1_1 |
| PDIA5 | TRANK1_1 |
| PDPN | VPS45_3 |
| PEX14 | miR137_3 |
| PFKM | STAT6_3 |
| PHF13 | NGEF_2 |
| PHF19 | NGEF_1 |
| PHLDA3 | LINC00637_1 |
| PIF1 | pos_SNAP91 |
| PITPNB | BCL11B_2 |
| PKIG | TCF4-ITF2_2 |
| PKN3 | TRANK1_1 |
| PLA2G12A | NGEF_2 |
| PLEKHJ1 | VPS45_3 |
| PLK3 | UBE2Q2P1_2 |
| PLXDC2 | LOC105376975_3 |
| PLXNA2 | pos_SNAP91 |
| POLE4 | UBE2Q2P1_1 |
| POU2F1 | pos_SNAP91 |
| PPFIBP1 | LOC105376975_2 |
| PPP1R35 | CHRNA3_3, TRANK1_2 |
| PPP1R9A | PBRM1_3 |
| PPP2R1B | pos_SNAP91 |
| PPP4C | NAB2_2 |
| PRKCI | BCL11B_3 |
| PSMB7 | CHRNA3_3 |
| PSME4 | SETD1A_1st |
| PTMA | LINC00637_1 |
| PTPRF | pos_SNAP91 |
| RAB33A | PCDHA123_2 |
| RAB6B | TCF4-ITF2_3 |
| RACGAP1 | pos_SNAP91 |
| RAP2A | PBRM1_1 |
| RIPK2 | PBRM1_1 |
| RNASEH2C | BCL11B_2 |
| RNF44 | UBE2Q2P1_2 |
| ROBO3 | CHRNA3_1 |
| RP11-14N7.2 | TRANK1_2 |
| RP11-398C13.8 | RERE_2 |
| RP11-539L10.3 | RERE_3 |
| RP11-706O15.1 | PBRM1_2 |
| RP11-835E18.2 | NGEF_2 |
| RPL22L1 | PBRM1_2 |
| RPL23A | NGEF_2 |
| RPP21 | PCDHA123_1 |
| RPRD2 | SETD1A_1st |
| RPS20 | NAB2_2 |
| RPS6KA4 | VPS45_3 |
| RRBP1 | NGEF_1 |
| RSBN1 | CHRNA3_1 |
| RTN2 | UBE2Q2P1_2 |
| RYBP | NAB2_2 |
| SAYSD1 | BCL11B_1 |
| SCAPER | CHRNA3_1 |
| SDHAF3 | SETD1A_1st |
| SEC11C | PBRM1_3 |
| SECISBP2L | UBE2Q2P1_2 |
| SEMA4C | KCTD13_3 |
| SEPT3 | PPP1R16B_1 |
| SEPT8 | KMT2E_3 |
| SFRP1 | PBRM1_1 |
| SFXN2 | PBRM1_1 |
| SGPL1 | LINC00637_2 |
| SHCBP1 | pos_SNAP91 |
| SHOC2 | CHRNA3_1 |
| SIRT1 | NGEF_1 |
| SLC11A2 | LOC105376975_3 |
| SLC25A51 | BCL11B_1 |
| SLC29A2 | VPS45_3 |
| SLC30A1 | NGEF_2 |
| SLC30A5 | KMT2E_2, STAT6_2, UBE2Q2P1_3 |
| SLC35A4 | NAB2_2 |
| SLC35C1 | PBRM1_1 |
| SLC35C2 | TCF4-ITF2_1 |
| SLC35F1 | CHRNA3_1 |
| SLX4IP | STAT6_1 |
| SMEK1 | BAG5_3 |
| SMG5 | NGEF_2 |
| SMG9 | LINC00637_1 |
| SMIM3 | PBRM1_3 |
| SNHG1 | CHRNA3_3 |
| SNTG1 | UBE2Q2P1_2 |
| SNX13 | TCF4-ITF2_3 |
| SNX5 | GALNT10_1 |
| SNX9 | KCTD13_2 |
| SOX1 | UBE2Q2P1_2 |
| SPP1 | PBRM1_3 |
| SRGAP1 | PBRM1_1 |
| SRR | LOC105376975_3 |
| SS18L1 | VPS45_1 |
| STAU2 | TRANK1_1 |
| STIM2 | LOC105376975_3 |
| STK16 | TRANK1_1 |
| STK33 | TRANK1_1 |
| STX3 | PBRM1_3 |
| SUPT6H | NGEF_1 |
| SUZ12 | UBE2Q2P1_3 |
| SYNE2 | STAT6_1 |
| TAF6 | CHRNA3_3 |
| TALDO1 | PBRM1_2, TCF4-ITF2_1 |
| TCN2 | CHRNA3_2 |
| TEAD2 | LOC105376975_3 |
| TELO2 | RERE_2 |
| THOC7 | LOC100507431_3 |
| TIMM17B | NGEF_2 |
| TIMM8A | LOC105376975_3 |
| TM2D2 | BCL11B_2, RERE_2, VPS45_3 |
| TMED5 | CHRNA3_3 |
| TMEM138 | CHRNA3_3 |
| TMEM223 | NGEF_2 |
| TMEM242 | KMT2E_1 |
| TMEM41A | BCL11B_3 |
| TMEM67 | TRANK1_1 |
| TMEM88 | LOC100507431_1 |
| TMOD3 | TRANK1_1 |
| TNKS1BP1 | UBE2Q2P1_2 |
| TNPO2 | UBE2Q2P1_2 |
| TNRC6C | KMT2E_1 |
| TOE1 | LOC105376975_3 |
| TOGARAM1 | DPYD_1 |
| TOX4 | CHRNA3_3 |
| TP53BP2 | BCL11B_1 |
| TPM1 | VPS45_3 |
| TPT1-AS1 | PBRM1_3 |
| TRIAP1 | CHRNA3_3 |
| TRIP6 | LINC00637_1 |
| TRMO | CHRNA3_1 |
| TRRAP | RERE_2 |
| TSC22D1 | CHRNA3_3 |
| TSPAN15 | NGEF_2 |
| TTC28 | CHRNA3_1 |
| TTC37 | NGEF_2 |
| TUBB2A | LINC00637_1 |
| TUBB2B | CHRNA3_1 |
| TUBGCP3 | pos_SNAP91 |
| TUNAR | NGEF_2 |
| TXNRD1 | PBRM1_2 |
| UBE2Z | LINC00637_1 |
| UBE4A | KMT2E_1 |
| UBL7 | RERE_2 |
| UFM1 | TRANK1_2 |
| UNC119B | PPP1R16B_3 |
| UPF1 | NGEF_1 |
| USP22 | CHRNA3_1 |
| UVRAG | RERE_3 |
| VEPH1 | miR137_1 |
| VGLL4 | PBRM1_2 |
| VRK1 | TRANK1_1 |
| WASHC3 | UBE2Q2P1_2 |
| WDR74 | CHRNA3_3 |
| WNK1 | TCF4-ITF2_3 |
| WSB2 | KMT2E_2 |
| YEATS4 | pos_SNAP91 |
| YIPF3 | UBE2Q2P1_1 |
| YY1 | CHRNA3_3 |
| ZBED1 | UBE2Q2P1_2 |
| ZBTB17 | CHRNA3_1 |
| ZFP14 | pos_SNAP91 |
| ZMYM3 | CHRNA3_1 |
| ZNF146 | NGEF_2 |
| ZNF317 | UBE2Q2P1_1 |
| ZNF516 | GALNT10_3 |
| ZNF521 | CHRNA3_1 |
| ZNF561 | DPYD_2 |
| ZNF599 | NGEF_2 |
| ZNF880 | BCL11B_1 |
| ZNF90 | GALNT10_2 |
| ZZZ3 | TRANK1_2 |
| num_cells | gRNA_target | signif_genes |
|---|---|---|
| 36 | BAG5_1 | KCNG1 |
| 31 | BAG5_3 | SMEK1 |
| 13 | BCL11B_1 | TP53BP2, SAYSD1, SLC25A51, HAGH, ZNF880 |
| 33 | BCL11B_2 | DAP3, CYP20A1, TM2D2, RNASEH2C, CTDNEP1, PITPNB |
| 13 | BCL11B_3 | PRKCI, TMEM41A, NINJ1 |
| 3 | CHRNA3_1 | ZBTB17, CEPT1, RSBN1, DNAH14, DCTN1, GAP43, TUBB2B, CDK19, SLC35F1, CCDC126, TRMO, CAMSAP1, MASTL, SHOC2, B4GALNT4, NUP98, GNG3, ROBO3, ENO2, CCDC85C, SCAPER, COG8, USP22, NAPG, ZNF521, KEAP1, DHX34, NNAT, GDAP1L1, TTC28, ZMYM3, ACSL4, DCX, CXorf56 |
| 23 | CHRNA3_2 | MRPL20, CAMK2N1, FOXP1, MAPK14, MLLT4, ASAP1, MAP6, TCN2 |
| 13 | CHRNA3_3 | TMED5, GOLGA4, MRPS18B, TAF6, PPP1R35, PSMB7, TMEM138, WDR74, SNHG1, TRIAP1, TSC22D1, TOX4, PCNX1, YY1, GABPB1, CDIP1, ATP5SL |
| 37 | DPYD_1 | TOGARAM1 |
| 68 | DPYD_2 | GALNT2, MCFD2, CEBPD, MAPK8, ZNF561 |
| 28 | DPYD_3 | MRPL46 |
| 38 | GALNT10_1 | SNX5 |
| 37 | GALNT10_2 | DDI2, GPATCH8, ZNF90 |
| 50 | GALNT10_3 | HECTD1, ZNF516 |
| 30 | KCTD13_1 | ARHGAP11A |
| 32 | KCTD13_2 | CHPF, FAT1, SNX9 |
| 15 | KCTD13_3 | SEMA4C, CDC25A, BRCA1, AMOT |
| 11 | KMT2E_1 | TMEM242, UBE4A, FZD10, DHRS4L2, GOLGA8B, TNRC6C |
| 19 | KMT2E_2 | C1orf35, SLC30A5, WSB2, OFD1 |
| 18 | KMT2E_3 | SEPT8, GBA2, FBXW2 |
| 45 | LINC00637_1 | PHLDA3, PTMA, IFT122, TUBB2A, TRIP6, FXYD6, CMAS, AJUBA, UBE2Z, SMG9 |
| 34 | LINC00637_2 | METAP1, SGPL1 |
| 53 | LINC00637_3 | COPS8, NDUFV1 |
| 15 | LOC100507431_1 | MRPL35, DCLK2, APPL2, CAB39L, DIAPH3, TMEM88 |
| 29 | LOC100507431_2 | DRAXIN, MGEA5, C12orf43 |
| 49 | LOC100507431_3 | FAM136A, THOC7 |
| 35 | LOC105376975_1 | MRPL20, CDKN2AIP, HIST1H1C, C9orf72, FAM60A, MTHFSD, KDSR |
| 50 | LOC105376975_2 | PPFIBP1 |
| 8 | LOC105376975_3 | TOE1, STIM2, AP3M2, PLXDC2, AMOTL1, SLC11A2, EFS, ITPK1, SRR, TEAD2, FIZ1, TIMM8A |
| 44 | miR137_1 | VEPH1 |
| 30 | miR137_2 | NR2F1, ARFGEF1, DCTN3, GFER, ENKD1 |
| 54 | miR137_3 | PEX14 |
| 36 | NAB2_1 | MYCBP, HPS5, GNB5 |
| 24 | NAB2_2 | EIF2AK2, MOB4, RYBP, SLC35A4, RPS20, CCAR1, KRR1, CTD-2270L9.4, PPP4C |
| 25 | NAB2_3 | PAXBP1 |
| 11 | NGEF_1 | CYTOR, HDLBP, KLHL7, PHF19, SIRT1, ANKRD1, C11orf68, SUPT6H, UPF1, RRBP1 |
| 2 | NGEF_2 | PHF13, CEPT1, ARNT, SMG5, SLC30A1, DNMT3A, CDC42EP3, ALS2, PLA2G12A, ELMOD2, IRX2, TTC37, NHP2, NUDT3, NUS1, C8orf33, TSPAN15, TMEM223, FGFR1OP2, FAM216A, MRPL52, TUNAR, MEG8, RPL23A, C17orf89, RP11-835E18.2, ZNF599, ZNF146, AC092296.1, DHX34, MTG2, MIR99AHG, IFNAR1, HIRA, MRPL40, TIMM17B, EDA2R |
| 4 | PBRM1_1 | CAMK2N1, ALPL, CCDC71L, SFRP1, RIPK2, SFXN2, SLC35C1, A2M, SRGAP1, RAP2A, CSNK2A2, HSD11B1L, C19orf60 |
| 12 | PBRM1_2 | ORC1, BZW1, VGLL4, RPL22L1, TALDO1, BCAT1, TXNRD1, DAZAP1, B4GALT5, RP11-706O15.1 |
| 4 | PBRM1_3 | OMA1, SPP1, SMIM3, PPP1R9A, MTUS1, MYC, STX3, CCND2, TPT1-AS1, MT1X, CLUH, SEC11C, CBR1, GLA |
| 53 | PCDHA123_1 | RPP21, BNIP3L |
| 64 | PCDHA123_2 | LATS1, RAB33A |
| 3 | pos_SNAP91 | CDCA8, PTPRF, NUF2, POU2F1, PLXNA2, AHCTF1, FRZB, KIF20A, MXD3, NUP153, LRRCC1, MKI67, PPP2R1B, MED21, RACGAP1, YEATS4, KMT5A, CKAP2, BORA, TUBGCP3, DNAL1, PIF1, SHCBP1, ATXN1L, MSL1, FZR1, ZFP14, FAM110A, AURKA, KIF4A, DLG3, LAMP2 |
| 44 | PPP1R16B_1 | CTNNBIP1, SEPT3 |
| 31 | PPP1R16B_2 | PAXBP1 |
| 35 | PPP1R16B_3 | GNA12, DYNC2H1, UNC119B |
| 46 | RERE_2 | TRRAP, TM2D2, RP11-398C13.8, FNDC3A, CLN5, UBL7, TELO2 |
| 25 | RERE_3 | RP11-539L10.3, COMMD9, UVRAG |
| 23 | SETD1A_1st | RPRD2, PSME4, SDHAF3, DPP3, FAM122B |
| 24 | STAT6_1 | GNG5, SYNE2, NDUFB1, SLX4IP |
| 38 | STAT6_2 | SLC30A5, CCDC85C, CHEK2 |
| 53 | STAT6_3 | EIF2AK2, ETV1, PFKM |
| 50 | TCF4-ITF2_1 | MTX1, TALDO1, ISCU, BRCA1, SLC35C2 |
| 45 | TCF4-ITF2_2 | PKIG |
| 12 | TCF4-ITF2_3 | KLHL20, RAB6B, FBXO45, MGST2, DAGLB, SNX13, CAPRIN1, WNK1, CCND2, CYB5D2, FAAP100 |
| 3 | TRANK1_1 | LTBP1, STK16, KIF15, PDIA5, EPHB4, STAU2, TMEM67, PKN3, STK33, AMN1, VRK1, TMOD3, PAXBP1 |
| 19 | TRANK1_2 | ATP6V0B, ZZZ3, LRIF1, RP11-14N7.2, DNAJB2, AIMP1, PPP1R35, ASTN2, UFM1, ATP6V1D, ISCA2, AEN, C19orf24 |
| 11 | UBE2Q2P1_1 | POLE4, PDCD6, YIPF3, OLMALINC, PARVA, IKBIP, ZNF317, LSS |
| 8 | UBE2Q2P1_2 | PLK3, NUAK2, FIGN, OTUD4, RNF44, SNTG1, JRK, ABHD17B, TNKS1BP1, ATN1, CAND1, WASHC3, GATC, SOX1, SECISBP2L, CMIP, CDK12, TNPO2, CLIP3, RTN2, NOVA2, BBC3, ZBED1 |
| 11 | UBE2Q2P1_3 | DCK, SLC30A5, SUZ12 |
| 14 | VPS45_1 | ERCC3, AC004951.6, AP4M1, GLI4, SS18L1 |
| 79 | VPS45_2 | GLTP |
| 9 | VPS45_3 | PDPN, DCAF6, AKT3, IGFBP2, TM2D2, ANKRD1, RPS6KA4, SLC29A2, TPM1, PLEKHJ1 |
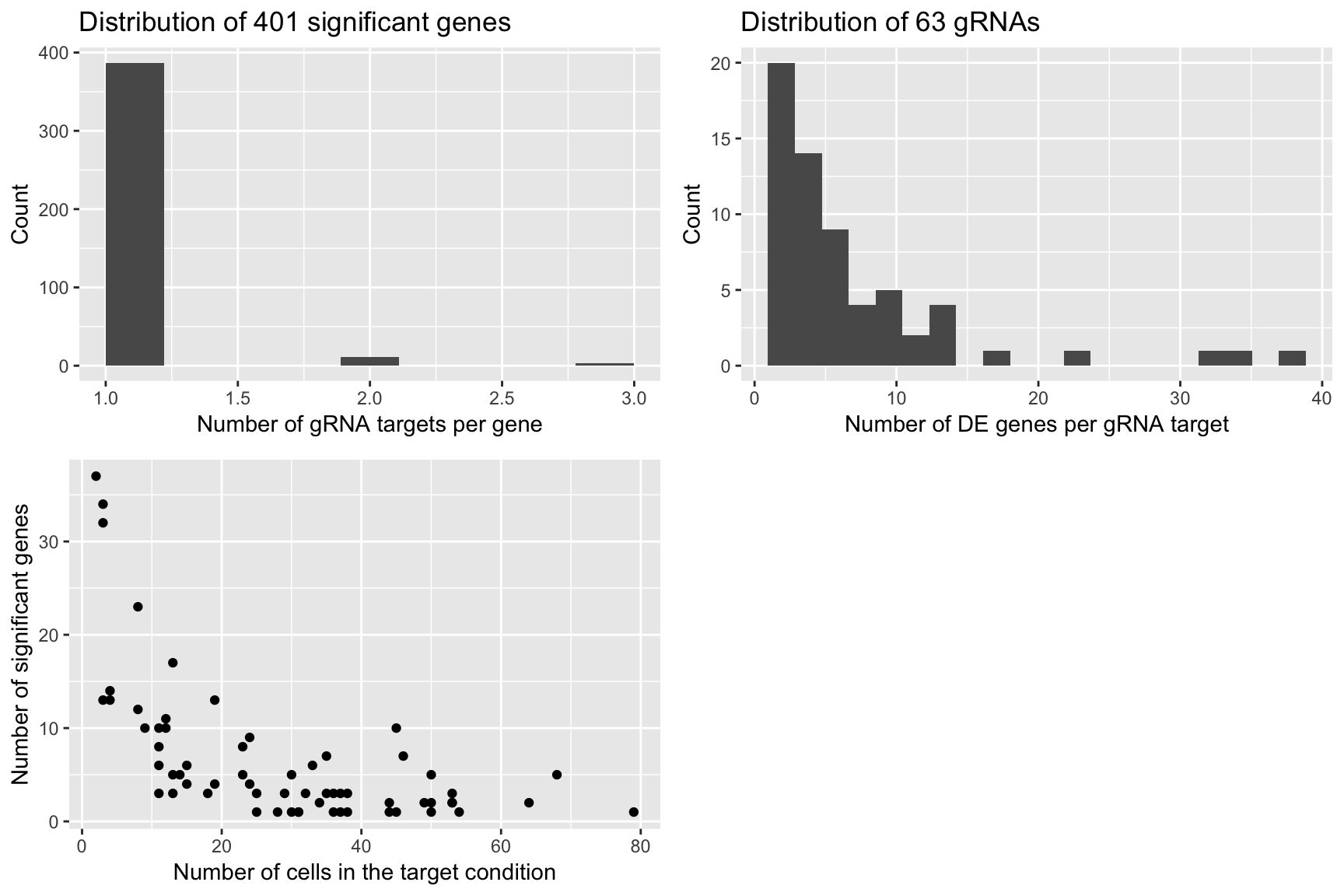
Session Information
sessionInfo()R version 3.5.2 (2018-12-20)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS Mojave 10.14
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid parallel stats4 stats graphics grDevices utils
[8] datasets methods base
other attached packages:
[1] MAST_1.8.2 SingleCellExperiment_1.4.1
[3] SummarizedExperiment_1.12.0 DelayedArray_0.8.0
[5] BiocParallel_1.16.6 matrixStats_0.54.0
[7] GenomicRanges_1.34.0 GenomeInfoDb_1.18.2
[9] edgeR_3.24.3 limma_3.38.3
[11] lattice_0.20-38 sctransform_0.2.0
[13] Matrix_1.2-16 clusterProfiler_3.10.1
[15] org.Hs.eg.db_3.7.0 AnnotationDbi_1.44.0
[17] IRanges_2.16.0 S4Vectors_0.20.1
[19] Biobase_2.42.0 BiocGenerics_0.28.0
[21] dplyr_0.8.0.1 gridExtra_2.3
[23] kableExtra_1.0.1 knitr_1.22
[25] ggplot2_3.1.0 ashr_2.2-32
loaded via a namespace (and not attached):
[1] fgsea_1.8.0 colorspace_1.4-0 ggridges_0.5.1
[4] qvalue_2.14.1 XVector_0.22.0 rstudioapi_0.9.0
[7] listenv_0.7.0 farver_1.1.0 urltools_1.7.2
[10] ggrepel_0.8.0 bit64_0.9-7 xml2_1.2.0
[13] codetools_0.2-16 splines_3.5.2 pscl_1.5.2
[16] doParallel_1.0.14 GOSemSim_2.8.0 polyclip_1.9-1
[19] jsonlite_1.6 GO.db_3.7.0 ggforce_0.2.1
[22] readr_1.3.1 compiler_3.5.2 httr_1.4.0
[25] rvcheck_0.1.3 assertthat_0.2.0 lazyeval_0.2.1
[28] tweenr_1.0.1 htmltools_0.3.6 prettyunits_1.0.2
[31] tools_3.5.2 igraph_1.2.4 gtable_0.2.0
[34] glue_1.3.0 GenomeInfoDbData_1.2.0 reshape2_1.4.3
[37] DO.db_2.9 fastmatch_1.1-0 Rcpp_1.0.0
[40] enrichplot_1.2.0 iterators_1.0.10 ggraph_1.0.2
[43] xfun_0.5 stringr_1.4.0 globals_0.12.4
[46] rvest_0.3.2 future_1.13.0 DOSE_3.8.2
[49] zlibbioc_1.28.0 europepmc_0.3 MASS_7.3-51.1
[52] scales_1.0.0 hms_0.4.2 RColorBrewer_1.1-2
[55] yaml_2.2.0 memoise_1.1.0 UpSetR_1.3.3
[58] triebeard_0.3.0 stringi_1.3.1 RSQLite_2.1.1
[61] SQUAREM_2017.10-1 highr_0.7 foreach_1.4.4
[64] truncnorm_1.0-8 rlang_0.3.1 pkgconfig_2.0.2
[67] bitops_1.0-6 evaluate_0.13 purrr_0.3.1
[70] labeling_0.3 cowplot_0.9.4 bit_1.1-14
[73] tidyselect_0.2.5 plyr_1.8.4 magrittr_1.5
[76] R6_2.4.0 DBI_1.0.0 pillar_1.3.1
[79] withr_2.1.2 abind_1.4-5 RCurl_1.95-4.12
[82] mixsqp_0.1-97 tibble_2.0.1 future.apply_1.2.0
[85] crayon_1.3.4 rmarkdown_1.11 viridis_0.5.1
[88] progress_1.2.0 locfit_1.5-9.1 data.table_1.12.0
[91] blob_1.1.1 digest_0.6.18 webshot_0.5.1
[94] tidyr_0.8.3 gridGraphics_0.3-0 munsell_0.5.0
[97] viridisLite_0.3.0 ggplotify_0.0.3